Classical and Emerging Regulatory Mechanisms of Cytokinesis in Animal Cells
Abstract
1. Introduction
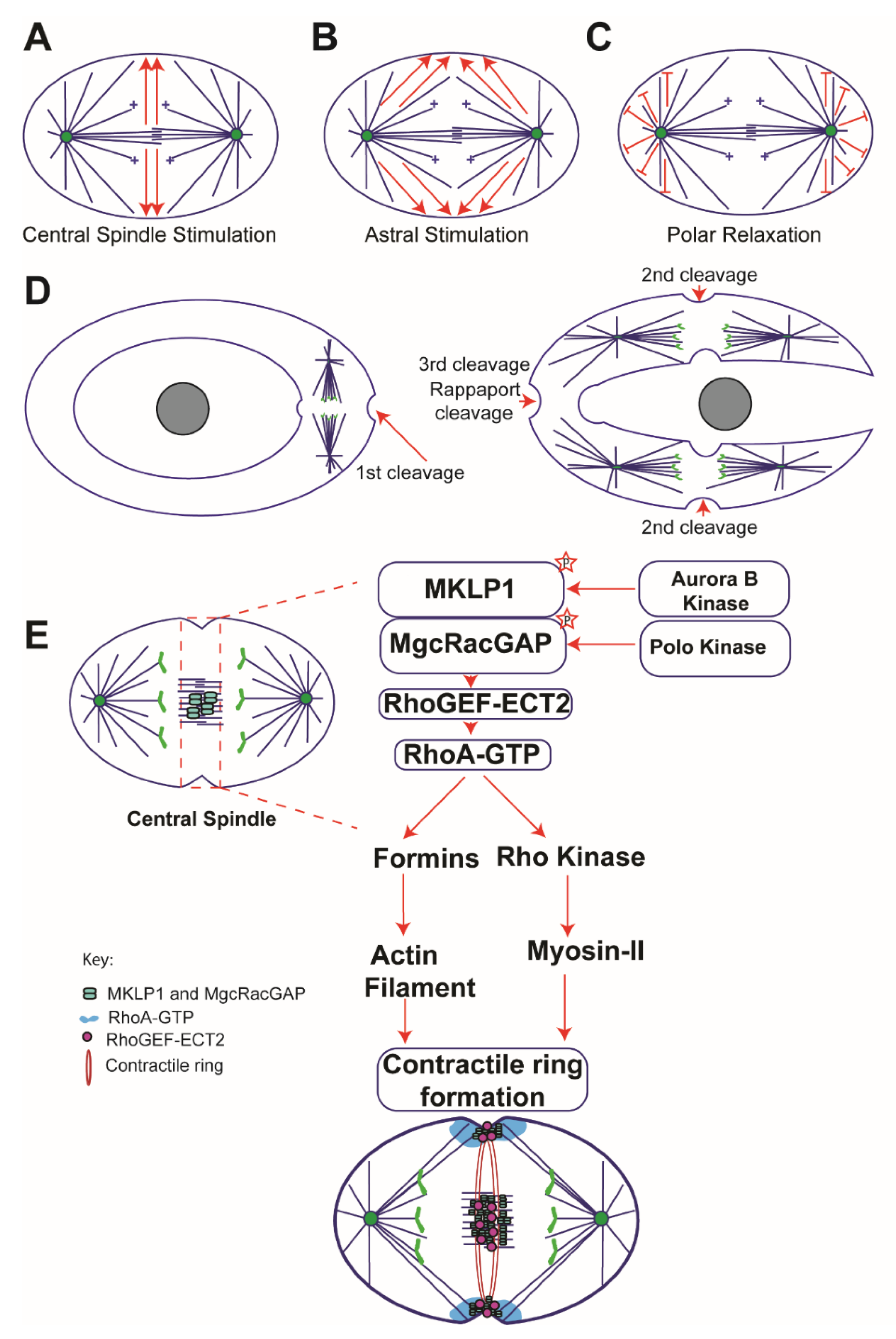
2. Classical Models of Cytokinesis
2.1. Central Spindle Stimulation Model
2.2. Astral Stimulation Model
2.3. Polar Relaxation Model
3. Spatiotemporal Regulation of Cytokinetic Events
3.1. Cdks
3.2. Polo-Like Kinase 1 (Plk1)
3.3. Aurora Kinases
4. Rho GTPases in Cytokinesis
5. Computational Modeling of Cytokinesis
5.1. What Specifies the Site of the Furrow?
5.2. How Does the Contractile Ring Assemble?
5.3. How Does the Ring Generate Force?
5.4. How Does this Force Form the Furrow?
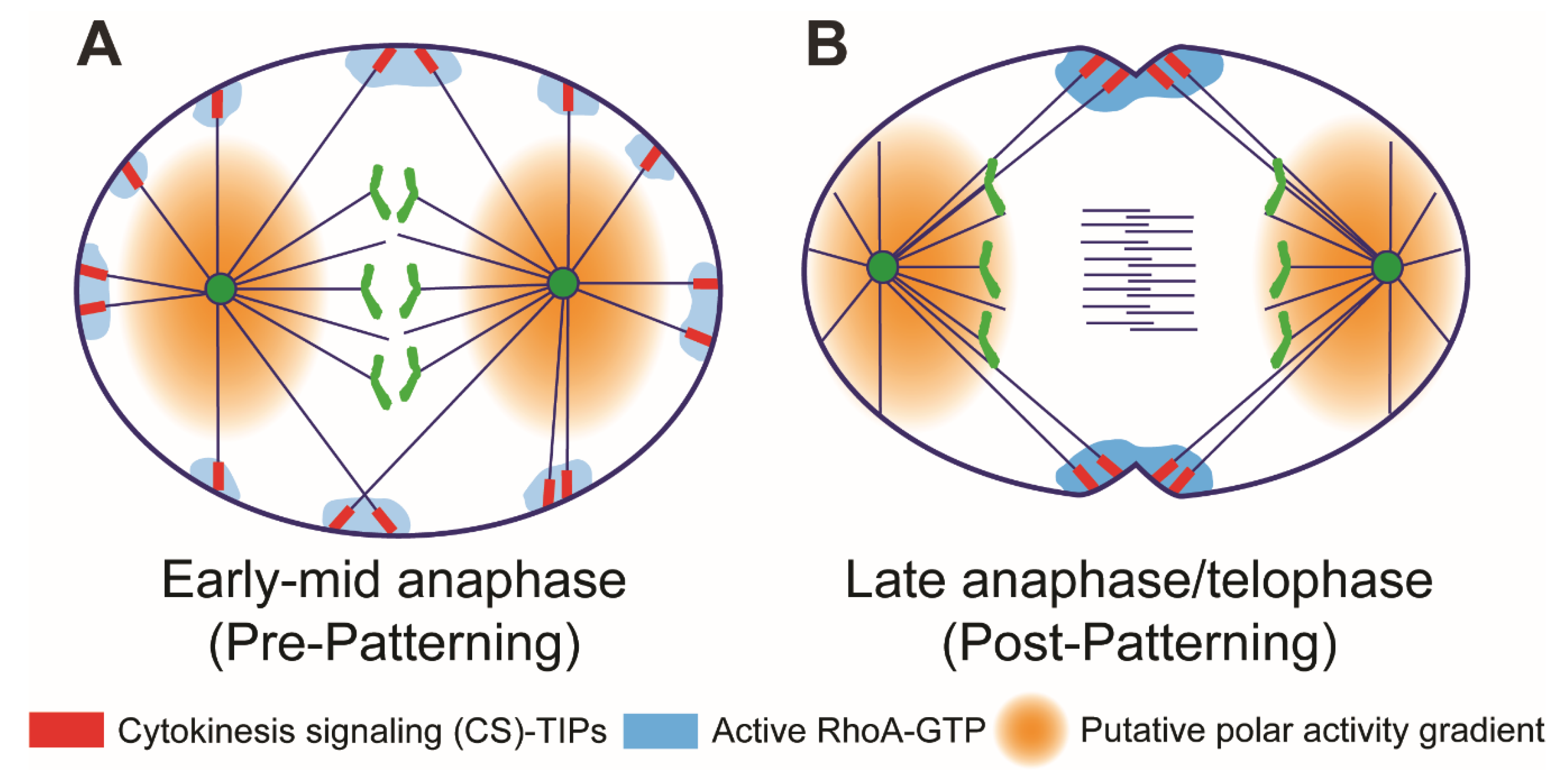
6. Midzone-Independent Signaling in Cleavage Plane Specification
7. Astral Stimulation and Polar Relaxation: Two Sides of the Same Coin?
8. Propagating Waves of Contractility in Cytokinesis
9. Architecture of the Contractile Ring, Ring Constriction, and Furrow Formation
10. Summary and Outlook
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Li, R. Cytokinesis in development and disease: Variations on a common theme. Cell. Mol. Life Sci. 2007, 64, 3044–3058. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, T.; Bandi, M.; Nitta, M.; Ivanova, E.V.; Bronson, R.T.; Pellman, D. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature 2005, 437, 1043–1047. [Google Scholar] [CrossRef] [PubMed]
- Pollard, T.D. Ray rappaport chronology: Twenty-five years of seminal papers on cytokinesis in the journal of experimental zoology. J. Exp. Zool Comp. Exp. Biol. 2004, 301, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Rappaport, R. Repeated furrow formation from a single mitotic apparatus in cylindrical sand dollar eggs. J. Exp. Zool. 1985, 234, 167–171. [Google Scholar] [CrossRef] [PubMed]
- Rappaport, R.; Ebstein, R.P. Duration of stimulus and latent periods preceding furrow formation in sand dollar eggs. J. Exp. Zool. 1965, 158, 373–382. [Google Scholar] [CrossRef]
- Rappaport, R. Cytokinesis in animal cells, 1st ed.; Cambridge University Press: Cambridge, UK, 1996; Volume 30, p. 386. [Google Scholar]
- Glotzer, M. Cleavage furrow positioning. J. Cell Biol. 2004, 164, 347–351. [Google Scholar] [CrossRef] [PubMed]
- Von Dassow, G. Concurrent cues for cytokinetic furrow induction in animal cells. Trends Cell Biol. 2009, 19, 165–173. [Google Scholar] [CrossRef]
- D’Avino, P.P.; Savoian, M.S.; Glover, D.M. Cleavage furrow formation and ingression during animal cytokinesis: A microtubule legacy. J. Cell Sci. 2005, 118, 1549–1558. [Google Scholar] [CrossRef]
- Hutterer, A.; Glotzer, M.; Mishima, M. Clustering of centralspindlin is essential for its accumulation to the central spindle and the midbody. Curr. Biol. 2009, 19, 2043–2049. [Google Scholar] [CrossRef]
- Canman, J.C.; Cameron, L.A.; Maddox, P.S.; Straight, A.; Tirnauer, J.S.; Mitchison, T.J.; Fang, G.; Kapoor, T.M.; Salmon, E.D. Determining the position of the cell division plane. Nature 2003, 424, 1074–1078. [Google Scholar] [CrossRef]
- Nguyen, P.A.; Groen, A.C.; Loose, M.; Ishihara, K.; Wuhr, M.; Field, C.M.; Mitchison, T.J. Spatial organization of cytokinesis signaling reconstituted in a cell-free system. Science 2014, 346, 244–247. [Google Scholar] [CrossRef] [PubMed]
- Bringmann, H.; Hyman, A.A. A cytokinesis furrow is positioned by two consecutive signals. Nature 2005, 436, 731–734. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.G.; Wang, Y.L. Signals from the spindle midzone are required for the stimulation of cytokinesis in cultured epithelial cells. Mol. Biol. Cell 1996, 7, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Crest, J.; Concha-Moore, K.; Sullivan, W. Rhogef and positioning of rappaport-like furrows in the early drosophila embryo. Curr. Biol. 2012, 22, 2037–2041. [Google Scholar] [CrossRef] [PubMed]
- Mogilner, A.; Manhart, A. Agent-based modeling: Case study in cleavage furrow models. Mol. Biol. Cell 2016, 27, 3379–3384. [Google Scholar] [CrossRef] [PubMed]
- Oliferenko, S.; Chew, T.G.; Balasubramanian, M.K. Positioning cytokinesis. Genes Dev. 2009, 23, 660–674. [Google Scholar] [CrossRef] [PubMed]
- Sellitto, C.; Kuriyama, R. Distribution of a matrix component of the midbody during the cell cycle in chinese hamster ovary cells. J. Cell Biol. 1988, 106, 431–439. [Google Scholar] [CrossRef]
- Adams, R.R.; Tavares, A.A.; Salzberg, A.; Bellen, H.J.; Glover, D.M. Pavarotti encodes a kinesin-like protein required to organize the central spindle and contractile ring for cytokinesis. Genes Dev. 1998, 12, 1483–1494. [Google Scholar] [CrossRef] [PubMed]
- Powers, J.; Bossinger, O.; Rose, D.; Strome, S.; Saxton, W. A nematode kinesin required for cleavage furrow advancement. Curr. Biol. 1998, 8, 1133–1136. [Google Scholar] [CrossRef]
- Toure, A.; Dorseuil, O.; Morin, L.; Timmons, P.; Jegou, B.; Reibel, L.; Gacon, G. Mgcracgap, a new human gtpase-activating protein for rac and cdc42 similar to drosophila rotundracgap gene product, is expressed in male germ cells. J. Biol. Chem. 1998, 273, 6019–6023. [Google Scholar] [CrossRef]
- Gonczy, P.; Schnabel, H.; Kaletta, T.; Amores, A.D.; Hyman, T.; Schnabel, R. Dissection of cell division processes in the one cell stage caenorhabditis elegans embryo by mutational analysis. J. Cell Biol. 1999, 144, 927–946. [Google Scholar] [CrossRef] [PubMed]
- Jantsch-Plunger, V.; Gonczy, P.; Romano, A.; Schnabel, H.; Hamill, D.; Schnabel, R.; Hyman, A.A.; Glotzer, M. Cyk-4: A rho family gtpase activating protein (gap) required for central spindle formation and cytokinesis. J. Cell Biol. 2000, 149, 1391–1404. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, T.; Kawashima, T.; Minoshima, Y.; Tonozuka, Y.; Hirose, K.; Nosaka, T. Role of mgcracgap/cyk4 as a regulator of the small gtpase rho family in cytokinesis and cell differentiation. Cell Struct. Funct. 2001, 26, 645–651. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, K. Microdissection studies on the dividing neuroblast of the grasshopper, with special reference to the mechanism of unequal cytokinesis. Exp. Cell Res. 1977, 106, 127–137. [Google Scholar] [CrossRef]
- Rappaport, R.; Rappaport, B.N. Establishment of cleavage furrows by the mitotic spindle. J. Exp. Zool. 1974, 189, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Wheatley, S.P.; Wang, Y. Midzone microtubule bundles are continuously required for cytokinesis in cultured epithelial cells. J. Cell. Biol. 1996, 135, 981–989. [Google Scholar] [CrossRef]
- Von Dassow, G.; Verbrugghe, K.J.C.; Miller, A.L.; Sider, J.R.; Bement, W.M. Action at a distance during cytokinesis. J. Cell Biol. 2010, 187, 831–845. [Google Scholar] [CrossRef]
- Carmena, M.; Wheelock, M.; Funabiki, H.; Earnshaw, W.C. The chromosomal passenger complex (cpc): From easy rider to the godfather of mitosis. Nat. Rev. Mol. Cell Biol. 2012, 13, 789–803. [Google Scholar] [CrossRef]
- Afonso, O.; Figueiredo, A.C.; Maiato, H. Late mitotic functions of aurora kinases. Chromosoma 2017, 126, 93–103. [Google Scholar] [CrossRef]
- Cooke, C.A.; Heck, M.M.; Earnshaw, W.C. The inner centromere protein (incenp) antigens: Movement from inner centromere to midbody during mitosis. J. Cell Biol. 1987, 105, 2053–2067. [Google Scholar] [CrossRef]
- Landino, J.; Norris, S.R.; Li, M.; Ballister, E.R.; Lampson, M.A.; Ohi, R. Two mechanisms coordinate the recruitment of the chromosomal passenger complex to the plane of cell division. Mol. Biol. Cell 2017, 28, 3634–3646. [Google Scholar] [CrossRef] [PubMed]
- Fuller, B.G.; Lampson, M.A.; Foley, E.A.; Rosasco-Nitcher, S.; Le, K.V.; Tobelmann, P.; Brautigan, D.L.; Stukenberg, P.T.; Kapoor, T.M. Midzone activation of aurora b in anaphase produces an intracellular phosphorylation gradient. Nature 2008, 453, 1132–1136. [Google Scholar] [CrossRef] [PubMed]
- Sampath, S.C.; Ohi, R.; Leismann, O.; Salic, A.; Pozniakovski, A.; Funabiki, H. The chromosomal passenger complex is required for chromatin-induced microtubule stabilization and spindle assembly. Cell 2004, 118, 187–202. [Google Scholar] [CrossRef] [PubMed]
- Kelly, A.E.; Sampath, S.C.; Maniar, T.A.; Woo, E.M.; Chait, B.T.; Funabiki, H. Chromosomal enrichment and activation of the aurora b pathway are coupled to spatially regulate spindle assembly. Dev. Cell 2007, 12, 31–43. [Google Scholar] [CrossRef] [PubMed]
- Afonso, O.; Matos, I.; Pereira, A.J.; Aguiar, P.; Lampson, M.A.; Maiato, H. Feedback control of chromosome separation by a midzone aurora b gradient. Science 2014, 345, 332–336. [Google Scholar] [CrossRef] [PubMed]
- Basant, A.; Lekomtsev, S.; Tse, Y.C.; Zhang, D.; Longhini, K.M.; Petronczki, M.; Glotzer, M. Aurora b kinase promotes cytokinesis by inducing centralspindlin oligomers that associate with the plasma membrane. Dev. Cell 2015, 33, 204–215. [Google Scholar] [CrossRef]
- Rappaport, R. Experiments concerning the cleavage stimulus in sand dollar eggs. J. Exp. Zool. 1961, 148, 81–89. [Google Scholar] [CrossRef]
- Foe, V.E.; von Dassow, G. Stable and dynamic microtubules coordinately shape the myosin activation zone during cytokinetic furrow formation. J. Cell Biol. 2008, 183, 457–470. [Google Scholar] [CrossRef]
- Odell, G.M.; Foe, V.E. An agent based model contrasts opposite effects of dynamic and stable microtubules on cleavage furrow positioning. J. Cell Biol. 2008, 183, 471–483. [Google Scholar] [CrossRef]
- Strickland, L.I.; Donnelly, E.J.; Burgess, D.R. Induction of cytokinesis is independent of precisely regulated microtubule dynamics. Mol. Biol. Cell 2005, 16, 4485–4494. [Google Scholar] [CrossRef]
- Minestrini, G.; Harley, A.S.; Glover, D.M. Localization of pavarotti-klp in living drosophila embryos suggests roles in reorganizing the cortical cytoskeleton during the mitotic cycle. Mol. Biol. Cell 2003, 14, 4028–4038. [Google Scholar] [CrossRef] [PubMed]
- Matuliene, J.; Kuriyama, R. Kinesin-like protein cho1 is required for the formation of midbody matrix and the completion of cytokinesis in mammalian cells. Mol. Biol. Cell 2002, 13, 1832–1845. [Google Scholar] [CrossRef] [PubMed]
- Murthy, K.; Wadsworth, P. Dual role for microtubules in regulating cortical contractility during cytokinesis. J. Cell Sci. 2008, 121, 2350–2359. [Google Scholar] [CrossRef] [PubMed]
- Mangal, S.; Sacher, J.; Kim, T.; Osorio, D.S.; Motegi, F.; Carvalho, A.X.; Oegema, K.; Zanin, E. Tpxl-1 activates aurora a to clear contractile ring components from the polar cortex during cytokinesis. J. Cell Biol. 2018, 217, 837–848. [Google Scholar] [CrossRef] [PubMed]
- Kunda, P.; Rodrigues, N.T.; Moeendarbary, E.; Liu, T.; Ivetic, A.; Charras, G.; Baum, B. Pp1-mediated moesin dephosphorylation couples polar relaxation to mitotic exit. Curr. Biol. 2012, 22, 231–236. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, N.T.; Lekomtsev, S.; Jananji, S.; Kriston-Vizi, J.; Hickson, G.R.; Baum, B. Kinetochore-localized pp1-sds22 couples chromosome segregation to polar relaxation. Nature 2015, 524, 489–492. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, M.; Fung, S.Y.; Hameed, U.F.; Goto, H.; Inagaki, M.; Lee, S.H. Cdk1 coordinates timely activation of mklp2 kinesin with relocation of the chromosome passenger complex for cytokinesis. Cell Rep. 2014, 7, 166–179. [Google Scholar] [CrossRef]
- Mishima, M.; Pavicic, V.; Gruneberg, U.; Nigg, E.A.; Glotzer, M. Cell cycle regulation of central spindle assembly. Nature 2004, 430, 908–913. [Google Scholar] [CrossRef]
- Mollinari, C.; Kleman, J.P.; Jiang, W.; Schoehn, G.; Hunter, T.; Margolis, R.L. Prc1 is a microtubule binding and bundling protein essential to maintain the mitotic spindle midzone. J. Cell Biol. 2002, 157, 1175–1186. [Google Scholar] [CrossRef]
- Hummer, S.; Mayer, T.U. Cdk1 negatively regulates midzone localization of the mitotic kinesin mklp2 and the chromosomal passenger complex. Curr. Biol. 2009, 19, 607–612. [Google Scholar] [CrossRef]
- Niiya, F.; Xie, X.; Lee, K.S.; Inoue, H.; Miki, T. Inhibition of cyclin-dependent kinase 1 induces cytokinesis without chromosome segregation in an ect2 and mgcracgap-dependent manner. J. Biol. Chem. 2005, 280, 36502–36509. [Google Scholar] [CrossRef] [PubMed]
- Mishima, M.; Mabuchi, I. Cell cycle-dependent phosphorylation of smooth muscle myosin light chain in sea urchin egg extracts. J. Biochem. 1996, 119, 906–913. [Google Scholar] [CrossRef] [PubMed]
- Satterwhite, L.L.; Lohka, M.J.; Wilson, K.L.; Scherson, T.Y.; Cisek, L.J.; Corden, J.L.; Pollard, T.D. Phosphorylation of myosin-ii regulatory light chain by cyclin-p34cdc2: A mechanism for the timing of cytokinesis. J. Cell Biol. 1992, 118, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Shuster, C.B.; Burgess, D.R. Transitions regulating the timing of cytokinesis in embryonic cells. Curr. Biol. 2002, 12, 854–858. [Google Scholar] [CrossRef]
- Yamakita, Y.; Yamashiro, S.; Matsumura, F. In vivo phosphorylation of regulatory light chain of myosin ii during mitosis of cultured cells. J Cell Biol. 1994, 124, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Petronczki, M.; Lenart, P.; Peters, J.M. Polo on the rise-from mitotic entry to cytokinesis with plk1. Dev. Cell 2008, 14, 646–659. [Google Scholar] [CrossRef]
- Sunkel, C.E.; Glover, D.M. Polo, a mitotic mutant of drosophila displaying abnormal spindle poles. J. Cell Sci. 1988, 89, 25–38. [Google Scholar] [PubMed]
- Hartwell, L.H.; Mortimer, R.K.; Culotti, J.; Culotti, M. Genetic control of the cell division cycle in yeast: V. Genetic analysis of cdc mutants. Genetics 1973, 74, 267–286. [Google Scholar]
- Petronczki, M.; Glotzer, M.; Kraut, N.; Peters, J.-M. Polo-like kinase1 triggers the initiation of cytokinesis in human cells by promoting recruitment of the rhogef ect2 to the central spindle. Devel. Cell 2007, 12, 713–725. [Google Scholar] [CrossRef]
- Burkard, M.E.; Maciejowski, J.; Rodriguez-Bravo, V.; Repka, M.; Lowery, D.M.; Clauser, K.R.; Zhang, C.; Shokat, K.M.; Carr, S.A.; Yaffe, M.B.; et al. Plk1 self-organization and priming phosphorylation of hscyk-4 at the spindle midzone regulate the onset of division in human cells. PLoS Biol. 2009, 7, e1000111. [Google Scholar] [CrossRef]
- Verma, V.; Maresca, T.J. Microtubule plus-ends act as physical signaling hubs to activate rhoa during cytokinesis. Elife 2019, 8, e38968. [Google Scholar] [CrossRef] [PubMed]
- Brennan, I.M.; Peters, U.; Kapoor, T.M.; Straight, A.F. Polo-like kinase controls vertebrate spindle elongation and cytokinesis. PLoS ONE 2007, 2, e409. [Google Scholar] [CrossRef] [PubMed]
- Sumara, I.; Gimenez-Abian, J.F.; Gerlich, D.; Hirota, T.; Kraft, C.; de la Torre, C.; Ellenberg, J.; Peters, J.M. Roles of polo-like kinase 1 in the assembly of functional mitotic spindles. Curr. Biol. 2004, 14, 1712–1722. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.W.; Erikson, E.; Li, C.; Maller, J.L. Activated polo-like kinase plx1 is required at multiple points during mitosis in xenopus laevis. Mol. Cell Biol. 1998, 18, 4262–4271. [Google Scholar] [CrossRef] [PubMed]
- Lenart, P.; Petronczki, M.; Steegmaier, M.; Di Fiore, B.; Lipp, J.J.; Hoffmann, M.; Rettig, W.J.; Kraut, N.; Peters, J.M. The small-molecule inhibitor bi 2536 reveals novel insights into mitotic roles of polo-like kinase 1. Curr. Biol. 2007, 17, 304–315. [Google Scholar] [CrossRef]
- Liu, D.; Davydenko, O.; Lampson, M.A. Polo-like kinase-1 regulates kinetochore-microtubule dynamics and spindle checkpoint silencing. J. Cell Biol. 2012, 198, 491–499. [Google Scholar] [CrossRef]
- Lane, H.A.; Nigg, E.A. Antibody microinjection reveals an essential role for human polo-like kinase 1 (plk1) in the functional maturation of mitotic centrosomes. J. Cell Biol. 1996, 135, 1701–1713. [Google Scholar] [CrossRef]
- Neef, R.; Gruneberg, U.; Kopajtich, R.; Li, X.; Nigg, E.A.; Sillje, H.; Barr, F.A. Choice of plk1 docking partners during mitosis and cytokinesis is controlled by the activation state of cdk1. Nat. Cell Biol. 2007, 9, 436–444. [Google Scholar] [CrossRef]
- Carmena, M.; Ruchaud, S.; Earnshaw, W.C. Making the auroras glow: Regulation of aurora a and b kinase function by interacting proteins. Curr. Opin. Cell Biol. 2009, 21, 796–805. [Google Scholar] [CrossRef]
- Hochegger, H.; Hegarat, N.; Pereira-Leal, J.B. Aurora at the pole and equator: Overlapping functions of aurora kinases in the mitotic spindle. Open Biol. 2013, 3, 120185. [Google Scholar] [CrossRef]
- Chan, C.S.; Botstein, D. Isolation and characterization of chromosome-gain and increase-in-ploidy mutants in yeast. Genetics 1993, 135, 677–691. [Google Scholar] [PubMed]
- Glover, D.M.; Leibowitz, M.H.; McLean, D.A.; Parry, H. Mutations in aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell 1995, 81, 95–105. [Google Scholar] [CrossRef]
- Carmena, M.; Earnshaw, W.C. The cellular geography of aurora kinases. Nat. Rev. Mol. Cell Biol. 2003, 4, 842–854. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Deng, Z.; Fu, J.; Xu, C.; Xin, G.; Wu, Z.; Luo, J.; Wang, G.; Zhang, S.; Zhang, B.; et al. Spatial compartmentalization specializes function of aurora-a and aurora-b. J. Biol. Chem. 2015, 290, 17546–17558. [Google Scholar] [CrossRef] [PubMed]
- Giet, R.; McLean, D.; Descamps, S.; Lee, M.J.; Raff, J.W.; Prigent, C.; Glover, D.M. Drosophila aurora a kinase is required to localize d-tacc to centrosomes and to regulate astral microtubules. J. Cell. Biol. 2002, 156, 437–451. [Google Scholar] [CrossRef] [PubMed]
- Hannak, E.; Kirkham, M.; Hyman, A.A.; Oegema, K. Aurora-a kinase is required for centrosome maturation in caenorhabditis elegans. J. Cell Biol. 2001, 155, 1109–1116. [Google Scholar] [CrossRef] [PubMed]
- Bakhoum, S.F.; Kabeche, L.; Murnane, J.P.; Zaki, B.I.; Compton, D.A. DNA-damage response during mitosis induces whole-chromosome missegregation. Cancer Discov. 2014, 4, 1281–1289. [Google Scholar] [CrossRef] [PubMed]
- Barisic, M.; Aguiar, P.; Geley, S.; Maiato, H. Kinetochore motors drive congression of peripheral polar chromosomes by overcoming random arm-ejection forces. Nat. Cell Biol. 2014, 16, 1249–1256. [Google Scholar] [CrossRef]
- Chmatal, L.; Yang, K.; Schultz, R.M.; Lampson, M.A. Spatial regulation of kinetochore microtubule attachments by destabilization at spindle poles in meiosis i. Curr. Biol. 2015, 25, 1835–1841. [Google Scholar] [CrossRef]
- Ye, A.A.; Deretic, J.; Hoel, C.M.; Hinman, A.W.; Cimini, D.; Welburn, J.P.; Maresca, T.J. Aurora a kinase contributes to a pole-based error correction pathway. Curr. Biol. 2015, 25, 1842–1851. [Google Scholar] [CrossRef]
- DeLuca, K.F.; Meppelink, A.; Broad, A.J.; Mick, J.E.; Peersen, O.B.; Pektas, S.; Lens, S.M.A.; DeLuca, J.G. Aurora a kinase phosphorylates hec1 to regulate metaphase kinetochore-microtubule dynamics. J. Cell Biol. 2018, 217, 163–177. [Google Scholar] [CrossRef] [PubMed]
- Hegarat, N.; Smith, E.; Nayak, G.; Takeda, S.; Eyers, P.A.; Hochegger, H. Aurora a and aurora b jointly coordinate chromosome segregation and anaphase microtubule dynamics. J. Cell. Biol. 2011, 195, 1103–1113. [Google Scholar] [CrossRef] [PubMed]
- Scrofani, J.; Sardon, T.; Meunier, S.; Vernos, I. Microtubule nucleation in mitosis by a rangtp-dependent protein complex. Curr. Biol. 2015, 25, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Lioutas, A.; Vernos, I. Aurora a kinase and its substrate tacc3 are required for central spindle assembly. EMBO Rep. 2013, 14, 829–836. [Google Scholar] [CrossRef] [PubMed]
- Reboutier, D.; Troadec, M.B.; Cremet, J.Y.; Chauvin, L.; Guen, V.; Salaun, P.; Prigent, C. Aurora a is involved in central spindle assembly through phosphorylation of ser 19 in p150glued. J. Cell Biol. 2013, 201, 65–79. [Google Scholar] [CrossRef] [PubMed]
- Ye, A.; Torabi, J.; Maresca, T.J. Aurora a kinase amplifies a midzone phosphorylation gradient to promote high-fidelity cytokinesis. Biol. Bull. 2016, 231, 61–72. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vale, R.D.; Spudich, J.A.; Griffis, E.R. Dynamics of myosin, microtubules, and kinesin-6 at the cortex during cytokinesis in drosophila s2 cells. J. Cell Biol. 2009, 186, 727–738. [Google Scholar] [CrossRef]
- Eggert, U.S.; Kiger, A.A.; Richter, C.; Perlman, Z.E.; Perrimon, N.; Mitchison, T.J.; Field, C.M. Parallel chemical genetic and genome-wide rnai screens identify cytokinesis inhibitors and targets. PLoS Biol. 2004, 2, e379. [Google Scholar] [CrossRef]
- Guse, A.; Mishima, M.; Glotzer, M. Phosphorylation of zen-4/mklp1 by aurora b regulates completion of cytokinesis. Curr. Biol. 2005, 15, 778–786. [Google Scholar] [CrossRef]
- Nunes Bastos, R.; Gandhi, S.R.; Baron, R.D.; Gruneberg, U.; Nigg, E.A.; Barr, F.A. Aurora b suppresses microtubule dynamics and limits central spindle size by locally activating kif4a. J. Cell Biol. 2013, 202, 605–621. [Google Scholar] [CrossRef]
- Gruneberg, U.; Neef, R.; Honda, R.; Nigg, E.A.; Barr, F.A. Relocation of aurora b from centromeres to the central spindle at the metaphase to anaphase transition requires mklp2. J. Cell Biol. 2004, 166, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Cesario, J.M.; Jang, J.K.; Redding, B.; Shah, N.; Rahman, T.; McKim, K.S. Kinesin 6 family member subito participates in mitotic spindle assembly and interacts with mitotic regulators. J. Cell Sci. 2006, 119, 4770–4780. [Google Scholar] [CrossRef] [PubMed]
- Basant, A.; Glotzer, M. Spatiotemporal regulation of rhoa during cytokinesis. Curr. Biol. 2018, 28, R570–R580. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.E.; Billadeau, D.D.; Chen, J. The tandem brct domains of ect2 are required for both negative and positive regulation of ect2 in cytokinesis. J. Biol. Chem. 2005, 280, 5733–5739. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, B.A.; Takaki, T.; Petronczki, M.; Glotzer, M. Polo-like kinase 1 directs assembly of the hscyk-4 rhogap/ect2 rhogef complex to initiate cleavage furrow formation. PLoS Biol. 2009, 7, e1000110. [Google Scholar] [CrossRef] [PubMed]
- Schonegg, S.; Constantinescu, A.T.; Hoege, C.; Hyman, A.A. The rho gtpase-activating proteins rga-3 and rga-4 are required to set the initial size of par domains in caenorhabditis elegans one-cell embryos. Proc. Natl. Acad. Sci. USA 2007, 104, 14976–14981. [Google Scholar] [CrossRef] [PubMed]
- Prokopenko, S.N.; Brumby, A.; O’Keefe, L.; Prior, L.; He, Y.; Saint, R.; Bellen, H.J. A putative exchange factor for rho1 gtpase is required for initiation of cytokinesis in drosophila. Genes Dev. 1999, 13, 2301–2314. [Google Scholar] [CrossRef]
- Kotynkova, K.; Su, K.-C.; West, S.C.; Petronczki, M. Plasma membrane association but not midzone recruitment of rhogef ect2 is essential for cytokinesis. Cell Rep. 2016, 17, 2672–2686. [Google Scholar] [CrossRef]
- Wagner, E.; Glotzer, M. Local rhoa actiation induces cytokinetic furrows independent of spindle position and cell cycle stage. J. Cell Biol. 2016, 213, 641–649. [Google Scholar] [CrossRef]
- Jordan, S.N.; Canman, J.C. Rho gtpases in animal cell cytokinesis: An occupation by the one percent. Cytoskeleton 2012, 69, 919–930. [Google Scholar] [CrossRef]
- White, E.A.; Glotzer, M. Centralspindlin: At the heart of cytokinesis. Cytoskeleton 2012, 69, 882–892. [Google Scholar] [CrossRef] [PubMed]
- Bastos, R.N.; Penate, X.; Bates, M.; Hammond, D.; Barr, F.A. Cyk4 inhibits rac1-dependent pak1 and arhgef7 effector pathways during cytokinesis. J. Cell Biol. 2012, 198, 865–880. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, A.Y.; Jan, Y.N.; Luo, L. Function and regulation of tumbleweed (racgap50c) in neuroblast proliferation and neuronal morphogenesis. Proc. Nat.l Acad. Sci. USA 2005, 102, 3834–3839. [Google Scholar] [CrossRef] [PubMed]
- Yamada, T.; Hikida, M.; Kurosaki, T. Regulation of cytokinesis by mgcracgap in b lymphocytes is independent of gap activity. Exp. Cell Res. 2006, 312, 3517–3525. [Google Scholar] [CrossRef] [PubMed]
- Loria, A.; Longhini, K.M.; Glotzer, M. The rhogap domain of cyk-4 has an essential role in rhoa activation. Curr. Biol. 2012, 22, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Zavortink, M.; Contreras, N.; Addy, T.; Bejsovec, A.; Saint, R. Tum/racgap50c provides a critical link between anaphase microtubules and the assembly of the contractile ring in drosophila melanogaster. J. Cell Sci. 2005, 118, 5381–5392. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Canman, J.C.; Lewellyn, L.; Laband, K.; Smerdon, S.J.; Desai, A.; Bowerman, B.; Oegema, K. Inhibition of rac by the gap activity of centralspindlin is essential for cytokinesis. Science 2008, 322, 1543–1546. [Google Scholar] [CrossRef] [PubMed]
- Minoshima, Y.; Kawashima, T.; Hirose, K.; Tonozuka, Y.; Kawajiri, A.; Bao, Y.C.; Deng, X.; Tatsuka, M.; Narumiya, S.; May, W.S., Jr.; et al. Phosphorylation by aurora b converts mgcracgap to a rhogap during cytokinesis. Dev. Cell 2003, 4, 549–560. [Google Scholar] [CrossRef]
- Zhang, D.; Glotzer, M. The rhogap activity of cyk-4/mgcracgap functions non-canonically by promoting rhoa activation during cytokinesis. Elife 2015, 4, e08898. [Google Scholar] [CrossRef]
- Pollard, T.D. Regulation of actin filament assembly by arp2/3 complex and formins. Annu. Rev. Biophys. Biomol. Struct. 2007, 36, 451–477. [Google Scholar] [CrossRef]
- Atkins, B.D.; Yoshida, S.; Saito, K.; Wu, C.F.; Lew, D.J.; Pellman, D. Inhibition of cdc42 during mitotic exit is required for cytokinesis. J. Cell Biol. 2013, 202, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Pollard, T.D. Nine unanswered questions about cytokinesis. J. Cell Biol. 2017, 216, 3007–3016. [Google Scholar] [CrossRef] [PubMed]
- Sept, D.; Carlsson, A.E. Modeling large-scale dynamic processes in the cell: Polarization, waves, and division. Q. Rev. Biophys. 2014, 47, 221–248. [Google Scholar] [CrossRef] [PubMed]
- Cortes, D.B.; Dawes, A.; Liu, J.; Nickaeen, M.; Strychalski, W.; Maddox, A.S. Unite to divide—How models and biological experimentation have come together to reveal mechanisms of cytokinesis. J. Cell Sci. 2018, 131. [Google Scholar] [CrossRef] [PubMed]
- White, J.G.; Borisy, G.G. On the mechanisms of cytokinesis in animal cells. J. Theor. Biol. 1983, 101, 289–316. [Google Scholar] [CrossRef]
- Devore, J.J.; Conrad, G.W.; Rappaport, R. A model for astral stimulation of cytokinesis in animal cells. J. Cell Biol. 1989, 109, 2225–2232. [Google Scholar] [CrossRef] [PubMed]
- Harris, A.K.; Gewalt, S.L. Simulation testing of mechanisms for inducing the formation of the contractile ring in cytokinesis. J. Cell Biol. 1989, 109, 2215–2223. [Google Scholar] [CrossRef]
- Yoshigaki, T. Why does a cleavage plane develop parallel to the spindle axis in conical sand dollar eggs? A key question for clarifying the mechanism of contractile ring positioning. J. Theor. Biol. 2003, 221, 229–244. [Google Scholar] [CrossRef]
- Vavylonis, D.; Wu, J.Q.; Hao, S.; O’Shaughnessy, B.; Pollard, T.D. Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science 2008, 319, 97–100. [Google Scholar] [CrossRef]
- Bidone, T.C.; Tang, H.; Vavylonis, D. Dynamic network morphology and tension buildup in a 3d model of cytokinetic ring assembly. Biophys. J. 2014, 107, 2618–2628. [Google Scholar] [CrossRef]
- Reymann, A.C.; Staniscia, F.; Erzberger, A.; Salbreux, G.; Grill, S.W. Cortical flow aligns actin filaments to form a furrow. Elife 2016, 5, e17807. [Google Scholar] [CrossRef] [PubMed]
- Bidone, T.C.; Jung, W.; Maruri, D.; Borau, C.; Kamm, R.D.; Kim, T. Morphological transformation and force generation of active cytoskeletal networks. PLoS Comput. Biol. 2017, 13, e1005277. [Google Scholar] [CrossRef] [PubMed]
- Mendes Pinto, I.; Rubinstein, B.; Kucharavy, A.; Unruh, J.R.; Li, R. Actin depolymerization drives actomyosin ring contraction during budding yeast cytokinesis. Dev. Cell 2012, 22, 1247–1260. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, T.E. The contractile ring. Ii. Determining its brief existence, volumetric changes, and vital role in cleaving arbacia eggs. J. Cell Biol. 1972, 53, 419–434. [Google Scholar] [CrossRef] [PubMed]
- Stachowiak, M.R.; Laplante, C.; Chin, H.F.; Guirao, B.; Karatekin, E.; Pollard, T.D.; O’Shaughnessy, B. Mechanism of cytokinetic contractile ring constriction in fission yeast. Dev. Cell 2014, 29, 547–561. [Google Scholar] [CrossRef] [PubMed]
- Belmonte, J.M.; Leptin, M.; Nedelec, F. A theory that predicts behaviors of disordered cytoskeletal networks. Mol. Syst. Biol. 2017, 13, 941. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Robinson, D.N. Balance of actively generated contractile and resistive forces controls cytokinesis dynamics. Proc. Natl. Acad. Sci. USA 2005, 102, 7186–7191. [Google Scholar] [CrossRef] [PubMed]
- Sedzinski, J.; Biro, M.; Oswald, A.; Tinevez, J.Y.; Salbreux, G.; Paluch, E. Polar actomyosin contractility destabilizes the position of the cytokinetic furrow. Nature 2011, 476, 462–466. [Google Scholar] [CrossRef] [PubMed]
- Poirier, C.C.; Ng, W.P.; Robinson, D.N.; Iglesias, P.A. Deconvolution of the cellular force-generating subsystems that govern cytokinesis furrow ingression. PLoS Comput. Biol. 2012, 8, e1002467. [Google Scholar] [CrossRef]
- Turlier, H.; Audoly, B.; Prost, J.; Joanny, J.F. Furrow constriction in animal cell cytokinesis. Biophys. J. 2014, 106, 114–123. [Google Scholar] [CrossRef]
- Gladilin, E.; Eils, R.; Peshkin, L. On the embryonic cell division beyond the contractile ring mechanism: Experimental and computational investigation of effects of vitelline confinement, temperature and egg size. PeerJ 2015, 3, e1490. [Google Scholar] [CrossRef]
- Li, Y.; He, L.; Gonzalez, N.A.P.; Graham, J.; Wolgemuth, C.; Wirtz, D.; Sun, S.X. Going with the flow: Water flux and cell shape during cytokinesis. Biophys. J. 2017, 113, 2487–2495. [Google Scholar] [CrossRef] [PubMed]
- Shlomovitz, R.; Gov, N.S. Physical model of contractile ring initiation in dividing cells. Biophys. J. 2008, 94, 1155–1168. [Google Scholar] [CrossRef] [PubMed]
- Dorn, J.F.; Zhang, L.; Phi, T.T.; Lacroix, B.; Maddox, P.S.; Liu, J.; Maddox, A.S. A theoretical model of cytokinesis implicates feedback between membrane curvature and cytoskeletal organization in asymmetric cytokinetic furrowing. Mol. Biol. Cell 2016, 27, 1286–1299. [Google Scholar] [CrossRef] [PubMed]
- Khaliullin, R.N.; Green, R.A.; Shi, L.Z.; Gomez-Cavazos, J.S.; Berns, M.W.; Desai, A.; Oegema, K. A positive-feedback-based mechanism for constriction rate acceleration during cytokinesis in caenorhabditis elegans. Elife 2018, 7, e36073. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Dembo, M. On the mechanics of the first cleavage division of the sea urchin egg. Exp. Cell Res. 1997, 233, 252–273. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Wang, Q. Modeling cytokinesis of eukaryotic cells driven by the actomyosin contractile ring. Int. J. Numer. Method. Biomed. Eng. 2016, 32, e02774. [Google Scholar] [CrossRef] [PubMed]
- Lee, S. Mathematical model of contractile ring-driven cytokinesis in a three-dimensional domain. Bull. Math. Biol. 2018, 80, 583–597. [Google Scholar] [CrossRef] [PubMed]
- Bement, W.M.; Leda, M.; Moe, A.M.; Kita, A.M.; Larson, M.E.; Golding, A.E.; Pfeuti, C.; Su, K.C.; Miller, A.L.; Goryachev, A.B.; et al. Activator-inhibitor coupling between rho signalling and actin assembly makes the cell cortex an excitable medium. Nat. Cell Biol. 2015, 17, 1471–1483. [Google Scholar] [CrossRef]
- Goryachev, A.B.; Leda, M.; Miller, A.L.; von Dassow, G.; Bement, W.M. How to make a static cytokinetic furrow out of traveling excitable waves. Small GTPases 2016, 7, 65–70. [Google Scholar] [CrossRef]
- Breznau, E.B.; Murt, M.; Blasius, T.L.; Verhey, K.J.; Miller, A.L. The mgcracgap sxip motif tethers centralspindlin to microtubule plus ends in xenopus laevis. J. Cell Sci. 2017, 130, 1809–1821. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, Y.; Yonemura, S. Centralspindlin regulates ect2 and rhoa accumulation at the equatorial cortex during cytokinesis. J. Cell Sci. 2006, 119, 104–114. [Google Scholar] [CrossRef] [PubMed]
- Tse, Y.C.; Werner, M.; Longhini, K.M.; Labbe, J.C.; Goldstein, B.; Glotzer, M. Rhoa activation during polarization and cytokinesis of the early caenorhabditis elegans embryo is differentially dependent on nop-1 and cyk-4. Mol. Biol. Cell 2012, 23, 4020–4031. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Jiang, W. Cell cycle-dependent translocation of prc1 on the spindle by kif4 is essential for midzone formation and cytokinesis. Proc. Natl. Acad. Sci. USA 2005, 102, 343–348. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Lau, E.; Schwarzenbacher, R.; Bossy-Wetzel, E.; Jiang, W. Spatiotemporal control of spindle midzone formation by prc1 in human cells. Proc. Natl. Acad. Sci. USA 2006, 103, 6196–6201. [Google Scholar] [CrossRef] [PubMed]
- Barr, F.A.; Sillje, H.H.; Nigg, E.A. Polo-like kinases and the orchestration of cell division. Nat. Rev. Mol. Cell Biol. 2004, 5, 429–440. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, R.; Ti, S.C.; Tan, L.; Darst, S.A.; Kapoor, T.M. Marking and measuring single microtubules by prc1 and kinesin-4. Cell 2013, 154, 377–390. [Google Scholar] [CrossRef] [PubMed]
- Bieling, P.; Telley, I.A.; Surrey, T. A minimal midzone protein module controls formation and length of antiparallel microtubule overlaps. Cell 2010, 142, 420–432. [Google Scholar] [CrossRef]
- Mollinari, C.; Kleman, J.P.; Saoudi, Y.; Jablonski, S.A.; Perard, J.; Yen, T.J.; Margolis, R.L. Ablation of prc1 by small interfering rna demonstrates that cytokinetic abscission requires a central spindle bundle in mammalian cells, whereas completion of furrowing does not. Mol. Biol. Cell 2005, 16, 1043–1055. [Google Scholar] [CrossRef]
- Adriaans, I.E.; Basant, A.; Ponsioen, B.; Glotzer, M.; Lens, S.M.A. Plk1 plays dual roles in centralspindlin regulation during cytokinesis. J. Cell Biol. 2019, 218, 1250–1264. [Google Scholar] [CrossRef]
- Kalab, P.; Heald, R. The rangtp gradient—A gps for the mitotic spindle. J. Cell Sci. 2008, 121, 1577–1586. [Google Scholar] [CrossRef] [PubMed]
- Kiyomitsu, T.; Cheeseman, I.M. Chromosome- and spindle-pole-derived signals generate an intrinsic code for spindle position and orientation. Nat. Cell Biol. 2012, 14, 311–317. [Google Scholar] [CrossRef] [PubMed]
- Lampson, M.A.; Cheeseman, I.M. Sensing centromere tension: Aurora b and the regulation of kinetochore function. Trends Cell Biol. 2011, 21, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Ye, A.A.; Maresca, T.J. It’s all relative: Centromere- versus pole-based error correction. Cell Cycle 2015, 14, 3777–3778. [Google Scholar] [CrossRef] [PubMed]
- Goshima, G. Identification of a tpx2-like microtubule-associated protein in drosophila. PLoS ONE 2011, 6, e28120. [Google Scholar] [CrossRef]
- Green, R.A.; Paluch, E.; Oegema, K. Cytokinesis in animal cells. Annu. Rev. Cell Dev. Biol. 2012, 28, 29–58. [Google Scholar] [CrossRef]
- Pollard, T.D.; O’Shaughnessy, B. Molecular mechanism of cytokinesis. Annu. Rev. Biochem. 2019, 88, 661–689. [Google Scholar] [CrossRef]
- Graessl, M.; Koch, J.; Calderon, A.; Kamps, D.; Banerjee, S.; Mazel, T.; Schulze, N.; Jungkurth, J.K.; Patwardhan, R.; Solouk, D.; et al. An excitable rho gtpase signaling network generates dynamic subcellular contraction patterns. J. Cell Biol. 2017, 216, 4271–4285. [Google Scholar] [CrossRef]
- Murray, A.W.; Kirschner, M.W. Cyclin synthesis drives the early embryonic cell cycle. Nature 1989, 339, 275–280. [Google Scholar] [CrossRef]
- Nishikawa, M.; Naganathan, S.R.; Julicher, F.; Grill, S.W. Controlling contractile instabilities in the actomyosin cortex. Elife 2017, 6, e19595. [Google Scholar] [CrossRef]
- Reinhard, N.R.; van Helden, S.F.; Anthony, E.C.; Yin, T.; Wu, Y.I.; Goedhart, J.; Gadella, T.W.; Hordijk, P.L. Spatiotemporal analysis of rhoa/b/c activation in primary human endothelial cells. Sci. Rep. 2016, 6, 25502. [Google Scholar] [CrossRef] [PubMed]
- Parent, C.A.; Devreotes, P.N. A cell’s sense of direction. Science 1999, 284, 765–770. [Google Scholar] [CrossRef] [PubMed]
- Levchenko, A.; Iglesias, P.A. Models of eukaryotic gradient sensing: Application to chemotaxis of amoebae and neutrophils. Biophys. J. 2002, 82, 50–63. [Google Scholar] [CrossRef]
- McDonald, N.A.; Lind, A.L.; Smith, S.E.; Li, R.; Gould, K.L. Nanoscale architecture of the schizosaccharomyces pombe contractile ring. Elife 2017, 6, e28865. [Google Scholar] [CrossRef] [PubMed]
- Laplante, C.; Huang, F.; Tebbs, I.R.; Bewersdorf, J.; Pollard, T.D. Molecular organization of cytokinesis nodes and contractile rings by super-resolution fluorescence microscopy of live fission yeast. Proc. Natl. Acad. Sci. USA 2016, 113, E5876–E5885. [Google Scholar] [CrossRef] [PubMed]
- Swulius, M.T.; Nguyen, L.T.; Ladinsky, M.S.; Ortega, D.R.; Aich, S.; Mishra, M.; Jensen, G.J. Structure of the fission yeast actomyosin ring during constriction. Proc. Natl. Acad. Sci. USA 2018, 115, E1455–E1464. [Google Scholar] [CrossRef] [PubMed]
- Beach, J.R.; Shao, L.; Remmert, K.; Li, D.; Betzig, E.; Hammer, J.A., 3rd. Nonmuscle myosin ii isoforms coassemble in living cells. Curr. Biol. 2014, 24, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Henson, J.H.; Ditzler, C.E.; Germain, A.; Irwin, P.M.; Vogt, E.T.; Yang, S.; Wu, X.; Shuster, C.B. The ultrastructural organization of actin and myosin ii filaments in the contractile ring: New support for an old model of cytokinesis. Mol. Biol. Cell 2017, 28, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Fenix, A.M.; Taneja, N.; Buttler, C.A.; Lewis, J.; Van Engelenburg, S.B.; Ohi, R.; Burnette, D.T. Expansion and concatenation of non-muscle myosin iia filaments drive cellular contractile system formation during interphase and mitosis. Mol. Biol. Cell 2016, 28, 613–623. [Google Scholar]
- Sun, L.; Guan, R.; Lee, I.J.; Liu, Y.; Chen, M.; Wang, J.; Wu, J.Q.; Chen, Z. Mechanistic insights into the anchorage of the contractile ring by anillin and mid1. Dev. Cell 2015, 33, 413–426. [Google Scholar] [CrossRef] [PubMed]
- D’Avino, P.P.; Takeda, T.; Capalbo, L.; Zhang, W.; Lilley, K.S.; Laue, E.D.; Glover, D.M. Interaction between anillin and racgap50c connects the actomyosin contractile ring with spindle microtubules at the cell division site. J. Cell Sci. 2008, 121, 1151–1158. [Google Scholar] [CrossRef] [PubMed]
- Frenette, P.; Haines, E.; Loloyan, M.; Kinal, M.; Pakarian, P.; Piekny, A. An anillin-ect2 complex stabilizes central spindle microtubules at the cortex during cytokinesis. PLoS ONE 2012, 7, e34888. [Google Scholar] [CrossRef] [PubMed]
- Piekny, A.J.; Glotzer, M. Anillin is a scaffold protein that links rhoa, actin, and myosin during cytokinesis. Curr. Biol. 2008, 18, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Straight, A.F.; Field, C.M.; Mitchison, T.J. Anillin binds nonmuscle myosin ii and regulates the contractile ring. Mol. Biol. Cell 2005, 16, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Field, C.M.; Alberts, B.M. Anillin, a contractile ring protein that cycles from the nucleus to the cell cortex. J. Cell Biol. 1995, 131, 165–178. [Google Scholar] [CrossRef] [PubMed]
- McDonald, N.A.; Vander Kooi, C.W.; Ohi, M.D.; Gould, K.L. Oligomerization but not membrane bending underlies the function of certain f-bar proteins in cell motility and cytokinesis. Dev. Cell 2015, 35, 725–736. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Verma, V.; Mogilner, A.; Maresca, T.J. Classical and Emerging Regulatory Mechanisms of Cytokinesis in Animal Cells. Biology 2019, 8, 55. https://doi.org/10.3390/biology8030055
Verma V, Mogilner A, Maresca TJ. Classical and Emerging Regulatory Mechanisms of Cytokinesis in Animal Cells. Biology. 2019; 8(3):55. https://doi.org/10.3390/biology8030055
Chicago/Turabian StyleVerma, Vikash, Alex Mogilner, and Thomas J. Maresca. 2019. "Classical and Emerging Regulatory Mechanisms of Cytokinesis in Animal Cells" Biology 8, no. 3: 55. https://doi.org/10.3390/biology8030055
APA StyleVerma, V., Mogilner, A., & Maresca, T. J. (2019). Classical and Emerging Regulatory Mechanisms of Cytokinesis in Animal Cells. Biology, 8(3), 55. https://doi.org/10.3390/biology8030055




