Anaphase B
Abstract
:1. Introduction and Historical Perspective
2. Dynamics of Anaphase B in Living Cells
3. Energetics of Anaphase B
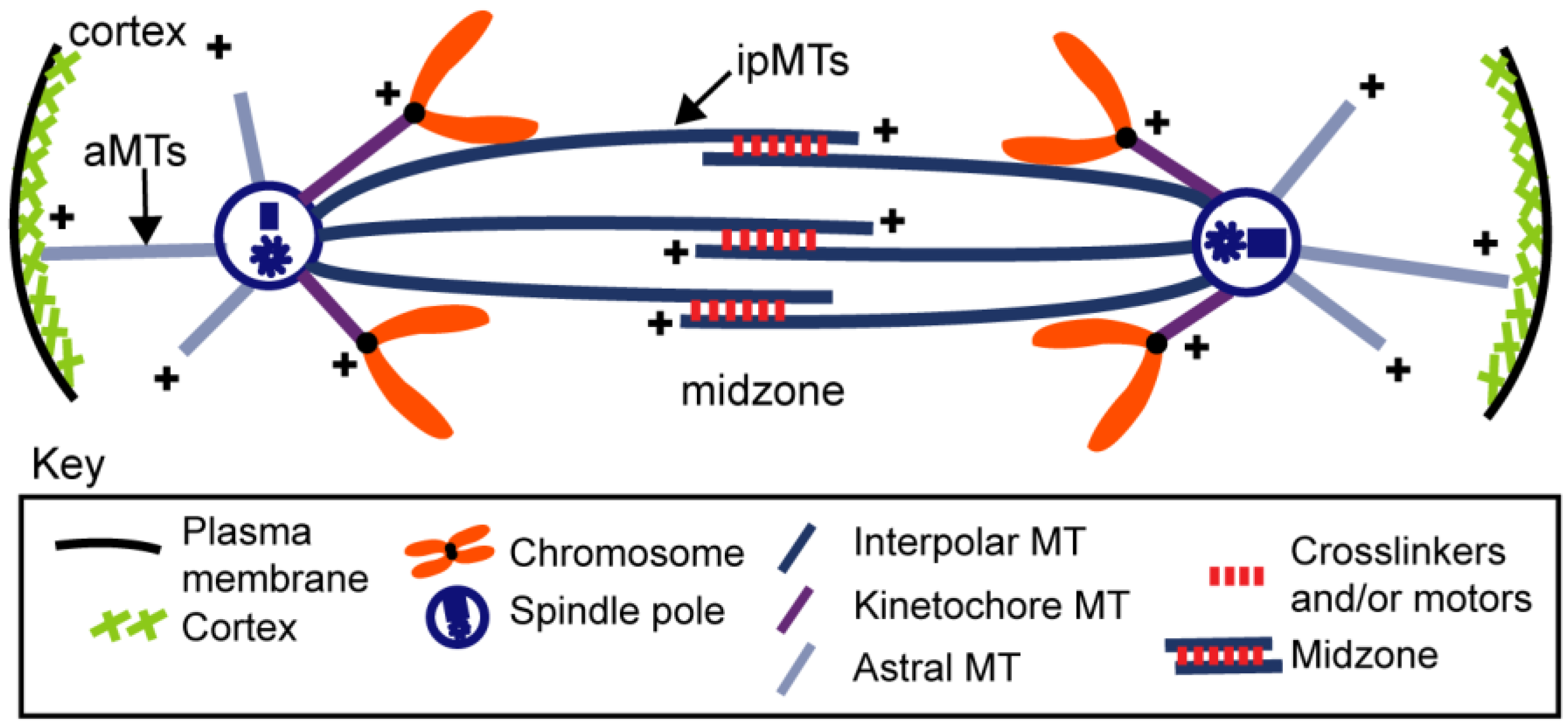
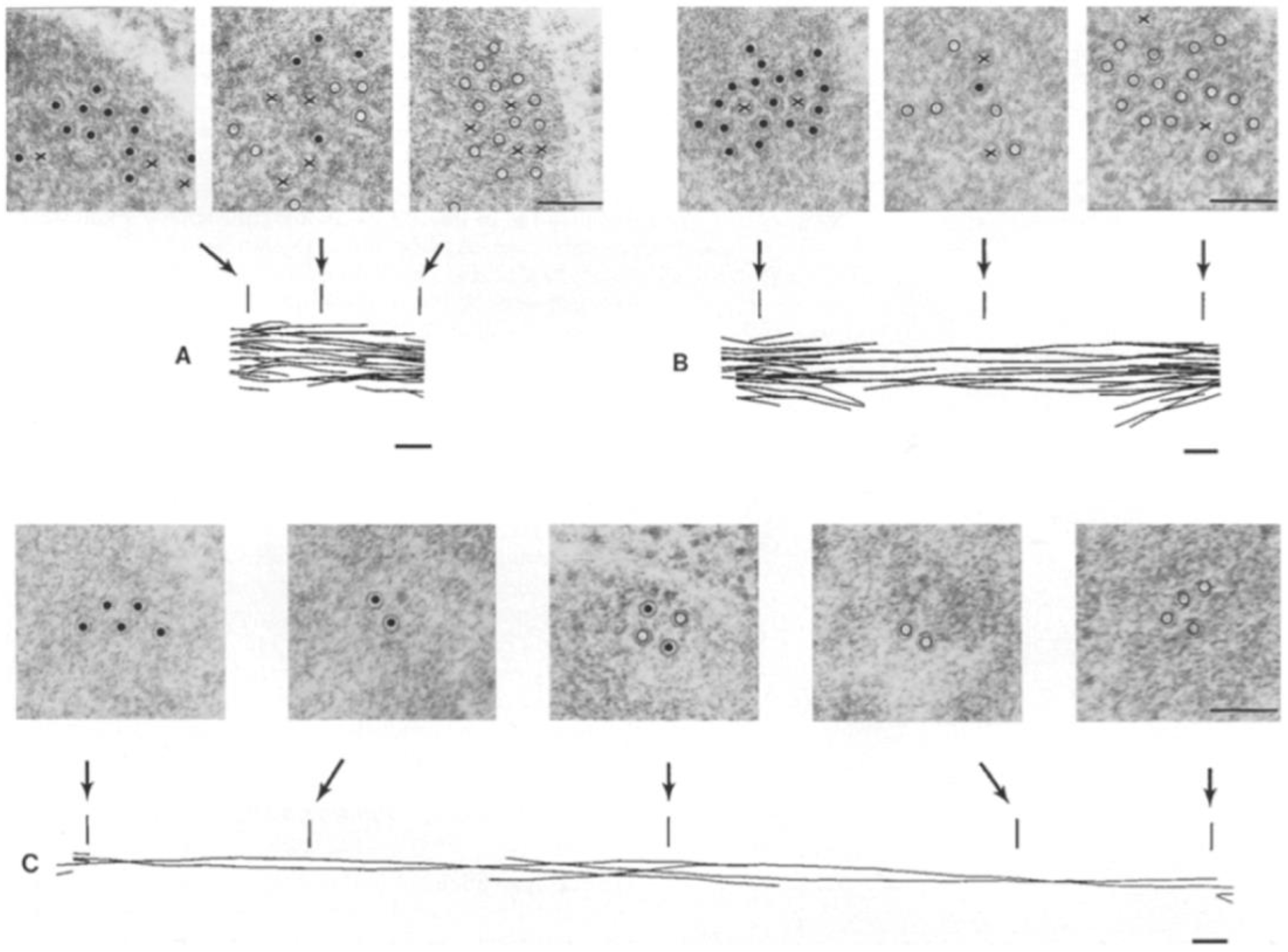
4. Structural Studies of the Anaphase B Spindle
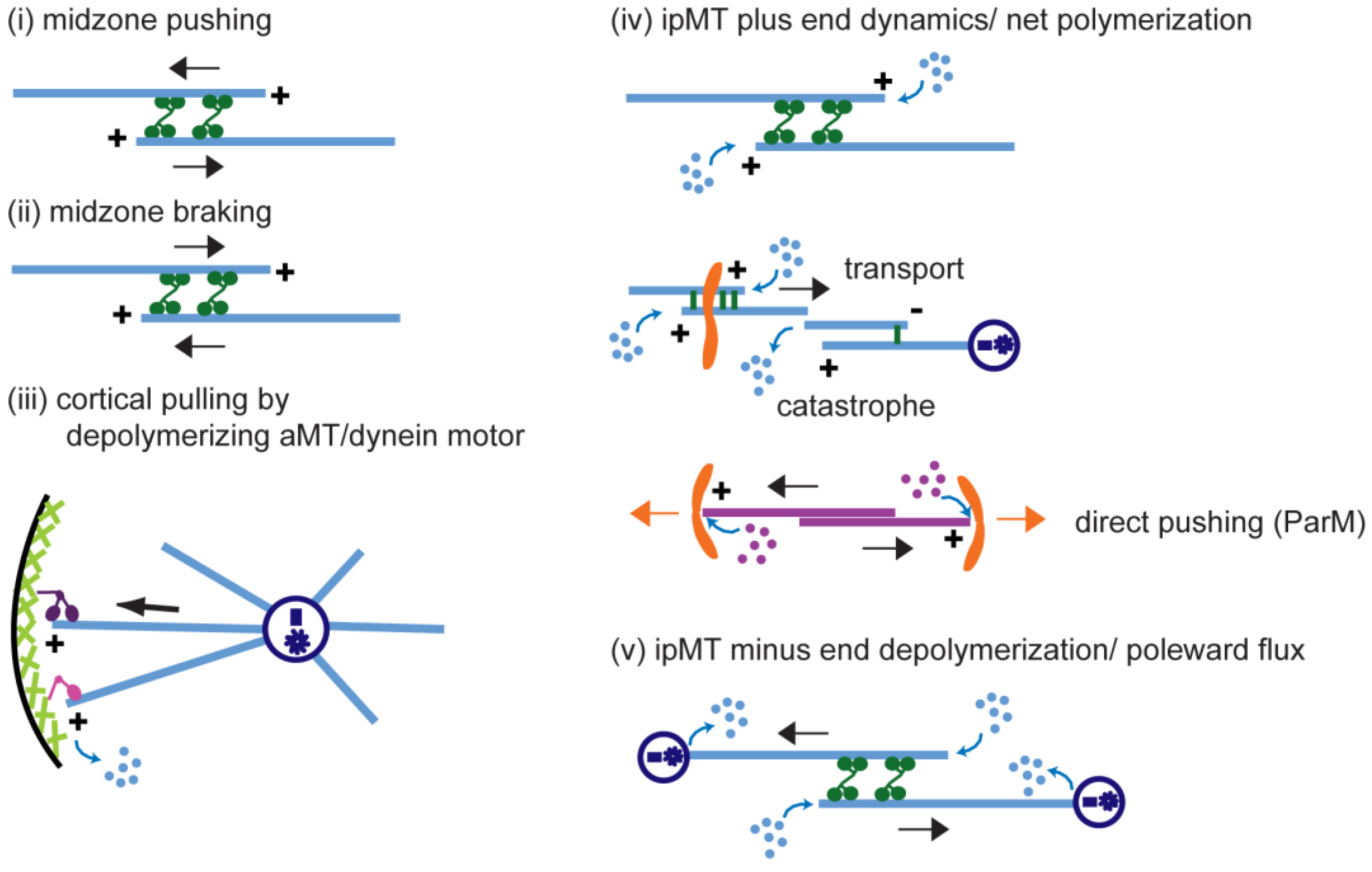
5. Conserved Biochemical Modules Involved in Anaphase B
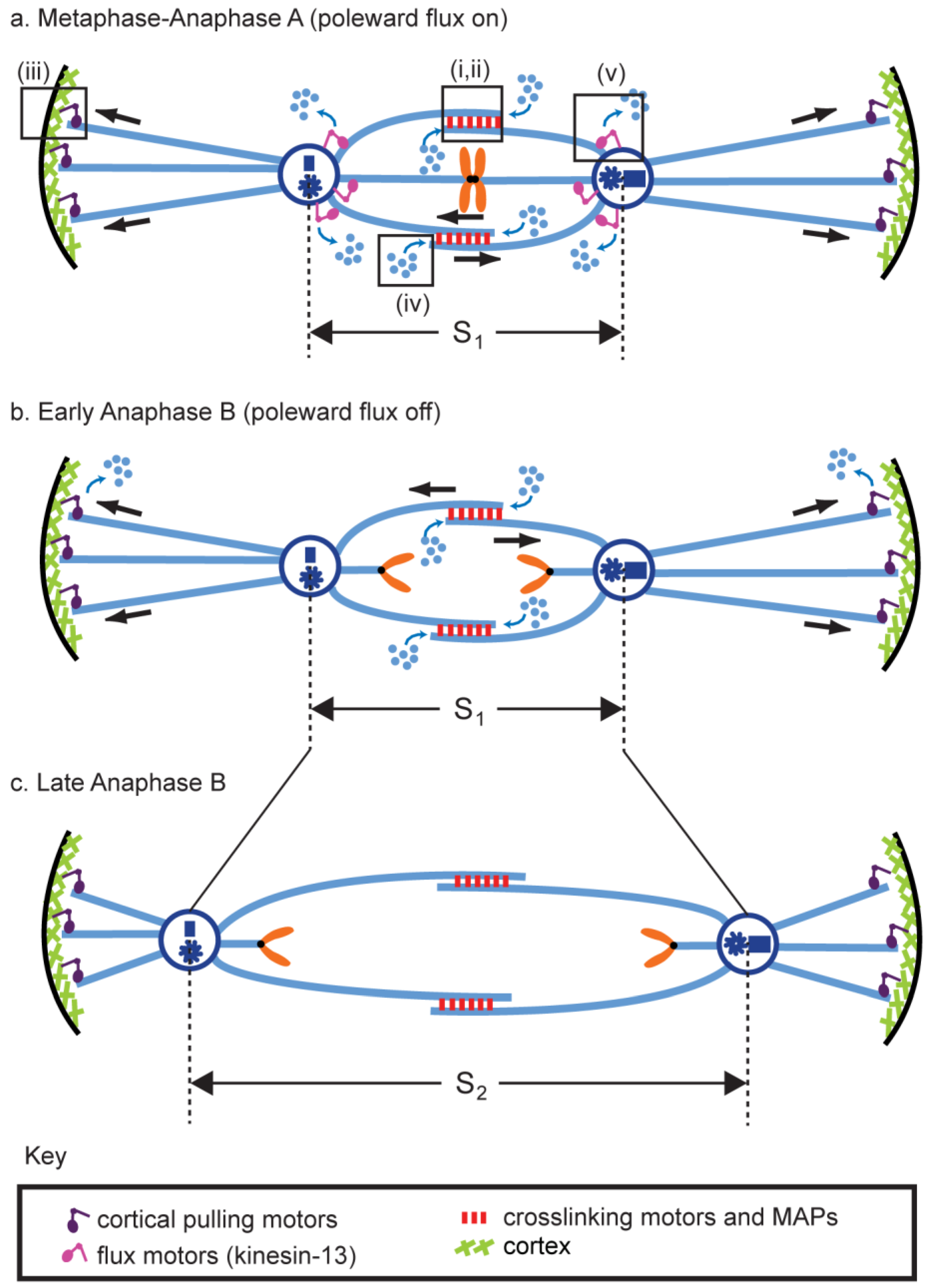
5.1. (Module i) Midzone Pushing: Pole–Pole Separation by Outward Sliding of Antiparallel ipMTs
5.2. (Module ii) Midzone Braking
5.3. (Module iii) Cortical Pulling Apart of the Anaphase B Spindle Poles
5.4. (Module iv) ipMT Plus End Dynamics and Net Polymerization
5.5. (Module v) ipMT Minus End Depolymerization: Poleward Flux as a Regulatory Switch for Anaphase B
5.6. Combination of Modules and the Force Balance Concept
6. Properties and Functions of the Molecular Nuts and Bolts of the Anaphase B Machinery
6.1. Molecules of the Central Spindle
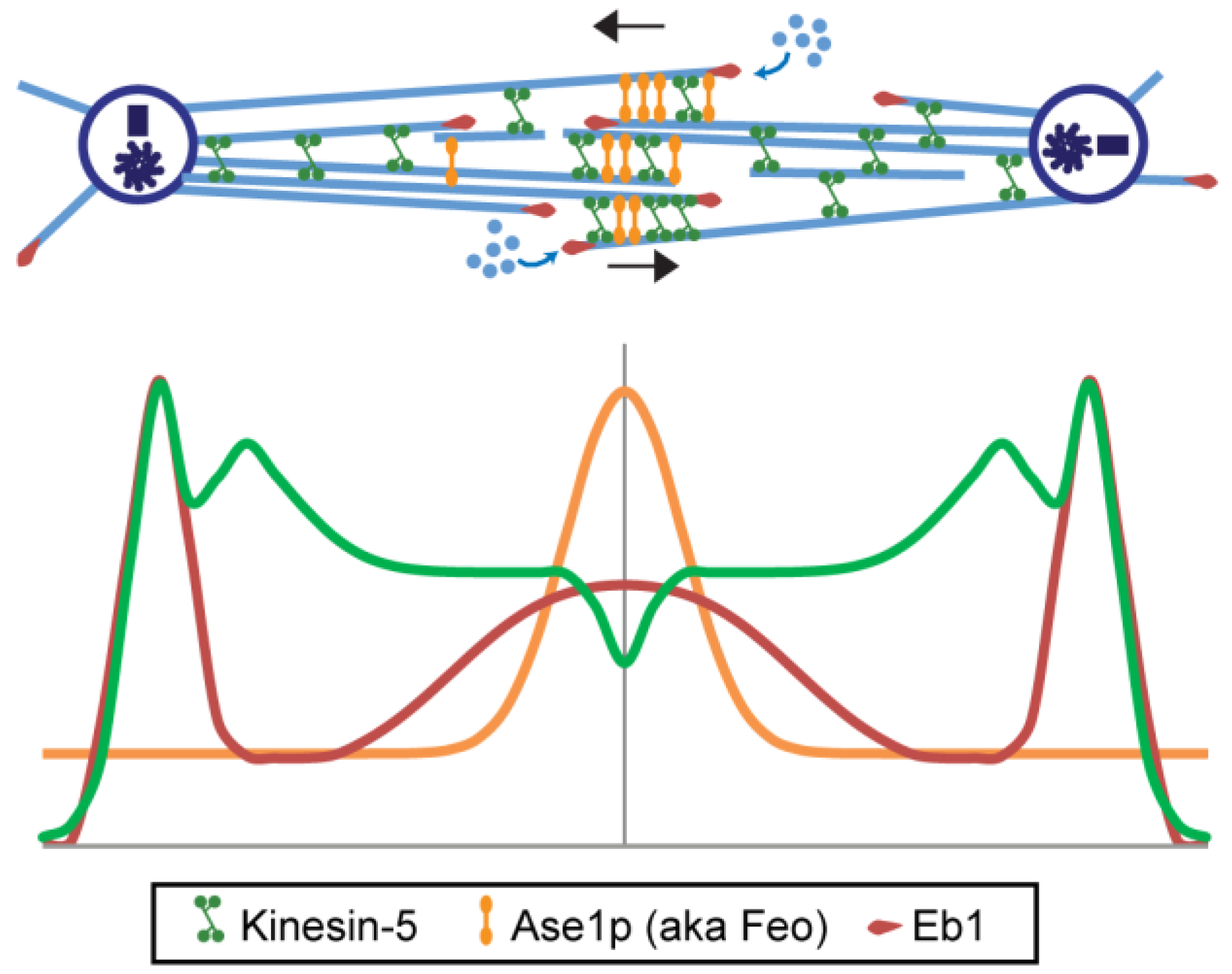
6.1.1. Antiparallel ipMT-Crosslinking MAPs of the Ase1p Family
6.1.2. MT Crosslinking and Sliding Motors; Kinesin-5 Plus Kinesins-4, -6, -8, and -12
6.1.3. Molecules Controlling MT Plus end Dynamics
6.1.4. Molecules Controlling ipMT Minus End Dynamics
6.1.5. Chromosomal Proteins Required for Anaphase B Spindle Elongation
6.2. Molecules of the Cortical Pulling Machinery
6.2.1. Attachment of MT Plus Ends to the Cortex
6.2.2. Cortical Force Generators
6.3. Molecules Involved in Prokaryotic Anaphase B
7. Cell Cycle Control of Anaphase B
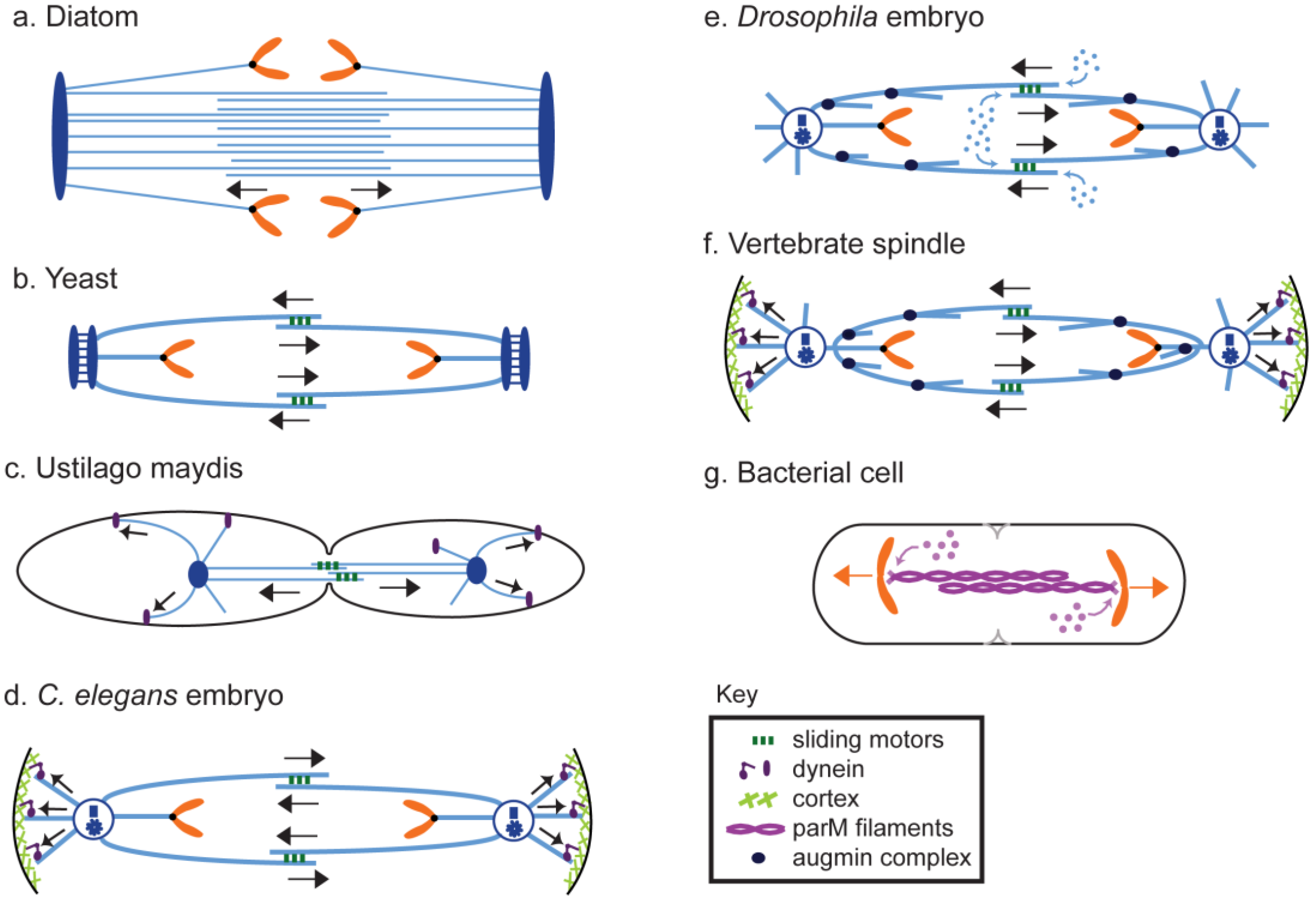
8. Anaphase B in Model Systems
8.1. Diatoms
8.2. Fungi
8.2.1. Yeast
8.2.2. Filamentous and Smut Fungi
8.3. Plants
8.4. Animals
8.4.1. Caenorhabditis Elegans
8.4.2. Drosophila
8.4.3. Vertebrates
8.5. Prokaryotes
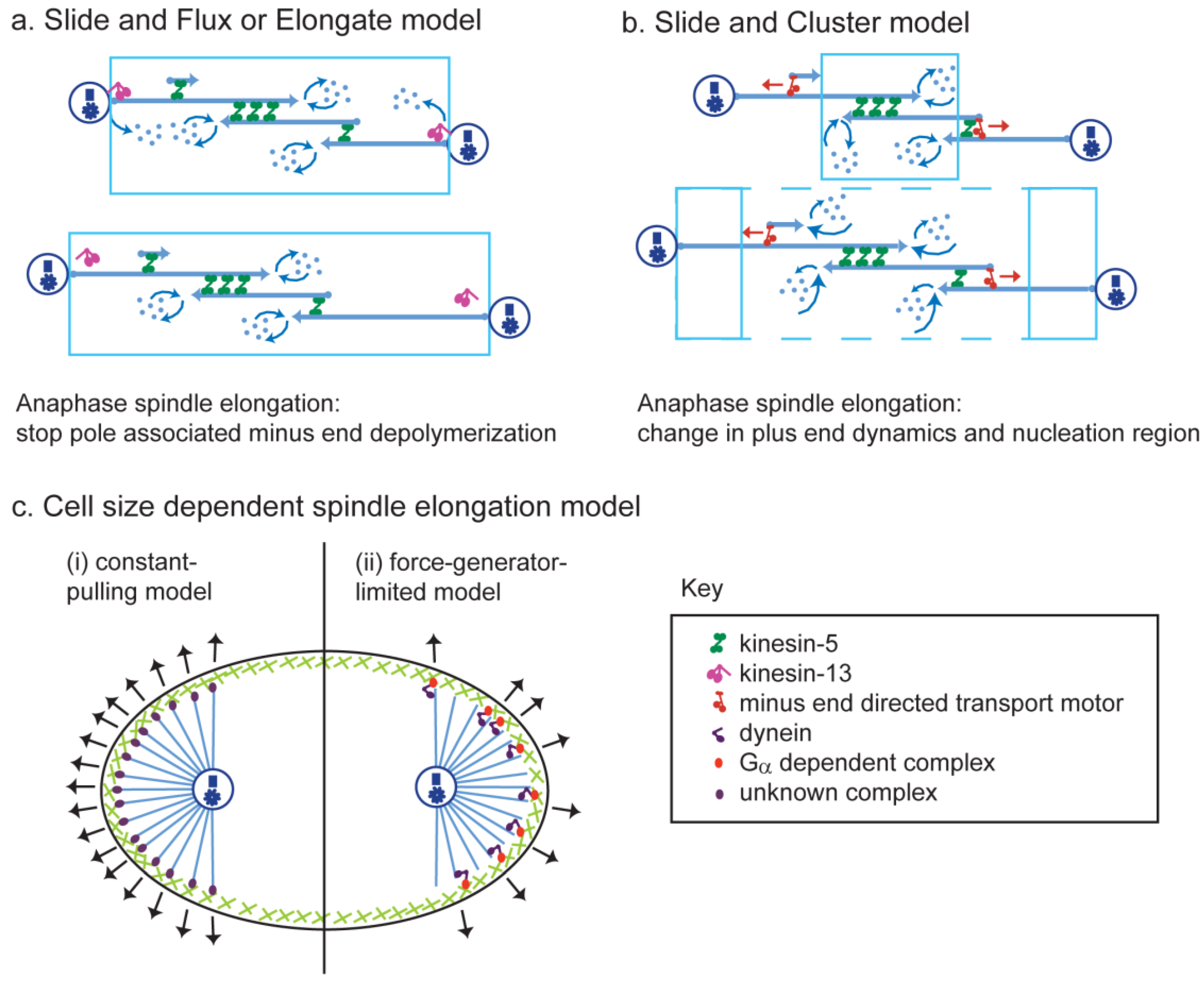
9. Theoretical Models of Anaphase B
10. Concluding Remarks
Acknowledgments
Author Contributions
Conflicts of Interest
References
- McIntosh, J.R.; Molodtsov, M.I.; Ataullakhanov, F.I. Biophysics of mitosis. Q. Rev. Biophys. 2012, 45, 147–207. [Google Scholar] [CrossRef] [PubMed]
- Walczak, C.E.; Cai, S.; Khodjakov, A. Mechanisms of chromosome behaviour during mitosis. Nat. Rev. Mol. Cell Biol. 2010, 11, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Goshima, G.; Scholey, J.M. Control of mitotic spindle length. Annu. Rev. Cell Dev. Biol. 2010, 26, 21–57. [Google Scholar] [CrossRef] [PubMed]
- Maiato, H.; Lince-Faria, M. The perpetual movements of anaphase. Cell Mol. Life Sci. 2010, 67, 2251–2269. [Google Scholar] [CrossRef] [PubMed]
- Gadde, S.; Heald, R. Mechanisms and molecules of the mitotic spindle. Curr. Biol. 2004, 14, R797–R805. [Google Scholar] [CrossRef] [PubMed]
- Cross, R.A.; McAinsh, A. Prime movers: The mechanochemistry of mitotic kinesins. Nat. Rev. Mol. Cell Biol. 2014, 15, 257–271. [Google Scholar] [CrossRef] [PubMed]
- Petry, S. Mechanisms of mitotic spindle assembly. Annu. Rev. Biochem. 2016, 85, 659–683. [Google Scholar] [CrossRef] [PubMed]
- Bouck, D.C.; Joglekar, A.P.; Bloom, K.S. Design features of a mitotic spindle: Balancing tension and compression at a single microtubule kinetochore interface in budding yeast. Annu. Rev. Genet. 2008, 42, 335–359. [Google Scholar] [CrossRef] [PubMed]
- Maddox, P.S.; Oegema, K.; Desai, A.; Cheeseman, I.M. “Holo”er than thou: Chromosome segregation and kinetochore function in C. elegans. Chromosome Res. 2004, 12, 641–653. [Google Scholar] [CrossRef] [PubMed]
- Gayathri, P.; Fujii, T.; Moller-Jensen, J.; van den Ent, F.; Namba, K.; Lowe, J. A bipolar spindle of antiparallel ParM filaments drives bacterial plasmid segregation. Science 2012, 338, 1334–1337. [Google Scholar] [CrossRef] [PubMed]
- Cimini, D.; Cameron, L.A.; Salmon, E.D. Anaphase spindle mechanics prevent mis-segregation of merotelically oriented chromosomes. Curr. Biol. 2004, 14, 2149–2155. [Google Scholar] [CrossRef] [PubMed]
- Courtheoux, T.; Gay, G.; Gachet, Y.; Tournier, S. Ase1/PRC1-dependent spindle elongation corrects merotely during anaphase in fission yeast. J. Cell Biol. 2009, 187, 399–412. [Google Scholar] [CrossRef] [PubMed]
- Pidoux, A.L.; Uzawa, S.; Perry, P.E.; Cande, W.Z.; Allshire, R.C. Live analysis of lagging chromosomes during anaphase and their effect on spindle elongation rate in fission yeast. J. Cell Sci. 2000, 113 Pt 23, 4177–4191. [Google Scholar] [PubMed]
- Ford, J.H. Protraction of anaphase B in lymphocyte mitosis with ageing: Possible contribution to age-related cancer risk. Mutagenesis 2013, 28, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Brust-Mascher, I.; Scholey, J.M. Mitotic motors and chromosome segregation: The mechanism of anaphase B. Biochem. Soc. Trans. 2011, 39, 1149–1153. [Google Scholar] [CrossRef] [PubMed]
- Cande, W.Z.; Hogan, C.J. The mechanism of anaphase spindle elongation. Bioessays 1989, 11, 5–9. [Google Scholar] [CrossRef] [PubMed]
- Roostalu, J.; Schiebel, E.; Khmelinskii, A. Cell cycle control of spindle elongation. Cell Cycle 2010, 9, 1084–1090. [Google Scholar] [CrossRef] [PubMed]
- Ris, H. A quantitative study of anaphase movement in the aphid tamalia. Biol. Bull. 1943, 85, 164–178. [Google Scholar] [CrossRef]
- Ris, H. The anaphase movement of chromosomes in the spermatocytes of the grasshopper. Biol. Bull. 1949, 96, 90–106. [Google Scholar] [CrossRef] [PubMed]
- Rapaport, R. Cytokinesis in Animal Cells; Cambridge University Press: Cambridge, UK, 1996; p. 386. [Google Scholar]
- Schrader, F. Mitosis. The Movement of Chromosomes during Cell Division, 3rd ed.; Columbia University Press: New York, NY, USA, 1949; p. 110. [Google Scholar]
- McIntosh, J.R.; Hepler, P.K.; van Wie, D.G. Model for mitosis. Nature 1969, 224, 659–663. [Google Scholar] [CrossRef]
- Huxley, H.; Hanson, J. Changes in the cross-striations of muscle during contraction and stretch and their structural interpretation. Nature 1954, 173, 973–976. [Google Scholar] [CrossRef] [PubMed]
- Euteneuer, U.; Jackson, W.T.; McIntosh, J.R. Polarity of spindle microtubules in haemanthus endosperm. J. Cell Biol. 1982, 94, 644–653. [Google Scholar] [CrossRef] [PubMed]
- Hepler, P.K.; McIntosh, J.R.; Cleland, S. Intermicrotubule bridges in mitotic spindle apparatus. J. Cell Biol. 1970, 45, 438–444. [Google Scholar] [CrossRef] [PubMed]
- McDonald, K.; Pickett-Heaps, J.D.; McIntosh, J.R.; Tippit, D.H. On the mechanism of anaphase spindle elongation in diatoma vulgare. J. Cell Biol. 1977, 74, 377–388. [Google Scholar] [CrossRef] [PubMed]
- McIntosh, J.R.; Landis, S.C. The distribution of spindle microtubules during mitosis in cultured human cells. J. Cell Biol. 1971, 49, 468–497. [Google Scholar] [CrossRef] [PubMed]
- Kashina, A.S.; Baskin, R.J.; Cole, D.G.; Wedaman, K.P.; Saxton, W.M.; Scholey, J.M. A bipolar kinesin. Nature 1996, 379, 270–272. [Google Scholar] [CrossRef] [PubMed]
- Scholey, J.M.; Porter, M.E.; Grissom, P.M.; McIntosh, J.R. Identification of kinesin in sea urchin eggs, and evidence for its localization in the mitotic spindle. Nature 1985, 318, 483–486. [Google Scholar] [CrossRef] [PubMed]
- Kapitein, L.C.; Peterman, E.J.; Kwok, B.H.; Kim, J.H.; Kapoor, T.M.; Schmidt, C.F. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature 2005, 435, 114–118. [Google Scholar] [CrossRef] [PubMed]
- Cole, D.G.; Saxton, W.M.; Sheehan, K.B.; Scholey, J.M. A “slow” homotetrameric kinesin-related motor protein purified from Drosophila embryos. J. Biol. Chem. 1994, 269, 22913–22916. [Google Scholar] [PubMed]
- Winey, M.; Mamay, C.L.; O’Toole, E.T.; Mastronarde, D.N.; Giddings, T.H., Jr.; McDonald, K.L.; McIntosh, J.R. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J. Cell Biol. 1995, 129, 1601–1615. [Google Scholar] [CrossRef] [PubMed]
- Saxton, W.M.; McIntosh, J.R. Interzone microtubule behavior in late anaphase and telophase spindles. J. Cell Biol. 1987, 105, 875–886. [Google Scholar] [CrossRef] [PubMed]
- Cande, W.Z.; McDonald, K. Physiological and ultrastructural analysis of elongating mitotic spindles reactivated in vitro. J. Cell Biol. 1986, 103, 593–604. [Google Scholar] [CrossRef] [PubMed]
- Cande, W.Z.; McDonald, K.L. In vitro reactivation of anaphase spindle elongation using isolated diatom spindles. Nature 1985, 316, 168–170. [Google Scholar] [CrossRef] [PubMed]
- Hogan, C.J.; Wein, H.; Wordeman, L.; Scholey, J.M.; Sawin, K.E.; Cande, W.Z. Inhibition of anaphase spindle elongation in vitro by a peptide antibody that recognizes kinesin motor domain. Proc. Natl. Acad. Sci. USA 1993, 90, 6611–6615. [Google Scholar] [CrossRef] [PubMed]
- Sharp, D.J.; McDonald, K.L.; Brown, H.M.; Matthies, H.J.; Walczak, C.; Vale, R.D.; Mitchison, T.J.; Scholey, J.M. The bipolar kinesin, KLP61F, cross-links microtubules within interpolar microtubule bundles of Drosophila embryonic mitotic spindles. J. Cell Biol. 1999, 144, 125–138. [Google Scholar] [CrossRef] [PubMed]
- Aist, J.R.; Bayles, C.J.; Tao, W.; Berns, M.W. Direct experimental evidence for the existence, structural basis and function of astral forces during anaphase B in vivo. J. Cell Sci. 1991, 100 Pt 2, 279–288. [Google Scholar] [PubMed]
- Fink, G.; Schuchardt, I.; Colombelli, J.; Stelzer, E.; Steinberg, G. Dynein-mediated pulling forces drive rapid mitotic spindle elongation in ustilago maydis. EMBO J. 2006, 25, 4897–4908. [Google Scholar] [CrossRef] [PubMed]
- Grill, S.W.; Gonczy, P.; Stelzer, E.H.; Hyman, A.A. Polarity controls forces governing asymmetric spindle positioning in the Caenorhabditis elegans embryo. Nature 2001, 409, 630–633. [Google Scholar] [CrossRef] [PubMed]
- Waters, J.C.; Cole, R.W.; Rieder, C.L. The force-producing mechanism for centrosome separation during spindle formation in vertebrates is intrinsic to each aster. J. Cell Biol. 1993, 122, 361–372. [Google Scholar] [CrossRef] [PubMed]
- FitzHarris, G. Anaphase B precedes anaphase A in the mouse egg. Curr. Biol. 2012, 22, 437–444. [Google Scholar] [CrossRef] [PubMed]
- Yanagida, M. The role of model organisms in the history of mitosis research. Cold Spring Harb. Perspect. Biol. 2014, 6, a015768. [Google Scholar] [CrossRef] [PubMed]
- Baskin, T.I.; Cande, W.Z. Kinetic analysis of mitotic spindle elongation in vitro. J. Cell Sci. 1990, 97 Pt 1, 79–89. [Google Scholar] [PubMed]
- Cohn, S.A.; Pickett-Heaps, J.D. The effects of colchicine and dinitrophenol on the in vivo rates of anaphase A and B in the diatom surirella. Eur. J. Cell Biol. 1988, 46, 523–530. [Google Scholar] [PubMed]
- Brust-Mascher, I.; Scholey, J.M. Mitotic spindle dynamics in Drosophila. Int. Rev. Cytol. 2007, 259, 139–172. [Google Scholar] [PubMed]
- Yeh, E.; Skibbens, R.V.; Cheng, J.W.; Salmon, E.D.; Bloom, K. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J. Cell Biol. 1995, 130, 687–700. [Google Scholar] [CrossRef] [PubMed]
- Desai, A.; Mitchison, T.J. Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 1997, 13, 83–117. [Google Scholar] [CrossRef] [PubMed]
- Inoue, S.; Salmon, E.D. Force generation by microtubule assembly/disassembly in mitosis and related movements. Mol. Biol. Cell 1995, 6, 1619–1640. [Google Scholar] [CrossRef] [PubMed]
- Rogers, G.C.; Rogers, S.L.; Sharp, D.J. Spindle microtubules in flux. J. Cell Sci. 2005, 118, 1105–1116. [Google Scholar] [CrossRef] [PubMed]
- Salmon, E.D.; Leslie, R.J.; Saxton, W.M.; Karow, M.L.; McIntosh, J.R. Spindle microtubule dynamics in sea urchin embryos: Analysis using a fluorescein-labeled tubulin and measurements of fluorescence redistribution after laser photobleaching. J. Cell Biol. 1984, 99, 2165–2174. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Y.; Kronebusch, P.J.; Simon, P.M.; Borisy, G.G. Microtubule dynamics at the g2/m transition: Abrupt breakdown of cytoplasmic microtubules at nuclear envelope breakdown and implications for spindle morphogenesis. J. Cell Biol. 1996, 135, 201–214. [Google Scholar] [CrossRef] [PubMed]
- Mitchison, T.J. Polewards microtubule flux in the mitotic spindle: Evidence from photoactivation of fluorescence. J. Cell Biol. 1989, 109, 637–652. [Google Scholar] [CrossRef] [PubMed]
- Mitchison, T.J.; Salmon, E.D. Poleward kinetochore fiber movement occurs during both metaphase and anaphase-A in newt lung cell mitosis. J. Cell Biol. 1992, 119, 569–582. [Google Scholar] [CrossRef] [PubMed]
- Waterman-Storer, C.M.; Salmon, E.D. Fluorescent speckle microscopy of microtubules: How low can you go? FASEB J. 1999, 13, S225–S230. [Google Scholar] [PubMed]
- Higuchi, T.; Uhlmann, F. Stabilization of microtubule dynamics at anaphase onset promotes chromosome segregation. Nature 2005, 433, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Maddox, P.S.; Bloom, K.S.; Salmon, E.D. The polarity and dynamics of microtubule assembly in the budding yeast Saccharomyces cerevisiae. Nat. Cell Biol. 2000, 2, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Mallavarapu, A.; Sawin, K.; Mitchison, T. A switch in microtubule dynamics at the onset of anaphase B in the mitotic spindle of Schizosaccharomyces pombe. Curr. Biol. 1999, 9, 1423–1426. [Google Scholar] [CrossRef]
- Zhai, Y.; Kronebusch, P.J.; Borisy, G.G. Kinetochore microtubule dynamics and the metaphase-anaphase transition. J. Cell Biol. 1995, 131, 721–734. [Google Scholar] [CrossRef] [PubMed]
- Brust-Mascher, I.; Civelekoglu-Scholey, G.; Kwon, M.; Mogilner, A.; Scholey, J.M. Model for anaphase B: Role of three mitotic motors in a switch from poleward flux to spindle elongation. Proc. Natl. Acad. Sci. USA 2004, 101, 15938–15943. [Google Scholar] [CrossRef] [PubMed]
- Brust-Mascher, I.; Scholey, J.M. Microtubule flux and sliding in mitotic spindles of Drosophila embryos. Mol. Biol. Cell 2002, 13, 3967–3975. [Google Scholar] [CrossRef] [PubMed]
- Cheerambathur, D.K.; Civelekoglu-Scholey, G.; Brust-Mascher, I.; Sommi, P.; Mogilner, A.; Scholey, J.M. Quantitative analysis of an anaphase B switch: Predicted role for a microtubule catastrophe gradient. J. Cell Biol. 2007, 177, 995–1004. [Google Scholar] [CrossRef] [PubMed]
- De Lartigue, J.; Brust-Mascher, I.; Scholey, J.M. Anaphase B spindle dynamics in Drosophila s2 cells: Comparison with embryo spindles. Cell Div. 2011. [Google Scholar] [CrossRef] [PubMed]
- Saunders, A.M.; Powers, J.; Strome, S.; Saxton, W.M. Kinesin-5 acts as a brake in anaphase spindle elongation. Curr. Biol. 2007, 17, R453–R454. [Google Scholar] [CrossRef] [PubMed]
- Collins, E.; Mann, B.J.; Wadsworth, P. Eg5 restricts anaphase B spindle elongation in mammalian cells. Cytoskeleton (Hoboken) 2014, 71, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Aist, J.R.; Liang, H.; Berns, M.W. Astral and spindle forces in PTK2 cells during anaphase B: A laser microbeam study. J. Cell Sci. 1993, 104 Pt 4, 1207–1216. [Google Scholar] [PubMed]
- Leslie, R.J.; Pickett-Heaps, J.D. Ultraviolet microbeam irradiations of mitotic diatoms: Investigation of spindle elongation. J. Cell Biol. 1983, 96, 548–561. [Google Scholar] [CrossRef] [PubMed]
- Khodjakov, A.; La Terra, S.; Chang, F. Laser microsurgery in fission yeast; role of the mitotic spindle midzone in anaphase B. Curr. Biol. 2004, 14, 1330–1340. [Google Scholar] [CrossRef] [PubMed]
- Verbrugghe, K.J.; White, J.G. SPD-1 is required for the formation of the spindle midzone but is not essential for the completion of cytokinesis in C. elegans embryos. Curr. Biol. 2004, 14, 1755–1760. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.Y.; Esmaeili, B.; Zealley, B.; Mishima, M. Direct interaction between centralspindlin and PRC1 reinforces mechanical resilience of the central spindle. Nat. Commun. 2015. [Google Scholar] [CrossRef] [PubMed]
- Tolic-Norrelykke, I.M.; Sacconi, L.; Thon, G.; Pavone, F.S. Positioning and elongation of the fission yeast spindle by microtubule-based pushing. Curr. Biol. 2004, 14, 1181–1186. [Google Scholar] [CrossRef] [PubMed]
- Nicklas, R.B. Chromosome velocity during mitosis as a function of chromosome size and position. J. Cell Biol. 1965, 25, 119–135. [Google Scholar] [CrossRef]
- Hiramoto, Y. Mechanical properties of the protoplasm of the sea urchin egg. II. Fertilized egg. Exp. Cell Res. 1969, 56, 209–218. [Google Scholar] [CrossRef]
- Scholey, J.M. Compare and contrast the reaction coordinate diagrams for chemical reactions and cytoskeletal force generators. Mol. Biol. Cell 2013, 24, 433–439. [Google Scholar] [CrossRef] [PubMed]
- Nicklas, R.B. Measurements of the force produced by the mitotic spindle in anaphase. J. Cell Biol. 1983, 97, 542–548. [Google Scholar] [CrossRef] [PubMed]
- Hiramoto, Y.; Nakano, Y. Micromanipulation studies of the mitotic apparatus in sand dollar eggs. Cell Motil. Cytoskelet. 1988, 10, 172–184. [Google Scholar] [CrossRef] [PubMed]
- Brust-Mascher, I.; Sommi, P.; Cheerambathur, D.K.; Scholey, J.M. Kinesin-5-dependent poleward flux and spindle length control in Drosophila embryo mitosis. Mol. Biol. Cell 2009, 20, 1749–1762. [Google Scholar] [CrossRef] [PubMed]
- Nicklas, R.B. A quantitative comparison of cellular motile systems. Cell Motil. 1984, 4, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Cande, W.Z. Nucleotide requirements for anaphase chromosome movements in permeabilized mitotic cells: Anaphase B but not anaphase A requires ATP. Cell 1982, 28, 15–22. [Google Scholar] [CrossRef]
- Cande, W.Z. Creatine kinase role in anaphase chromosome movement. Nature 1983, 304, 557–558. [Google Scholar] [CrossRef] [PubMed]
- Koons, S.J.; Eckert, B.S.; Zobel, C.R. Immunofluorescence and inhibitor studies on creatine kinase and mitosis. Exp. Cell Res. 1982, 140, 401–409. [Google Scholar] [CrossRef]
- Palazzo, R.E.; Lutz, D.A.; Rebhun, L.I. Reactivation of isolated mitotic apparatus: Metaphase versus anaphase spindles. Cell Motil. Cytoskelet. 1991, 18, 304–318. [Google Scholar] [CrossRef] [PubMed]
- Civelekoglu-Scholey, G.; Scholey, J.M. Mitotic force generators and chromosome segregation. Cell Mol. Life Sci. 2010, 67, 2231–2250. [Google Scholar] [CrossRef] [PubMed]
- Janson, M.E.; Loughlin, R.; Loiodice, I.; Fu, C.; Brunner, D.; Nedelec, F.J.; Tran, P.T. Crosslinkers and motors organize dynamic microtubules to form stable bipolar arrays in fission yeast. Cell 2007, 128, 357–368. [Google Scholar] [CrossRef] [PubMed]
- Lansky, Z.; Braun, M.; Ludecke, A.; Schlierf, M.; ten Wolde, P.R.; Janson, M.E.; Diez, S. Diffusible crosslinkers generate directed forces in microtubule networks. Cell 2015, 160, 1159–1168. [Google Scholar] [CrossRef] [PubMed]
- Heidemann, S.R.; McIntosh, J.R. Visualization of the structural polarity of microtubules. Nature 1980, 286, 517–519. [Google Scholar] [CrossRef] [PubMed]
- Ding, R.; McDonald, K.L.; McIntosh, J.R. Three-dimensional reconstruction and analysis of mitotic spindles from the yeast, Schizosaccharomyces pombe. J. Cell Biol. 1993, 120, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Ward, J.J.; Roque, H.; Antony, C.; Nedelec, F. Mechanical design principles of a mitotic spindle. Elife 2014, 3, e03398. [Google Scholar] [CrossRef] [PubMed]
- Huxley, H.E. Electron microscope studies on the structure of natural and synthetic protein filaments from striated muscle. J. Mol. Biol. 1963, 7, 281–308. [Google Scholar] [CrossRef]
- Mastronarde, D.N.; McDonald, K.L.; Ding, R.; McIntosh, J.R. Interpolar spindle microtubules in PTK cells. J. Cell Biol. 1993, 123, 1475–1489. [Google Scholar] [CrossRef] [PubMed]
- Wilson, H.J. Arms and bridges on microtubules in the mitotic apparatus. J. Cell Biol. 1969, 40, 854–859. [Google Scholar] [CrossRef] [PubMed]
- Cheerambathur, D.K.; Brust-Mascher, I.; Civelekoglu-Scholey, G.; Scholey, J.M. Dynamic partitioning of mitotic kinesin-5 cross-linkers between microtubule-bound and freely diffusing states. J. Cell Biol. 2008, 182, 429–436. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Brust-Mascher, I.; Scholey, J.M. The microtubule cross-linker Feo controls the midzone stability, motor composition, and elongation of the anaphase B spindle in Drosophila embryos. Mol. Biol. Cell 2015, 26, 1452–1462. [Google Scholar] [CrossRef] [PubMed]
- Acar, S.; Carlson, D.B.; Budamagunta, M.S.; Yarov-Yarovoy, V.; Correia, J.J.; Ninonuevo, M.R.; Jia, W.; Tao, L.; Leary, J.A.; Voss, J.C.; et al. The bipolar assembly domain of the mitotic motor kinesin-5. Nat. Commun. 2013. [Google Scholar] [CrossRef] [PubMed]
- Glotzer, M. The 3ms of central spindle assembly: Microtubules, motors and maps. Nat. Rev. Mol. Cell Biol. 2009, 10, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Peterman, E.J.; Scholey, J.M. Mitotic microtubule crosslinkers: Insights from mechanistic studies. Curr. Biol. 2009, 19, R1089–R1094. [Google Scholar] [CrossRef] [PubMed]
- Rozelle, D.K.; Hansen, S.D.; Kaplan, K.B. Chromosome passenger complexes control anaphase duration and spindle elongation via a kinesin-5 brake. J. Cell Biol. 2011, 193, 285–294. [Google Scholar] [CrossRef] [PubMed]
- Shimamoto, Y.; Forth, S.; Kapoor, T.M. Measuring pushing and braking forces generated by ensembles of kinesin-5 crosslinking two microtubules. Dev. Cell 2015, 34, 669–681. [Google Scholar] [CrossRef] [PubMed]
- Tikhonenko, I.; Nag, D.K.; Martin, N.; Koonce, M.P. Kinesin-5 is not essential for mitotic spindle elongation in Dictyostelium. Cell Motil. Cytoskelet. 2008, 65, 853–862. [Google Scholar] [CrossRef] [PubMed]
- Grill, S.W.; Howard, J.; Schaffer, E.; Stelzer, E.H.; Hyman, A.A. The distribution of active force generators controls mitotic spindle position. Science 2003, 301, 518–521. [Google Scholar] [CrossRef] [PubMed]
- Grill, S.W.; Hyman, A.A. Spindle positioning by cortical pulling forces. Dev. Cell 2005, 8, 461–465. [Google Scholar] [CrossRef] [PubMed]
- Grishchuk, E.L.; Molodtsov, M.I.; Ataullakhanov, F.I.; McIntosh, J.R. Force production by disassembling microtubules. Nature 2005, 438, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Kozlowski, C.; Srayko, M.; Nedelec, F. Cortical microtubule contacts position the spindle in C. elegans embryos. Cell 2007, 129, 499–510. [Google Scholar] [CrossRef] [PubMed]
- Telley, I.A.; Gaspar, I.; Ephrussi, A.; Surrey, T. Aster migration determines the length scale of nuclear separation in the Drosophila syncytial embryo. J. Cell Biol. 2012, 197, 887–895. [Google Scholar] [CrossRef] [PubMed]
- Masuda, H.; Cande, W.Z. The role of tubulin polymerization during spindle elongation in vitro. Cell 1987, 49, 193–202. [Google Scholar] [CrossRef]
- Masuda, H.; McDonald, K.L.; Cande, W.Z. The mechanism of anaphase spindle elongation: Uncoupling of tubulin incorporation and microtubule sliding during in vitro spindle reactivation. J. Cell Biol. 1988, 107, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Uehara, R.; Goshima, G. Functional central spindle assembly requires de novo microtubule generation in the interchromosomal region during anaphase. J. Cell Biol. 2010, 191, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Nahaboo, W.; Zouak, M.; Askjaer, P.; Delattre, M. Chromatids segregate without centrosomes during Caenorhabditis elegans mitosis in a ran- and clasp-dependent manner. Mol. Biol. Cell 2015, 26, 2020–2029. [Google Scholar] [CrossRef] [PubMed]
- Dumont, J.; Oegema, K.; Desai, A. A kinetochore-independent mechanism drives anaphase chromosome separation during acentrosomal meiosis. Nat. Cell Biol. 2010, 12, 894–901. [Google Scholar] [CrossRef] [PubMed]
- Burbank, K.S.; Mitchison, T.J.; Fisher, D.S. Slide-and-cluster models for spindle assembly. Curr. Biol. 2007, 17, 1373–1383. [Google Scholar] [CrossRef] [PubMed]
- Hays, T.S.; Wise, D.; Salmon, E.D. Traction force on a kinetochore at metaphase acts as a linear function of kinetochore fiber length. J. Cell Biol. 1982, 93, 374–389. [Google Scholar] [CrossRef] [PubMed]
- Ostergren, G. Considerations on some elementary features of mitosis. Hereditas 1950, 36, 1–19. [Google Scholar] [CrossRef]
- Wang, H.; Brust-Mascher, I.; Civelekoglu-Scholey, G.; Scholey, J.M. Patronin mediates a switch from kinesin-13-dependent poleward flux to anaphase B spindle elongation. J. Cell Biol. 2013, 203, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Akhmanova, A.; Steinmetz, M.O. Control of microtubule organization and dynamics: Two ends in the limelight. Nat. Rev. Mol. Cell Biol. 2015, 16, 711–726. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, L.S. Functional redundancy in mitotic force generation. J. Cell Biol. 1993, 120, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Pellman, D.; Bagget, M.; Tu, Y.H.; Fink, G.R.; Tu, H. Two microtubule-associated proteins required for anaphase spindle movement in saccharomyces cerevisiae. J. Cell Biol. 1995, 130, 1373–1385. [Google Scholar] [CrossRef] [PubMed]
- Schuyler, S.C.; Liu, J.Y.; Pellman, D. The molecular function of Ase1p: Evidence for a map-dependent midzone-specific spindle matrix. Microtubule-associated proteins. J. Cell Biol. 2003, 160, 517–528. [Google Scholar] [CrossRef] [PubMed]
- Kapitein, L.C.; Janson, M.E.; van den Wildenberg, S.M.; Hoogenraad, C.C.; Schmidt, C.F.; Peterman, E.J. Microtubule-driven multimerization recruits Ase1p onto overlapping microtubules. Curr. Biol. 2008, 18, 1713–1717. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, R.; Ti, S.C.; Tan, L.; Darst, S.A.; Kapoor, T.M. Marking and measuring single microtubules by PRC1 and kinesin-4. Cell 2013, 154, 377–390. [Google Scholar] [CrossRef] [PubMed]
- Khmelinskii, A.; Roostalu, J.; Roque, H.; Antony, C.; Schiebel, E. Phosphorylation-dependent protein interactions at the spindle midzone mediate cell cycle regulation of spindle elongation. Dev. Cell 2009, 17, 244–256. [Google Scholar] [CrossRef] [PubMed]
- Wein, H.; Bass, H.W.; Cande, W.Z. Dsk1, a kinesin-related protein involved in anaphase spindle elongation, is a component of a mitotic spindle matrix. Cell Motil. Cytoskelet. 1998, 41, 214–224. [Google Scholar] [CrossRef]
- Odde, D.J. Mitosis, diffusible crosslinkers, and the ideal gas law. Cell 2015, 160, 1041–1043. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Wang, S.; Huang, Y.; He, X.; Cui, H.; Zhu, X.; Zheng, Y. Phase transition of spindle-associated protein regulate spindle apparatus assembly. Cell 2015, 163, 108–122. [Google Scholar] [CrossRef] [PubMed]
- Civelekoglu-Scholey, G.; Tao, L.; Brust-Mascher, I.; Wollman, R.; Scholey, J.M. Prometaphase spindle maintenance by an antagonistic motor-dependent force balance made robust by a disassembling lamin-b envelope. J. Cell Biol. 2010, 188, 49–68. [Google Scholar] [CrossRef] [PubMed]
- Scholey, J.E.; Nithianantham, S.; Scholey, J.M.; Al-Bassam, J. Structural basis for the assembly of the mitotic motor kinesin-5 into bipolar tetramers. Elife 2014, 3, e02217. [Google Scholar] [CrossRef] [PubMed]
- Sawin, K.E.; Le Guellec, K.; Philippe, M.; Mitchison, T.J. Mitotic spindle organization by a plus-end-directed microtubule motor. Nature 1992, 359, 540–543. [Google Scholar] [CrossRef] [PubMed]
- Valentine, M.T.; Gilbert, S.P. To step or not to step? How biochemistry and mechanics influence processivity in kinesin and Eg5. Curr. Opin. Cell Biol. 2007, 19, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Valentine, M.T.; Fordyce, P.M.; Krzysiak, T.C.; Gilbert, S.P.; Block, S.M. Individual dimers of the mitotic kinesin motor Eg5 step processively and support substantial loads in vitro. Nat. Cell Biol. 2006, 8, 470–476. [Google Scholar] [CrossRef] [PubMed]
- Hildebrandt, E.R.; Gheber, L.; Kingsbury, T.; Hoyt, M.A. Homotetrameric form of cin8p, a Saccharomyces cerevisiae kinesin-5 motor, is essential for its in vivo function. J. Biol. Chem. 2006, 281, 26004–26013. [Google Scholar] [CrossRef] [PubMed]
- Van den Wildenberg, S.M.; Tao, L.; Kapitein, L.C.; Schmidt, C.F.; Scholey, J.M.; Peterman, E.J. The homotetrameric kinesin-5 KLP61F preferentially crosslinks microtubules into antiparallel orientations. Curr. Biol. 2008, 18, 1860–1864. [Google Scholar] [CrossRef] [PubMed]
- Enos, A.P.; Morris, N.R. Mutation of a gene that encodes a kinesin-like protein blocks nuclear division in A. nidulans. Cell 1990, 60, 1019–1027. [Google Scholar] [CrossRef]
- Straight, A.F.; Sedat, J.W.; Murray, A.W. Time-lapse microscopy reveals unique roles for kinesins during anaphase in budding yeast. J. Cell Biol. 1998, 143, 687–694. [Google Scholar] [CrossRef] [PubMed]
- Saunders, W.S.; Koshland, D.; Eshel, D.; Gibbons, I.R.; Hoyt, M.A. Saccharomyces cerevisiae kinesin- and dynein-related proteins required for anaphase chromosome segregation. J. Cell Biol. 1995, 128, 617–624. [Google Scholar] [CrossRef] [PubMed]
- Avunie-Masala, R.; Movshovich, N.; Nissenkorn, Y.; Gerson-Gurwitz, A.; Fridman, V.; Koivomagi, M.; Loog, M.; Hoyt, M.A.; Zaritsky, A.; Gheber, L. Phospho-regulation of kinesin-5 during anaphase spindle elongation. J. Cell Sci. 2011, 124, 873–878. [Google Scholar] [CrossRef] [PubMed]
- Sharp, D.J.; Brown, H.M.; Kwon, M.; Rogers, G.C.; Holland, G.; Scholey, J.M. Functional coordination of three mitotic motors in Drosophila embryos. Mol. Biol. Cell 2000, 11, 241–253. [Google Scholar] [CrossRef] [PubMed]
- Roostalu, J.; Hentrich, C.; Bieling, P.; Telley, I.A.; Schiebel, E.; Surrey, T. Directional switching of the kinesin cin8 through motor coupling. Science 2011, 332, 94–99. [Google Scholar] [CrossRef] [PubMed]
- Gerson-Gurwitz, A.; Thiede, C.; Movshovich, N.; Fridman, V.; Podolskaya, M.; Danieli, T.; Lakamper, S.; Klopfenstein, D.R.; Schmidt, C.F.; Gheber, L. Directionality of individual kinesin-5 cin8 motors is modulated by loop 8, ionic strength and microtubule geometry. EMBO J. 2011, 30, 4942–4954. [Google Scholar] [CrossRef] [PubMed]
- Edamatsu, M. Bidirectional motility of the fission yeast kinesin-5, cut7. Biochem. Biophys. Res. Commun. 2014, 446, 231–234. [Google Scholar] [CrossRef] [PubMed]
- Kwon, M.; Morales-Mulia, S.; Brust-Mascher, I.; Rogers, G.C.; Sharp, D.J.; Scholey, J.M. The chromokinesin, KLP3a, dives mitotic spindle pole separation during prometaphase and anaphase and facilitates chromatid motility. Mol. Biol. Cell 2004, 15, 219–233. [Google Scholar] [CrossRef] [PubMed]
- Nislow, C.; Lombillo, V.A.; Kuriyama, R.; McIntosh, J.R. A plus-end-directed motor enzyme that moves antiparallel microtubules in vitro localizes to the interzone of mitotic spindles. Nature 1992, 359, 543–547. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Arellano-Santoyo, H.; Portran, D.; Gaillard, J.; Vantard, M.; Thery, M.; Pellman, D. Microtubule-sliding activity of a kinesin-8 promotes spindle assembly and spindle-length control. Nat. Cell Biol. 2013, 15, 948–957. [Google Scholar] [CrossRef] [PubMed]
- Varga, V.; Leduc, C.; Bormuth, V.; Diez, S.; Howard, J. Kinesin-8 motors act cooperatively to mediate length-dependent microtubule depolymerization. Cell 2009, 138, 1174–1183. [Google Scholar] [CrossRef] [PubMed]
- Rizk, R.S.; Discipio, K.A.; Proudfoot, K.G.; Gupta, M.L., Jr. The kinesin-8 kip3 scales anaphase spindle length by suppression of midzone microtubule polymerization. J. Cell Biol. 2014, 204, 965–975. [Google Scholar] [CrossRef] [PubMed]
- Mishima, M.; Kaitna, S.; Glotzer, M. Central spindle assembly and cytokinesis require a kinesin-like protein/RhoGAP complex with microtubule bundling activity. Dev. Cell 2002, 2, 41–54. [Google Scholar] [CrossRef]
- Tao, L.; Fasulo, B.; Warecki, B.; Sullivan, W. Tum/RacGAP functions as a switch activating the Pav/kinesin-6 motor. Nat. Commun. 2016. [Google Scholar] [CrossRef] [PubMed]
- Mishima, M. Centralspindlin in rappaport’s cleavage signaling. Semin. Cell Dev. Biol. 2016, 53, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.; Ward, J.J.; Loiodice, I.; Velve-Casquillas, G.; Nedelec, F.J.; Tran, P.T. Phospho-regulated interaction between kinesin-6 KLP9p and microtubule bundler Ase1p promotes spindle elongation. Dev. Cell 2009, 17, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Drechsler, H.; McAinsh, A.D. Kinesin-12 motors cooperate to suppress microtubule catastrophes and drive the formation of parallel microtubule bundles. Proc. Natl. Acad. Sci. USA 2016, 113, E1635–E1644. [Google Scholar] [CrossRef] [PubMed]
- McNally, K.P.; Panzica, M.T.; Kim, T.; Cortes, D.B.; McNally, F.J. A novel chromosome segregation mechanism during female meiosis. Mol. Biol. Cell 2016, 27, 2576–2589. [Google Scholar] [CrossRef] [PubMed]
- Bieling, P.; Telley, I.A.; Surrey, T. A minimal midzone protein module controls formation and length of antiparallel microtubule overlaps. Cell 2010, 142, 420–432. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.K.; Coughlin, M.; Field, C.M.; Mitchison, T.J. Kif4 regulates midzone length during cytokinesis. Curr. Biol. 2011, 21, 815–824. [Google Scholar] [CrossRef] [PubMed]
- Nunes Bastos, R.; Gandhi, S.R.; Baron, R.D.; Gruneberg, U.; Nigg, E.A.; Barr, F.A. Aurora b suppresses microtubule dynamics and limits central spindle size by locally activating Kif4a. J. Cell Biol. 2013, 202, 605–621. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Zhao, J.; Bibikova, M.; Leverson, J.D.; Bossy-Wetzel, E.; Fan, J.B.; Abraham, R.T.; Jiang, W. Functional analysis of human microtubule-based motor proteins, the kinesins and dyneins, in mitosis/cytokinesis using rna interference. Mol. Biol. Cell 2005, 16, 3187–3199. [Google Scholar] [CrossRef] [PubMed]
- Bieling, P.; Laan, L.; Schek, H.; Munteanu, E.L.; Sandblad, L.; Dogterom, M.; Brunner, D.; Surrey, T. Reconstitution of a microtubule plus-end tracking system in vitro. Nature 2007, 450, 1100–1105. [Google Scholar] [CrossRef] [PubMed]
- Pereira, A.L.; Pereira, A.J.; Maia, A.R.; Drabek, K.; Sayas, C.L.; Hergert, P.J.; Lince-Faria, M.; Matos, I.; Duque, C.; Stepanova, T.; et al. Mammalian clasp1 and clasp2 cooperate to ensure mitotic fidelity by regulating spindle and kinetochore function. Mol. Biol. Cell 2006, 17, 4526–4542. [Google Scholar] [CrossRef] [PubMed]
- Maton, G.; Edwards, F.; Lacroix, B.; Stefanutti, M.; Laband, K.; Lieury, T.; Kim, T.; Espeut, J.; Canman, J.C.; Dumont, J. Kinetochore components are required for central spindle assembly. Nat. Cell Biol. 2015, 17, 697–705. [Google Scholar] [CrossRef] [PubMed]
- Maiato, H.; Khodjakov, A.; Rieder, C.L. Drosophila clasp is required for the incorporation of microtubule subunits into fluxing kinetochore fibres. Nat. Cell Biol. 2005, 7, 42–47. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Hancock, W.O. Kinesin-5 is a microtubule polymerase. Nat. Commun. 2015. [Google Scholar] [CrossRef] [PubMed]
- Cole, D.G.; Chinn, S.W.; Wedaman, K.P.; Hall, K.; Vuong, T.; Scholey, J.M. Novel heterotrimeric kinesin-related protein purified from sea urchin eggs. Nature 1993, 366, 268–270. [Google Scholar] [CrossRef] [PubMed]
- Craft, J.M.; Harris, J.A.; Hyman, S.; Kner, P.; Lechtreck, K.F. Tubulin transport by IFT is upregulated during ciliary growth by a cilium-autonomous mechanism. J. Cell Biol. 2015, 208, 223–237. [Google Scholar] [CrossRef] [PubMed]
- Henson, J.H.; Cole, D.G.; Terasaki, M.; Rashid, D.; Scholey, J.M. Immunolocalization of the heterotrimeric kinesin-related protein KRP(85/95) in the mitotic apparatus of sea urchin embryos. Dev. Biol. 1995, 171, 182–194. [Google Scholar] [CrossRef] [PubMed]
- Lecland, N.; Luders, J. The dynamics of microtubule minus ends in the human mitotic spindle. Nat. Cell Biol. 2014, 16, 770–778. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, S.S.; Vale, R.D. Patronin regulates the microtubule network by protecting microtubule minus ends. Cell 2010, 143, 263–274. [Google Scholar] [CrossRef] [PubMed]
- Hendershott, M.C.; Vale, R.D. Regulation of microtubule minus-end dynamics by camsaps and patronin. Proc. Natl. Acad. Sci. USA 2014, 111, 5860–5865. [Google Scholar] [CrossRef] [PubMed]
- Jiang, K.; Hua, S.; Mohan, R.; Grigoriev, I.; Yau, K.W.; Liu, Q.; Katrukha, E.A.; Altelaar, A.F.; Heck, A.J.; Hoogenraad, C.C.; et al. Microtubule minus-end stabilization by polymerization-driven camsap deposition. Dev. Cell 2014, 28, 295–309. [Google Scholar] [CrossRef] [PubMed]
- Rogers, G.C.; Rogers, S.L.; Schwimmer, T.A.; Ems-McClung, S.C.; Walczak, C.E.; Vale, R.D.; Scholey, J.M.; Sharp, D.J. Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature 2004, 427, 364–370. [Google Scholar] [CrossRef] [PubMed]
- Carmena, M.; Wheelock, M.; Funabiki, H.; Earnshaw, W.C. The chromosomal passenger complex (CPC): From easy rider to the godfather of mitosis. Nat. Rev. Mol. Cell Biol. 2012, 13, 789–803. [Google Scholar] [CrossRef] [PubMed]
- Pavin, N.; Tolic-Norrelykke, I.M. Dynein, microtubule and cargo: A menage a trois. Biochem. Soc. Trans. 2013, 41, 1731–1735. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, H.; Carter, A.P. Review: Structure and mechanism of the dynein motor ATPase. Biopolymers 2016, 105, 557–567. [Google Scholar] [CrossRef] [PubMed]
- Ananthanarayanan, V.; Schattat, M.; Vogel, S.K.; Krull, A.; Pavin, N.; Tolic-Norrelykke, I.M. Dynein motion switches from diffusive to directed upon cortical anchoring. Cell 2013, 153, 1526–1536. [Google Scholar] [CrossRef] [PubMed]
- Laan, L.; Pavin, N.; Husson, J.; Romet-Lemonne, G.; van Duijn, M.; Lopez, M.P.; Vale, R.D.; Julicher, F.; Reck-Peterson, S.L.; Dogterom, M. Cortical dynein controls microtubule dynamics to generate pulling forces that position microtubule asters. Cell 2012, 148, 502–514. [Google Scholar] [CrossRef] [PubMed]
- Gonczy, P.; Pichler, S.; Kirkham, M.; Hyman, A.A. Cytoplasmic dynein is required for distinct aspects of mtoc positioning, including centrosome separation, in the one cell stage Caenorhabditis elegans embryo. J. Cell Biol. 1999, 147, 135–150. [Google Scholar] [CrossRef] [PubMed]
- Vaisberg, E.A.; Koonce, M.P.; McIntosh, J.R. Cytoplasmic dynein plays a role in mammalian mitotic spindle formation. J. Cell Biol. 1993, 123, 849–858. [Google Scholar] [CrossRef] [PubMed]
- Gerdes, K.; Howard, M.; Szardenings, F. Pushing and pulling in prokaryotic DNA segregation. Cell 2010, 141, 927–942. [Google Scholar] [CrossRef] [PubMed]
- Bharat, T.A.; Murshudov, G.N.; Sachse, C.; Lowe, J. Structures of actin-like ParM filaments show architecture of plasmid-segregating spindles. Nature 2015, 523, 106–110. [Google Scholar] [CrossRef] [PubMed]
- Garner, E.C.; Campbell, C.S.; Weibel, D.B.; Mullins, R.D. Reconstitution of DNA segregation driven by assembly of a prokaryotic actin homolog. Science 2007, 315, 1270–1274. [Google Scholar] [CrossRef] [PubMed]
- Bastos, R.N.; Cundell, M.J.; Barr, F.A. Kif4a and pp2a-b56 form a spatially restricted feedback loop opposing aurora b at the anaphase central spindle. J. Cell Biol. 2014, 207, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Musacchio, A. Spindle assembly checkpoint: The third decade. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2011, 366, 3595–3604. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, M.; Morgan, D.O. Finishing mitosis, one step at a time. Nat. Rev. Mol. Cell Biol. 2007, 8, 894–903. [Google Scholar] [CrossRef] [PubMed]
- Parry, D.H.; O’Farrell, P.H. The schedule of destruction of three mitotic cyclins can dictate the timing of events during exit from mitosis. Curr. Biol. 2001, 11, 671–683. [Google Scholar] [CrossRef]
- Yuan, K.; O’Farrell, P.H. Cyclin b3 is a mitotic cyclin that promotes the metaphase-anaphase transition. Curr. Biol. 2015, 25, 811–816. [Google Scholar] [CrossRef] [PubMed]
- Afonso, O.; Matos, I.; Pereira, A.J.; Aguiar, P.; Lampson, M.A.; Maiato, H. Feedback control of chromosome separation by a midzone Aurora B gradient. Science 2014, 345, 332–336. [Google Scholar] [CrossRef] [PubMed]
- Drechsler, H.; McAinsh, A.D. Exotic mitotic mechanisms. Open Biol. 2012. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, D.S.; Huffaker, T.C. Astral microtubules are not required for anaphase B in Saccharomyces cerevisiae. J. Cell Biol. 1992, 119, 379–388. [Google Scholar] [CrossRef] [PubMed]
- Yeh, E.; Yang, C.; Chin, E.; Maddox, P.; Salmon, E.D.; Lew, D.J.; Bloom, K. Dynamic positioning of mitotic spindles in yeast: Role of microtubule motors and cortical determinants. Mol. Biol. Cell 2000, 11, 3949–3961. [Google Scholar] [CrossRef] [PubMed]
- Bannigan, A.; Lizotte-Waniewski, M.; Riley, M.; Baskin, T.I. Emerging molecular mechanisms that power and regulate the anastral mitotic spindle of flowering plants. Cell. Motil. Cytoskelet. 2008, 65, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T.; Sano, T.; Kutsuna, N.; Kumagai-Sano, F.; Hasezawa, S. Contribution of anaphase B to chromosome separation in higher plant cells estimated by image processing. Plant Cell Physiol. 2007, 48, 1509–1513. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.R.; Qiu, W.; Liu, B. Kinesin motors in plants: From subcellular dynamics to motility regulation. Curr. Opin. Plant Biol. 2015, 28, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Struk, S.; Dhonukshe, P. Maps: Cellular navigators for microtubule array orientations in arabidopsis. Plant Cell Rep. 2014, 33, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Miki, T.; Naito, H.; Nishina, M.; Goshima, G. Endogenous localizome identifies 43 mitotic kinesins in a plant cell. Proc. Natl. Acad. Sci. USA 2014, 111, E1053–E1061. [Google Scholar] [CrossRef] [PubMed]
- Cowan, C.R.; Hyman, A.A. Asymmetric cell division in C. elegans: Cortical polarity and spindle positioning. Annu. Rev. Cell Dev. Biol. 2004, 20, 427–453. [Google Scholar] [CrossRef] [PubMed]
- Hara, Y.; Kimura, A. Cell-size-dependent spindle elongation in the Caenorhabditis elegans early embryo. Curr. Biol. 2009, 19, 1549–1554. [Google Scholar] [CrossRef] [PubMed]
- Nguyen-Ngoc, T.; Afshar, K.; Gonczy, P. Coupling of cortical dynein and G alpha proteins mediates spindle positioning in Caenorhabditis elegans. Nat. Cell Biol. 2007, 9, 1294–1302. [Google Scholar] [CrossRef] [PubMed]
- Hayward, D.; Metz, J.; Pellacani, C.; Wakefield, J.G. Synergy between multiple microtubule-generating pathways confers robustness to centrosome-driven mitotic spindle formation. Dev. Cell 2014, 28, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Brugues, J.; Nuzzo, V.; Mazur, E.; Needleman, D.J. Nucleation and transport organize microtubules in metaphase spindles. Cell 2012, 149, 554–564. [Google Scholar] [CrossRef] [PubMed]
- Walczak, C.E.; Vernos, I.; Mitchison, T.J.; Karsenti, E.; Heald, R. A model for the proposed roles of different microtubule-based motor proteins in establishing spindle bipolarity. Curr. Biol. 1998, 8, 903–913. [Google Scholar] [CrossRef]
- Murray, A.W.; Desai, A.B.; Salmon, E.D. Real time observation of anaphase in vitro. Proc. Natl. Acad. Sci. USA 1996, 93, 12327–12332. [Google Scholar] [CrossRef] [PubMed]
- Civelekoglu-Scholey, G.; Cimini, D. Modelling chromosome dynamics in mitosis: A historical perspective on models of metaphase and anaphase in eukaryotic cells. Interface Focus 2014. [Google Scholar] [CrossRef] [PubMed]
- Brust-Mascher, I.; Civelekoglu-Scholey, G.; Scholey, J.M. Mechanism for anaphase B: Evaluation of “slide-and-cluster” versus “slide-and-flux-or-elongate” models. Biophys. J. 2015, 108, 2007–2018. [Google Scholar] [CrossRef] [PubMed]
- Loughlin, R.; Heald, R.; Nedelec, F. A computational model predicts Xenopus meiotic spindle organization. J. Cell Biol. 2010, 191, 1239–1249. [Google Scholar] [CrossRef] [PubMed]
- Uehara, R.; Tsukada, Y.; Kamasaki, T.; Poser, I.; Yoda, K.; Gerlich, D.W.; Goshima, G. Aurora B and Kif2a control microtubule length for assembly of a functional central spindle during anaphase. J. Cell Biol. 2013, 202, 623–636. [Google Scholar] [CrossRef] [PubMed]
- Kellogg, E.H.; Howes, S.; Ti, S.C.; Ramirez-Aportela, E.; Kapoor, T.M.; Chacon, P.; Nogales, E. Near-atomic cryo-EM structure of PRC1 bound to the microtubule. Proc. Natl. Acad. Sci. USA 2016, 113, 9430–9439. [Google Scholar] [CrossRef] [PubMed]
- Chen, I.A. Cell division: Breaking up is easy to do. Curr. Biol. 2009, 19, R327–R328. [Google Scholar] [CrossRef] [PubMed]
| Organism | Rate of Spindle Elongation | Extent of Spindle Elongation from Metaphase to Telophase | Reference(s) |
|---|---|---|---|
| Diatom | Live: 0.038 ± 0.005 μm/s | ~2 μm | [45] |
| Isolated: 0.015 ± 0.002 μm/s * | 1.9 ± 0.17 μm | [44] | |
| Ustilago maydis | Slow: 0.02 ± 0.003 μm/s | ~0.5 μm | [39] |
| Fast: 0.09 ± 0.003 μm/s | ~4.5 μm | ||
| Schizosaccharomyces pombe | ~0.013 μm/s | 7–10 μm | [58] |
| Saccharomyces cerevisiae | Fast: 0.018 ± 0.005 μm/s | ~4 μm | [47] |
| Slow: ~0.006 μm/s | 2–6 μm | ||
| Drosophila syncytial embryo | 0.08 ± 0.015 μm/s | 5 μm | [61] |
| (cycle 12) | |||
| S2 cell | 0.017 μm/s | 5 μm | [63] |
| C. elegans | 0.107 ± 0.008 μm/s | 8.33 ± 0.29 μm | [64] |
| LLC-Pk1 epithelial cells | 0.049 ± 0.017 μm/s | 8.73 ± 2.4 μm | [65] |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scholey, J.M.; Civelekoglu-Scholey, G.; Brust-Mascher, I. Anaphase B. Biology 2016, 5, 51. https://doi.org/10.3390/biology5040051
Scholey JM, Civelekoglu-Scholey G, Brust-Mascher I. Anaphase B. Biology. 2016; 5(4):51. https://doi.org/10.3390/biology5040051
Chicago/Turabian StyleScholey, Jonathan M., Gul Civelekoglu-Scholey, and Ingrid Brust-Mascher. 2016. "Anaphase B" Biology 5, no. 4: 51. https://doi.org/10.3390/biology5040051
APA StyleScholey, J. M., Civelekoglu-Scholey, G., & Brust-Mascher, I. (2016). Anaphase B. Biology, 5(4), 51. https://doi.org/10.3390/biology5040051





