Use of a Fluorescent Aptamer RNA as an Exonic Sequence to Analyze Self-Splicing Ability of a Group I Intron from Structured RNAs
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plasmid Construction and RNA Preparation
2.2. In Vitro Self-Splicing Assay with Denaturing Polyacrylamide Gel Electrophoresis
2.3. In Vitro Self-Splicing Monitored by Fluorescence of Spinach RNA–DFHBI
2.4. Cotranscriptional Self-Splicing Assay
2.5. Electrophoretic Mobility Shift (EMS) Assay
3. Results
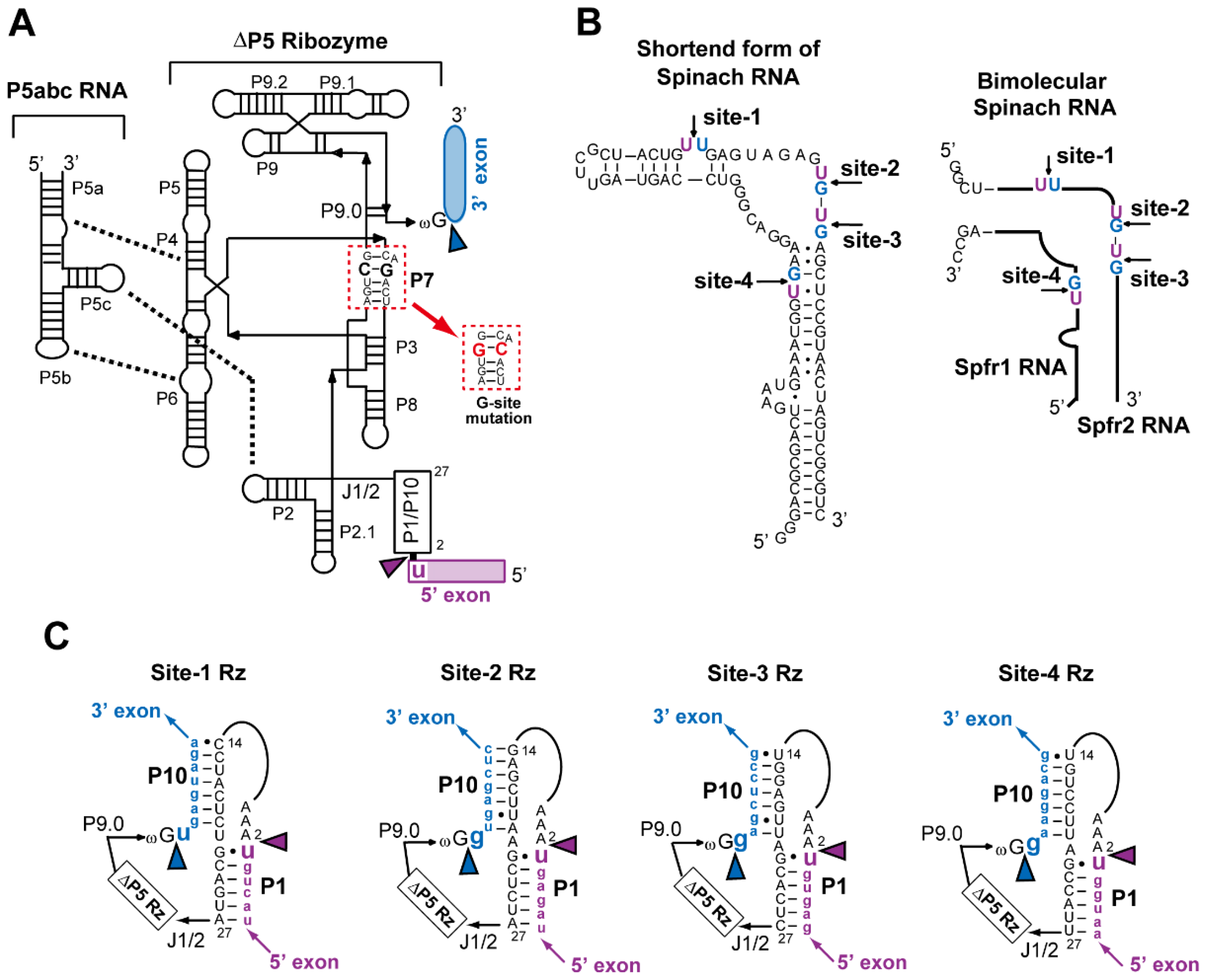
3.1. Experimental Design
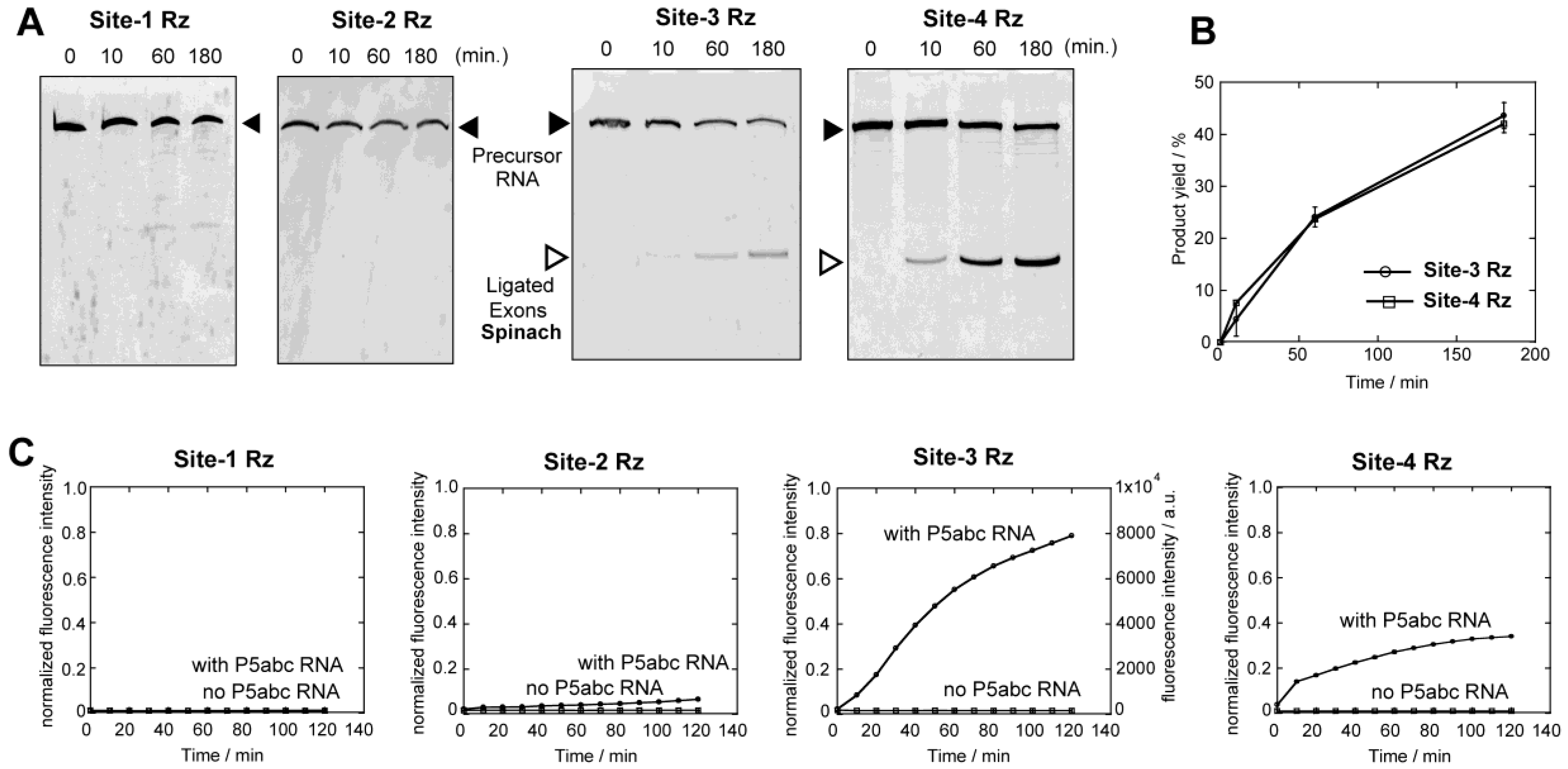
3.2. Analyses of Self-Splicing to Produce the Fluorescent RNA Aptamer
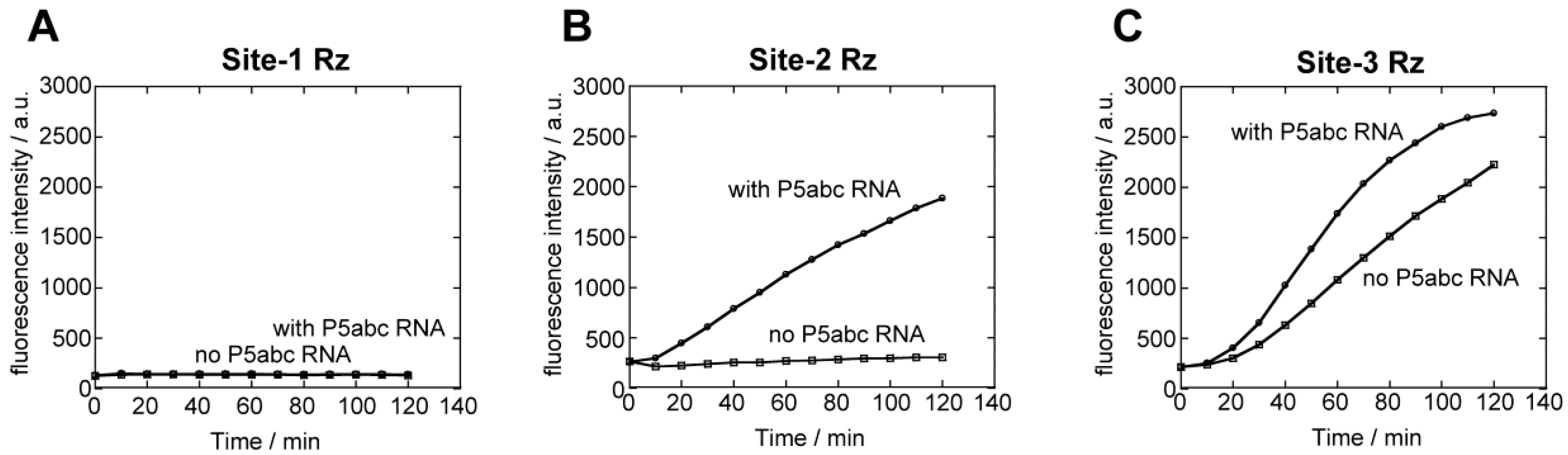
3.3. Tertiary Folding of ∆P5 Ribozyme Probed by an Electrophoretic Mobility Shift (EMS) Assay with P5abc RNA
3.4. Physical Dissection of Spinach RNA to Resolve Folding Problems of ∆P5 Ribozyme
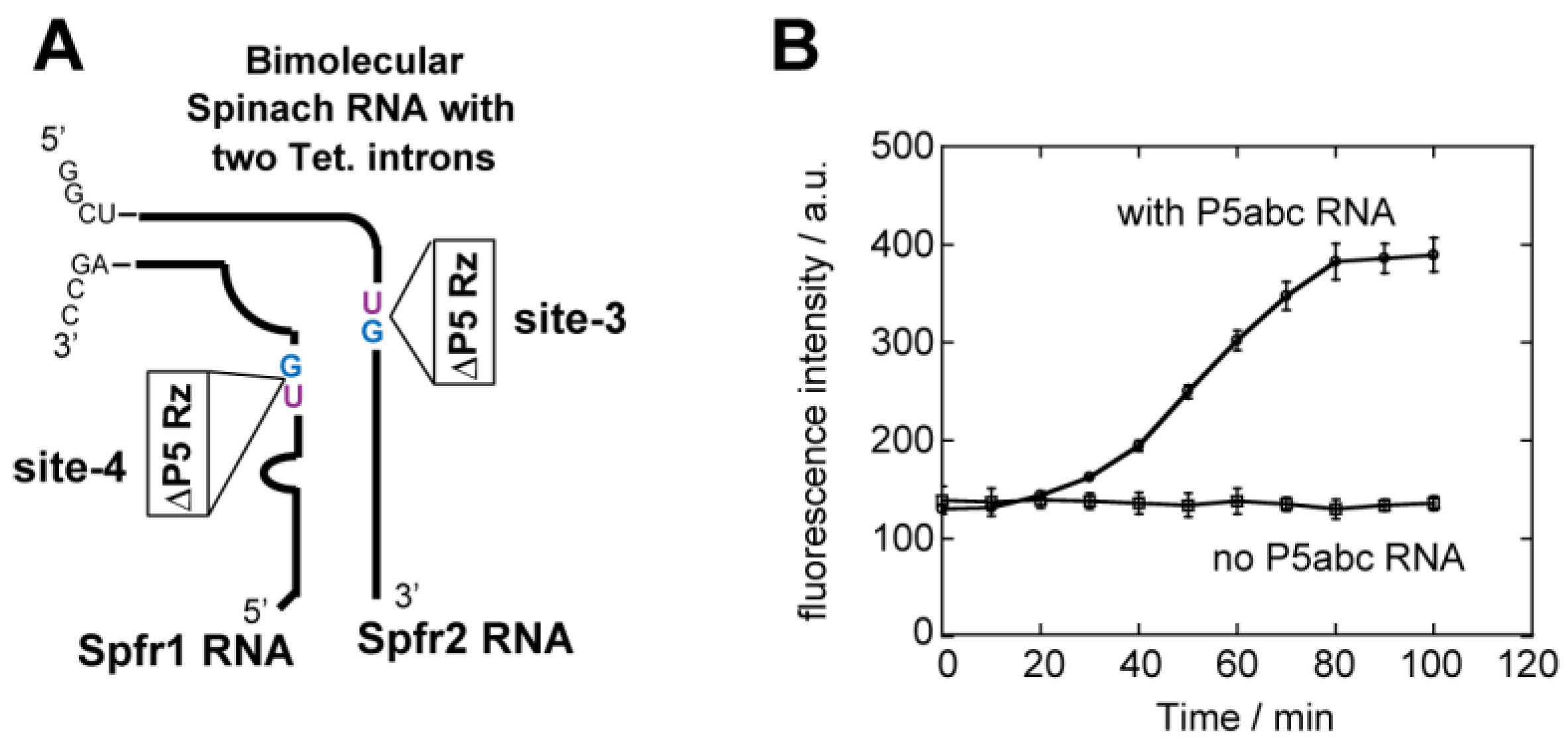
3.5. Application of Bimolecular Spinach RNA to Analyze Engineered Tetrahymena Ribozymes
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lilley, D.M.; Eckstein, F. Ribozymes and RNA Catalysis; RSC publishing: London, UK, 2007. [Google Scholar]
- Elliott, D.; Ladomery, M. Molecular Biology of RNA, 2nd ed.; Oxford University Press: Oxford, UK, 2016. [Google Scholar]
- Cech, T.R. Nobel lecture. Self-splicing and enzymatic activity of an intervening sequence RNA from Tetrahymena. Biosci. Rep. 1990, 10, 239–261. [Google Scholar] [CrossRef] [PubMed]
- Phizicky, E.M.; Greer, C.L. Pre-tRNA splicing: Variation on a theme or exception to the rule? Trends Biochem. Sci. 1993, 18, 31–34. [Google Scholar] [CrossRef]
- Golden, B.L. Group I introns: Biochemical and crystallographic characterization of the active site structure. In Ribozymes and RNA Catalysis; RSC publishing: London, UK, 2007; pp. 178–200. [Google Scholar]
- Woodson, S.A. Structure and assembly of group I introns. Curr. Opin. Struct. Biol. 2005, 15, 324–330. [Google Scholar] [CrossRef] [PubMed]
- Vicens, Q.; Cech, T.R. Atomic level architecture of group I introns revealed. Trends Biochem. Sci. 2006, 31, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Stahley, M.R.; Strobel, S.A. RNA splicing: Group I intron crystal structures reveal the basis of splice site selection and metal ion catalysis. Curr. Opin. Struct. Biol. 2006, 16, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Adams, P.L.; Stahley, M.R.; Kosek, A.B.; Wang, J.; Strobel, S.A. Crystal structure of a self-splicing group I intron with both exons. Nature 2004, 430, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Golden, B.L.; Kim, H.; Chase, E. Crystal structure of a phage Twort group I ribozyme-product complex. Nat. Struct. Mol. Biol. 2005, 12, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Gooding, A.R.; Cech, T.R. Structure of the Tetrahymena ribozyme: Base triple sandwich and metal ion at the active site. Mol. Cell 2004, 16, 351–362. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, L.B.; Jones, B.C.; Cosstick, R.; Cech, T.R. A second catalytic metal ion in group I ribozyme. Nature 1997, 388, 805–808. [Google Scholar] [PubMed]
- Michel, F.; Hanna, M.; Green, R.; Bartel, D.P.; Szostak, J.W. The guanosine binding site of the Tetrahymena ribozyme. Nature 1989, 342, 391–395. [Google Scholar] [CrossRef] [PubMed]
- Been, M.D.; Perrotta, A.T. Group I intron self-splicing with adenosine: Evidence for a single nucleoside-binding site. Science 1991, 252, 434–437. [Google Scholar] [CrossRef] [PubMed]
- Haugen, P.; Simon, D.M.; Bhattacharya, D. The natural history of group I introns. Trends Genet. 2005, 21, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Phylactou, L.A.; Darrah, C.; Wood, M.J. Ribozyme-mediated trans-splicing of a trinucleotide repeat. Nat. Genet. 1998, 18, 378–381. [Google Scholar] [CrossRef] [PubMed]
- Sullenger, B.A.; Cech, T.R. Ribozyme-mediated repair of defective mRNA by targeted, trans-splicing. Nature 1994, 371, 619–622. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.T.; Sullenger, B.A. Evaluating and enhancing ribozyme reaction efficiency in mammalian cells. Nat. Biotechnol. 1997, 15, 902–905. [Google Scholar] [CrossRef] [PubMed]
- Campbell, T.B.; Cech, T.R. Identification of ribozymes within a ribozyme library that efficiently cleave a long substrate RNA. RNA 1995, 1, 598–609. [Google Scholar] [PubMed]
- Dotson, P.P., 2nd; Hart, J.; Noe, C.; Testa, S.M. Ribozyme-mediated trans insertion-splicing into target RNAs. Methods Mol. Biol. 2012, 848, 385–394. [Google Scholar] [PubMed]
- Amini, Z.N.; Olson, K.E.; Müller, U.F. Spliceozymes: Ribozymes that remove introns from pre-mRNAs in trans. PLoS ONE 2014, 9, e101932. [Google Scholar] [CrossRef] [PubMed]
- Olson, K.E.; Müller, U.F. An in vivo selection method to optimize trans-splicing ribozymes. RNA 2012, 18, 581–589. [Google Scholar] [CrossRef] [PubMed]
- Satterwhite, L.E.; Yeates, J.A.; Lehman, N. Group I intron internal guide sequence binding strength as a component of ribozyme network formation. Molecules 2016, 21. [Google Scholar] [CrossRef] [PubMed]
- Paige, J.S.; Wu, K.Y.; Jaffrey, S.R. RNA mimics of green fluorescent protein. Science 2011, 333, 642–646. [Google Scholar] [CrossRef] [PubMed]
- Warner, K.D.; Chen, M.C.; Song, W.; Strack, R.L.; Thorn, A.; Jaffrey, S.R.; Ferré-D'Amaré, A.R. Structural basis for activity of highly efficient RNA mimics of green fluorescent protein. Nat. Struct. Mol. Biol. 2014, 21, 658–663. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Suslov, N.B.; Li, N.S.; Shelke, S.A.; Evans, M.E.; Koldobskaya, Y.; Rice, P.A.; Piccirilli, J.A. A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore. Nat. Chem. Biol. 2014, 10, 686–691. [Google Scholar] [CrossRef] [PubMed]
- Williams, K.P.; Fujimoto, D.N.; Inoue, T. A region of group I introns that contains universally conserved residues but is not essential for self-splicing. Proc. Natl. Acad. Sci. USA 1992, 89, 10400–10404. [Google Scholar] [CrossRef] [PubMed]
- Ikawa, Y.; Moriyama, S.; Furuta, H. Facile syntheses of BODIPY derivatives for fluorescent labeling of the 3′ and 5′ ends of RNAs. Anal. Biochem. 2008, 378, 166–170. [Google Scholar] [CrossRef] [PubMed]
- Sargueil, B.; Tanner, N.K. A shortened form of the Tetrahymena thermophila group I intron can catalyze the complete splicing reaction in trans. J. Mol. Biol. 1993, 233, 629–643. [Google Scholar] [CrossRef] [PubMed]
- Ikawa, Y.; Yoshimura, T.; Hara, H.; Shiraishi, H.; Inoue, T. Two conserved structural components, A-rich bulge and P4 XJ6/7 base-triples, in activating the group I ribozymes. Genes Cells 2002, 7, 1205–1215. [Google Scholar] [CrossRef] [PubMed]
- Donghi, D.; Schnabl, J. Multiple roles of metal ions in large ribozymes. Met. Ions Life Sci. 2011, 9, 197–234. [Google Scholar] [PubMed]
- Joyce, G.F.; van der Horst, G.; Inoue, T. Catalytic activity is retained in the Tetrahymena group I intron despite removal of the large extension of element P5. Nucleic Acids Res. 1989, 17, 7879–7889. [Google Scholar] [CrossRef] [PubMed]
- Van der Horst, G.; Christian, A.; Inoue, T. Reconstitution of a group I intron self-splicing reaction with an activator RNA. Proc. Natl. Acad. Sci. USA 1991, 88, 184–188. [Google Scholar] [CrossRef] [PubMed]
- Engelhardt, M.A.; Doherty, E.A.; Knitt, D.S.; Doudna, J.A.; Herschlag, D. The P5abc peripheral element facilitates preorganization of the Tetrahymena group I ribozyme for catalysis. Biochemistry 2000, 39, 2639–2651. [Google Scholar] [CrossRef] [PubMed]
- Johnson, T.H.; Tijerina, P.; Chadee, A.B.; Herschlag, D.; Russell, R. Structural specificity conferred by a group I RNA peripheral element. Proc. Natl. Acad. Sci. USA 2005, 102, 10176–10181. [Google Scholar] [CrossRef] [PubMed]
- Couture, S.; Ellington, A.D.; Gerber, A.S.; Cherry, J.M.; Doudna, J.A.; Green, R.; Hanna, M.; Pace, U.; Rajagopal, J.; Szostak, J.W. Mutational analysis of conserved nucleotides in a self-splicing group I intron. J. Mol. Biol. 1990, 215, 345–358. [Google Scholar] [CrossRef]
- Che, A.J.; Knight, T.F., Jr. Engineering a family of synthetic splicing ribozymes. Nucleic Acids Res. 2010, 38, 2748–2755. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Matsumura, S.; Furuta, H.; Ikawa, Y. Tecto-GIRz: Engineered group I ribozyme the catalytic ability of which can be controlled by self-dimerization. ChemBioChem. 2016, 17, 1448–1455. [Google Scholar] [CrossRef] [PubMed]


© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Furukawa, A.; Tanaka, T.; Furuta, H.; Matsumura, S.; Ikawa, Y. Use of a Fluorescent Aptamer RNA as an Exonic Sequence to Analyze Self-Splicing Ability of a Group I Intron from Structured RNAs. Biology 2016, 5, 43. https://doi.org/10.3390/biology5040043
Furukawa A, Tanaka T, Furuta H, Matsumura S, Ikawa Y. Use of a Fluorescent Aptamer RNA as an Exonic Sequence to Analyze Self-Splicing Ability of a Group I Intron from Structured RNAs. Biology. 2016; 5(4):43. https://doi.org/10.3390/biology5040043
Chicago/Turabian StyleFurukawa, Airi, Takahiro Tanaka, Hiroyuki Furuta, Shigeyoshi Matsumura, and Yoshiya Ikawa. 2016. "Use of a Fluorescent Aptamer RNA as an Exonic Sequence to Analyze Self-Splicing Ability of a Group I Intron from Structured RNAs" Biology 5, no. 4: 43. https://doi.org/10.3390/biology5040043
APA StyleFurukawa, A., Tanaka, T., Furuta, H., Matsumura, S., & Ikawa, Y. (2016). Use of a Fluorescent Aptamer RNA as an Exonic Sequence to Analyze Self-Splicing Ability of a Group I Intron from Structured RNAs. Biology, 5(4), 43. https://doi.org/10.3390/biology5040043





