Differentiation of Crohn’s Disease-Associated Isolates from Other Pathogenic Escherichia coli by Fimbrial Adhesion under Shear Force
Abstract
:1. Introduction
2. Materials and Methods
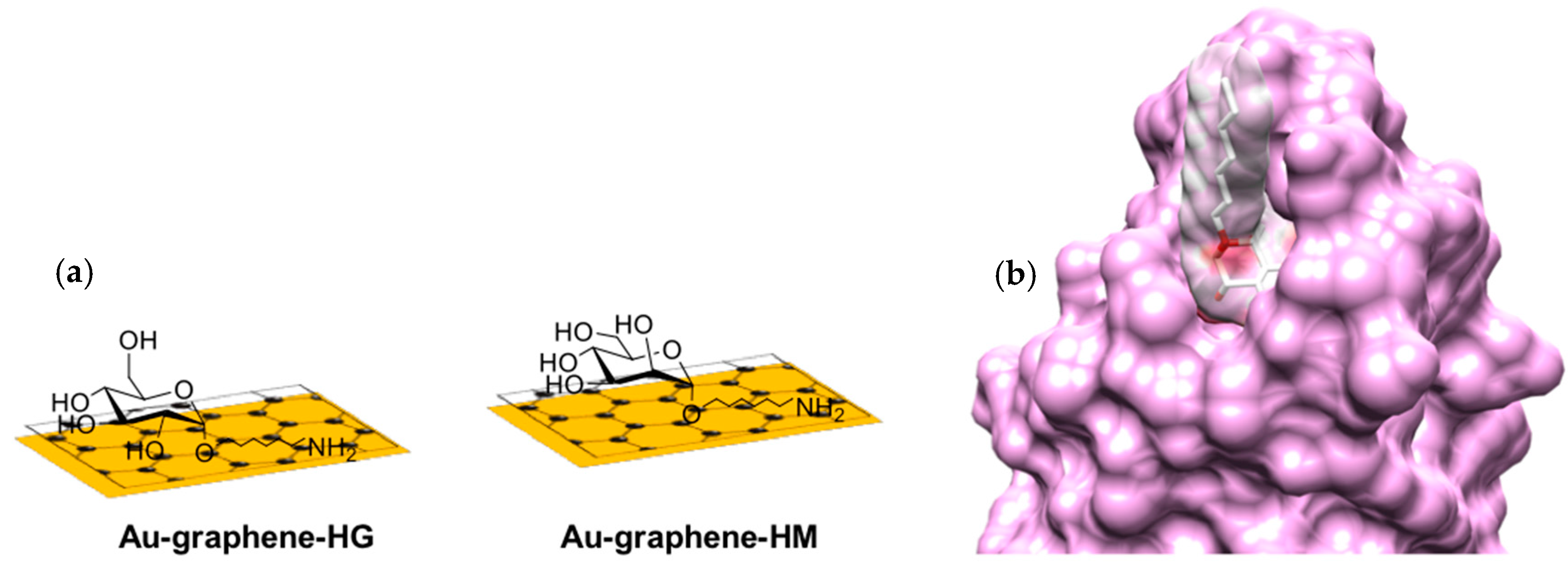
2.1. Functionalization of Au-Graphene SPR Interfaces with Glycans
2.2. Selection and Growth of Escherichia coli Pathovars
2.3. Bacterial Hemagglutination
2.4. SPR Measurements at Different Flow Rates
2.5. fimH Sequencing of Novel Bacterial Clones
2.6. Structural Analyses
3. Results
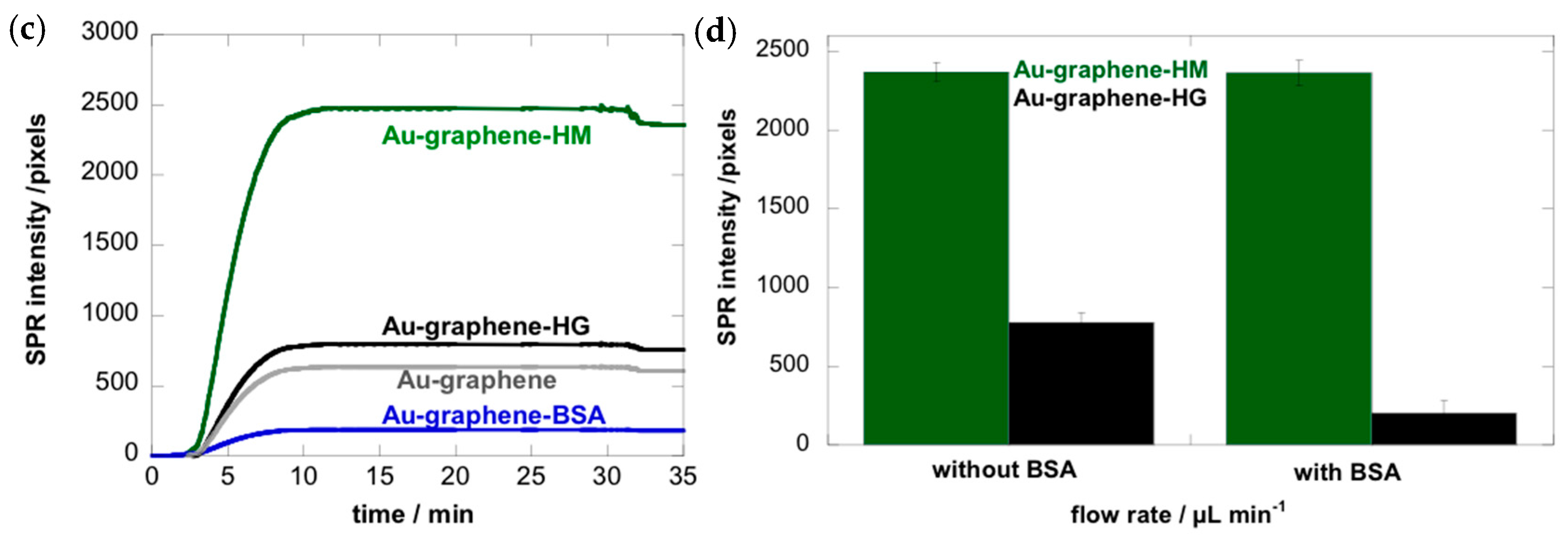
3.1. Mannose-Specific Adhesion of E. coli UTI89 to Graphene-on-Gold Surfaces under Flow
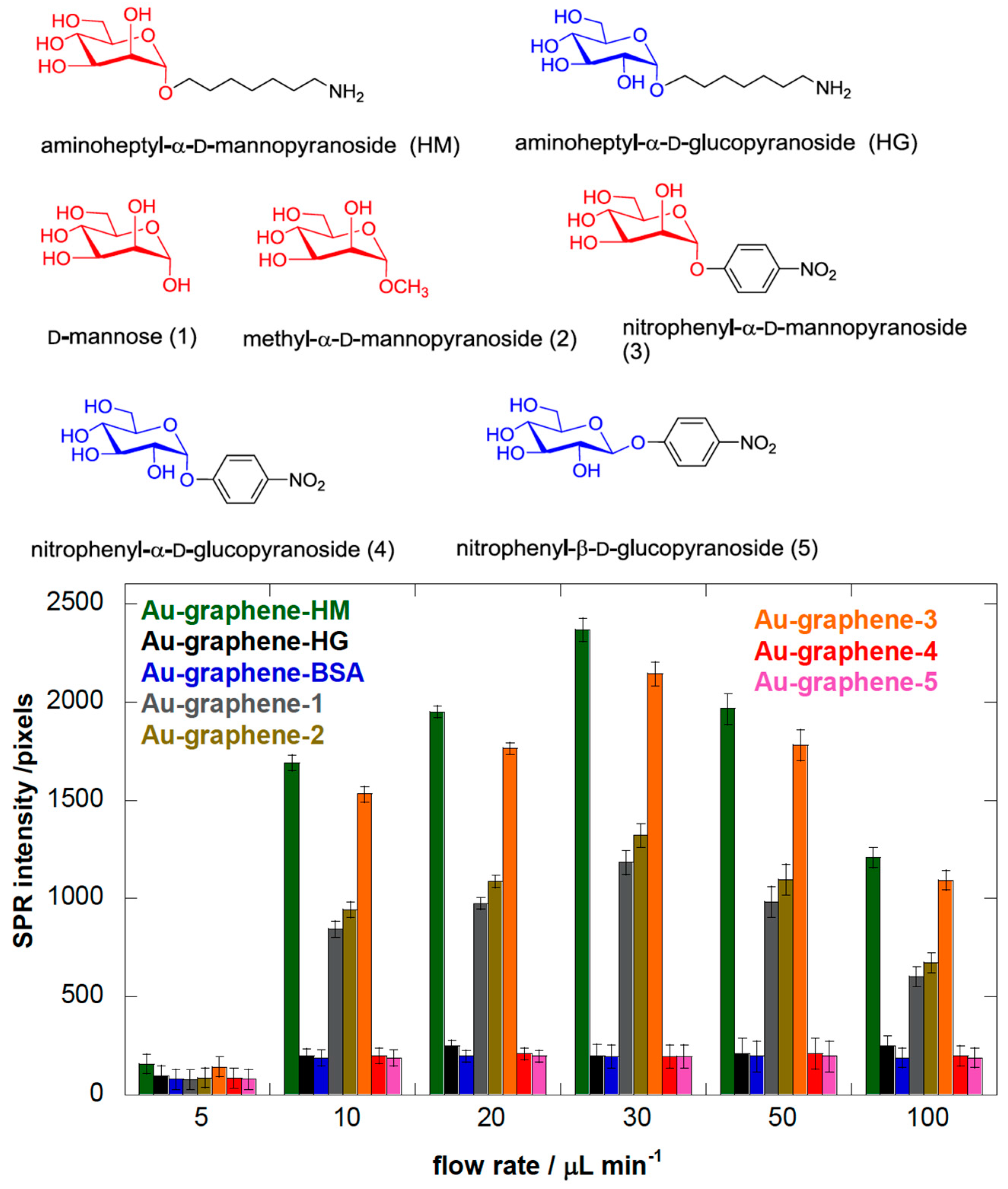
3.2. Heptyl α-d-Mannose Integrates into Graphene in a Way that Is Very Favorable for the Specific Detection of the FimH Adhesin from E. coli
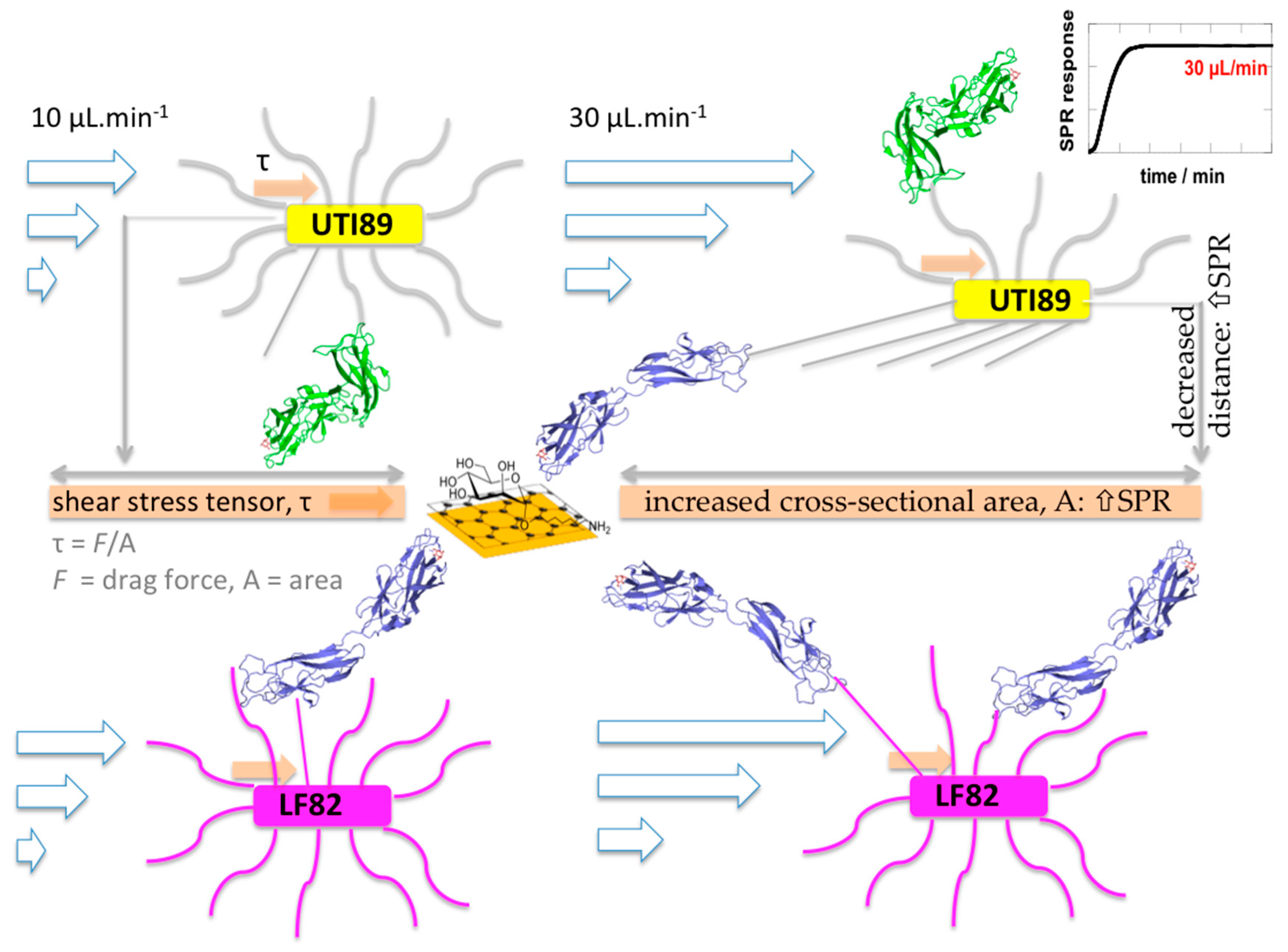
3.3. Adhesion of E. coli UTI89 to Mannosylated Surfaces Is Enhanced under Shear Flow
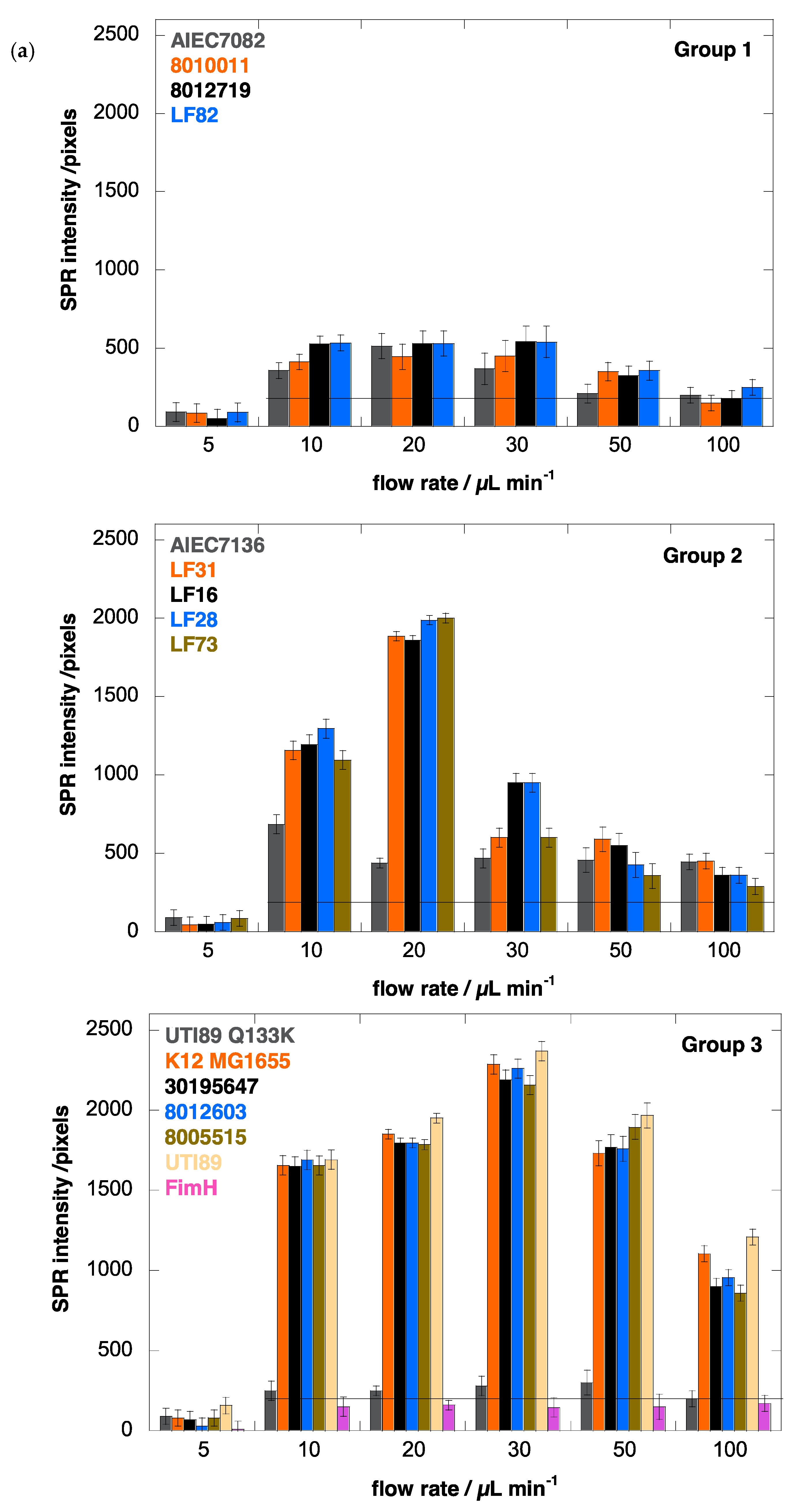
3.4. E. coli Strains UTI89 and K12 Display the Strongest Shear Force-Enhanced Affinity
3.5. The AIEC Strain LF82 Displays the Highest Affinity under Static Conditions
3.6. Human E. coli Pathovars Display a Different Optimum Flow Rate for Enhanced Adhesion
3.7. Amino Acid Variations in FimH from AIEC Strains Moderate Shear Force Regulation
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| AIEC | adherent-invasive Escherichia coli |
| BSA | bovine serum albumin |
| HG | aminoheptyl α-d-glucopyranoside |
| HM | aminoheptyl α-d-mannopyranoside |
| OD | optical density |
| PBS | phosphate-buffered saline |
| SPR | surface plasmon resonance |
| TDA | two-domain adhesin |
References
- Knight, S.D.; Bouckaert, J. Structure, function, and assembly of type-1 fimbriae. Top. Curr. Chem. 2009, 288, 67–107. [Google Scholar] [PubMed]
- Jones, C.H.; Pinkner, J.S.; Roth, R.; Heuser, J.; Nicholoes, A.V.; Abraham, S.N.; Hultgren, S.J. FimH adhesin of type-1 pili is assembled into a fibrillar tip structure in the Enterobacteriaceae. Proc. Natl. Acad. Sci. USA 1995, 92, 2081–2085. [Google Scholar] [CrossRef] [PubMed]
- Sokurenko, E.V.; Chesnokova, V.; Dykhuizen, D.E.; Ofek, I.; Wu, X.R.; Krogfelt, K.A.; Struve, C.; Schembri, M.A.; Hasty, D.L. Pathogenic adaptation of Escherichia coli by natural variation of the FimH adhesin. Proc. Natl. Acad. Sci. USA 1998, 95, 8922–8926. [Google Scholar] [CrossRef] [PubMed]
- Le Trong, I.; Aprikian, P.; Kidd, B.A.; Forero-Shelton, M.; Tchesnokova, V.; Rajagopal, P.; Rodriguez, V.; Interlandi, G.; Klevit, R.; Vogel, V.; et al. Structural basis for mechanical force regulation of the adhesin FimH via finger trap-like beta sheet twisting. Cell 2010, 141, 645–655. [Google Scholar] [CrossRef] [PubMed]
- Aprikian, P.; Tchesnokova, V.; Kidd, B.; Yakovenko, O.; Yarov-Yarovoy, V.; Trinchina, E.; Vogel, V.; Thomas, W.; Sokurenko, E. Interdomain interaction in the FimH adhesin of Escherichia coli regulates the affinity to mannose. J. Biol. Chem. 2007, 282, 23437–23446. [Google Scholar] [CrossRef] [PubMed]
- Bouckaert, J.; Berglund, J.; Schembri, M.; De Genst, E.; Cools, L.; Wuhrer, M.; Hung, C.S.; Pinkner, J.; Slattegard, R.; Zavialov, A.; et al. Receptor binding studies disclose a novel class of high-affinity inhibitors of the Escherichia coli FimH adhesin. Mol. Microbiol. 2005, 55, 441–455. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, D.; Thompson, A.; Stojanoff, V.; Langermann, S.; Pinkner, J.; Hultgren, S.J.; Knight, S.D. X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science 1999, 285, 1061–1066. [Google Scholar] [CrossRef] [PubMed]
- Hung, C.S.; Bouckaert, J.; Hung, D.; Pinkner, J.; Widberg, C.; DeFusco, A.; Auguste, C.G.; Strouse, R.; Langermann, S.; Waksman, G.; et al. Structural basis of tropism of Escherichia coli to the bladder during urinary tract infection. Mol. Microbiol. 2002, 44, 903–915. [Google Scholar] [CrossRef] [PubMed]
- Aprikian, P.; Interlandi, G.; Kidd, B.A.; Le Trong, I.; Tchesnokova, V.; Yakovenko, O.; Whitfield, M.J.; Bullitt, E.; Stenkamp, R.E.; Thomas, W.E.; et al. The bacterial fimbrial tip acts as a mechanical force sensor. PLoS. Biol. 2011, 9, e1000617. [Google Scholar] [CrossRef] [PubMed]
- Thomas, W.E.; Trintchina, E.; Forero, M.; Vogel, V.; Sokurenko, E.V. Bacterial adhesion to target cells enhanced by shear force. Cell 2002, 109, 913–923. [Google Scholar] [CrossRef]
- Thomas, W.; Forero, M.; Yakovenko, O.; Nilsson, L.; Vicini, P.; Sokurenko, E.; Vogel, V. Catch-bond model derived from allostery explains force-activated bacterial adhesion. Biophys. J. 2006, 90, 753–764. [Google Scholar] [CrossRef] [PubMed]
- Thomas, W. For catch bonds, it all hinges on the interdomain region. J. Cell Biol. 2006, 174, 911–913. [Google Scholar] [CrossRef] [PubMed]
- Yakovenko, O.; Sharma, S.; Forero, M.; Tchesnokova, V.; Aprikian, P.; Kidd, B.; March, A.; Vogel, V.; Sokurenko, E.; Thomas, W. Fimh forms catch bonds that are enhanced by mechanical force due to allosteric regulation. J. Biol. Chem. 2008, 283, 11596–11605. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, L.M.; Thomas, W.E.; Sokurenko, E.V.; Vogel, V. Beyond induced-fit receptor-ligand interactions: Structural changes that can significantly extend bond lifetimes. Structure 2008, 16, 1047–1058. [Google Scholar] [CrossRef] [PubMed]
- Sauer, M.M.; Jakob, R.P.; Eras, J.; Baday, S.; Eris, D.; Navarra, G.; Berneche, S.; Ernst, B.; Maier, T.; Glockshuber, R. Catch-bond mechanism of the bacterial adhesin FimH. Nat. Commun. 2016. [Google Scholar] [CrossRef] [PubMed]
- Sokurenko, E.V.; Vogel, V.; Thomas, W.E. Catch bond mechanism of force-enhanced adhesion: Counter-intuitive, elusive but … widespread? Cell Host Microbe 2008, 16, 314–323. [Google Scholar] [CrossRef] [PubMed]
- Thomas, W.E.; Nilsson, L.M.; Forero, M.; Sokurenko, E.V.; Vogel, V. Shear-dependent ‘stick-and-roll’ adhesion of type-1 fimbriated Escherichia coli. Mol. Microbiol. 2004, 53, 1545–1557. [Google Scholar] [CrossRef] [PubMed]
- Busscher, H.J.; van der Mei, H.C. Microbial adhesion in flow displacement systems. Clin. Microbiol. Rev. 2006, 19, 127–141. [Google Scholar] [CrossRef] [PubMed]
- Lecuyer, S.; Rusconi, R.; Shen, Y.R.; Forsyth, A.; Vlamakis, H.; Kolter, R.; Stone, H.A. Shear stress increases the residence time of adhesion of Pseudomonas aeruginosa. Biophys. J. 2011, 100, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Klinth, J.E.; Castelain, M.; Uhlin, B.E.; Axner, O. The influence of ph on the specific adhesion of p piliated Escherichia coli. PLoS ONE 2012, 7, e38548. [Google Scholar] [CrossRef] [PubMed]
- Tchesnokova, V.; Aprikian, P.; Yakovenko, O.; Larock, C.; Kidd, B.A.; Vogel, V.; Thomas, W.; Sokurenko, E. Integrin-like allosteric properties of the catch bond-forming fimh adhesin of Escherichia coli. J. Biol. Chem. 2008, 283, 7823–7833. [Google Scholar] [CrossRef] [PubMed]
- Bulard, E.; Bouchet-Spinelli, A.; Chaud, P.; Roget, A.; Calemczuk, R.; Fort, S.; Livache, T. Carbohydrates as new probes for the identification of closely related Escherichia coli strains using surface plasmon resonance imaging. Anal. Chem. 2015, 87, 1804–1811. [Google Scholar] [CrossRef] [PubMed]
- Zagorodko, O.; Bouckaert, J.; Dumych, T.; Bilyy, R.; Larroulet, I.; Serrano, A.Y.; Dorta, D.A.; Gouin, S.G.; Dima, S.O.; Oancea, F.; et al. Surface plasmon resonance (SPR) for the evaluation of shear-force-dependent bacterial adhesion. Biosensors 2015, 5, 276–287. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, P.; Barka-Bouaifel, F.; Bouckaert, J.; Yamakawa, N.; Boukerroub, R.; Szunerits, S. Graphene-coated surface plasmon resonance interfaces for studying the interactions between bacteria and surfaces. ACS Appl. Mater. Interfaces 2014, 6, 5422–5431. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Nayak, T.R.; Hong, H.; Cai, W. Graphene: A versatile nanoplatform for biomedical applications. Nanoscale 2012, 4, 3833–3842. [Google Scholar] [CrossRef] [PubMed]
- Penezic, A.; Deokar, G.; Vignaud, D.; Pichonat, E.; Happy, H.; Subramanian, P.; Gašparovic, B.; Boukherroub, R.; Szunerits, S. Carbohydrate-lectin interaction on graphene-coated surface plasmon resonance (SPR) interfaces. Plasmonics 2014, 9, 677–683. [Google Scholar] [CrossRef]
- Zagorodko, O.; Spadavecchia, J.; Yanguas Serrano, A.; Larroulet, I.; Pesquera, A.; Zurutuza, A.; Boukherroub, R.; Szunerits, S. Highly sensitive detection of DNA hybridization on commercialized graphene coated surface plasmon resonance interfaces. Anal. Chem. 2014, 86, 11211–11216. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, P.; Lesniewski, A.; Kaminska, I.; Vlandas, A.; Vasilescu, A.; Niedziolka-Jonsson, J.; Pichonat, E.; Happy, H.; Boukherroub, R.; Szunerits, S. Lysozyme detection on aptamer functionalized graphene-coated SPR interfaces. Biosens. Bioelectron. 2013, 50, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Nayak, T.R.; Andersen, H.; Makam, V.S.; Khaw, C.; Bae, S.; Xu, X.; Ee, P.-L.R.; Ahn, J.-H.; Hong, B.H.; Pastorin, G. Graphene for controlled and accelerated osteogenic differentiation of human mesenchymal stem cells. ACS Nano 2011, 5, 4670–4678. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, O.N.; Fernando, K.A.S.; Wang, B.; Brown, N.A.; Luo, P.G.; McNamara, N.D.; Vangsness, M.; Sun, Y.-P.; Bunker, C.E. Graphene oxide: A nonspecific enhancer of cellular growth. ACS Nano 2011, 5, 8100–8107. [Google Scholar] [CrossRef] [PubMed]
- Ryoo, S.-R.; Kim, Y.-K.; Kim, M.-H.; Min, D.-H. Behaviors of nih-3t3 fibroblasts on graphene/carbon nanotubes: Proliferation, focal adhesion, and gene transfection studies. ACS Nano 2010, 4, 6587–6598. [Google Scholar] [CrossRef] [PubMed]
- Dreux, N.; Denizot, J.; Martinez-Medina, M.; Mellmann, A.; Billig, M.; Kisiela, D.; Chattopadhyay, S.; Sokurenko, E.; Neut, C.; Gower-Rousseau, C.; et al. Point mutations in FimH adhesin of Crohn’s disease-associated adherent-invasive Escherichia coli enhance intestinal inflammatory response. PLoS Pathog. 2013, 9, e1003141. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Sivignon, A.; Yamakawa, N.; Crepet, A.; Travelet, C.; Borsali, R.; Dumych, T.; Li, Z.; Bilyy, R.; Deniaud, D.; et al. Glycopolymers as antiadhesives of E. coli strains inducing inflammatory bowel diseases. Biomacromolecules 2015, 16, 1827–1836. [Google Scholar] [CrossRef] [PubMed]
- Jayasundara, D.R. Carbohydrate coatings via aryldiazonium chemistry for surface biomimicry. Chem. Mater. 2013, 25, 4122–4128. [Google Scholar] [CrossRef]
- Barras, A.; Martin, F.A.; Bande, O.; Baumann, J.S.; Ghigo, J.M.; Boukherroub, R.; Beloin, C.; Siriwardena, A.; Szunerits, S. Glycan-functionalized diamond nanoparticles as potent E. coli anti-adhesives. Nanoscale 2013, 5, 2307–2316. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.L.; Hung, C.S.; Pinkner, J.S.; N., W.J.; Cusumano, C.K.; Li, Z.; Bouckaert, J.; Gordon, J.I.; Hultgren, S. Positive selection identifies an in vivo role for FimH during urinary tract infection in addition to mannose binding. Proc. Natl. Acad. Sci. USA 2009, 109, 22439–22444. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Schwartz, S.; Wagner, L.; Miller, W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000, 7, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of COOT. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [PubMed]
- Delano, W.L. The PyMOL Molecular Graphics System; Version 1.8; DeLano Scientific: Palo Alto, CA, USA.
- Liang, M.N.; Smith, S.P.; Metallo, S.J.; Choi, I.S.; Prentiss, M.; Whitesides, G.M. Measuring the forces involved in polyvalent adhesion of uropathogenic Escherichia coli to mannose-presenting surfaces. Proc. Natl. Acad. Sci. USA 2000, 97, 13092–13096. [Google Scholar] [CrossRef] [PubMed]
- Touaibia, M.; Wellens, A.; Shiao, T.C.; Wang, Q.; Sirois, S.; Bouckaert, J.; Roy, R. Mannosylated G(0) dendrimers with nanomolar affinities to Escherichia coli FimH. Chem. Med. Chem. 2007, 2, 1190–1201. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Fierens, K.; Zhang, Z.; Vanparijs, N.; Schuijs, M.; van Steendam, K.; Feiner Gracia, N.; de Rycke, R.; de Beer, T.; de Beuckelaer, A.; et al. Spontaneous protein adsorption on graphene oxide nanosheets allows efficient intracellular vaccine protein delivery. ACS Appl. Mater. Interfaces 2016, 8, 1147–1155. [Google Scholar] [CrossRef] [PubMed]
- Kaminska, I.; Barras, A.; Coffinier, Y.; Lisowski, W.; Niedziolka-Jonsson, J.; Woisel, P.; Lyskawa, J.; Opallo, M.; Siriwardena, A.; Boukherroub, R.; et al. Preparation of a responsive carbohydrate-coated biointerface based on graphene/azido-terminated tetrathiafulvalene nanohybrid material. ACS Appl. Mater. Interfaces 2012, 4, 5386–5393. [Google Scholar] [CrossRef] [PubMed]
- Björk, J.; Hanke, F.; Palma, C.-A.; Samori, P.; Cecchini, M.; Persson, M. Adsorption of aromatic and anti-aromatic systems on graphene through π−π stacking. J. Phys. Chem. Lett. 2010, 1, 3407–3412. [Google Scholar] [CrossRef]
- Rochefort, A.; Wuest, J.D. Interaction of substituted aromatic compounds with graphene. Langmuir 2008, 25, 210–215. [Google Scholar] [CrossRef] [PubMed]
- Bakker, D.P.; Busccher, H.J.; Van der Mei, H.C. Bacterial deposition in a parallel plate and a stagnation point flow chamber: Microbial adhesion mechanisms depend on the mass transport conditions. Microbiology 2002, 148, 597–603. [Google Scholar] [CrossRef] [PubMed]
- Boks, N.P.; Norde, W.; Van der Mei, H.C.; Busscher, H.J. Forces involved in bacterial adhesion to hydrophilic and hydrophobic surfaces. Microbiology 2008, 154, 3122–3133. [Google Scholar] [CrossRef] [PubMed]
- Lonardi, E.; Moonens, K.; Buts, L.; de Boer, A.R.; Olsson, J.D.; Weiss, M.W.S.; Fabre, E.; Guerardel, Y.; Deelder, A.M.; Oscarson, S.; et al. Structural sampling of glycan interaction profiles reveals mucosal receptors for fimbrial adhesins of enterotoxigenic Escherichia coli. Biology 2013, 2, 894–917. [Google Scholar] [CrossRef] [PubMed]
- Rich, R.L.; Cannon, M.J.; Jenkins, J.; Pandian, P.; Sundaram, S.; Magyar, R.; Brockman, J.; Lambert, J.; Myszka, D.G. Extracting kinetic rate constants from surface plasmon resonance array systems. Anal. Biochem. 2008, 373, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Hultgren, S.J.; Schwan, W.R.; Schaeffer, A.J.; Duncan, J.L. Regulation of production of type-1 pili among urinary tract isolates of Escherichia coli. Infect. Immun. 1986, 54, 613–620. [Google Scholar] [PubMed]
- Brument, S.; Sivignon, A.; Dumych, T.I.; Moreau, N.; Roos, G.; Guerardel, Y.; Chalopin, T.; Deniaud, D.; Bilyy, R.O.; Darfeuille-Michaud, A.; et al. Thiazolylaminomannosides as potent anti-adhesives of type-1 piliated Escherichia coli isolated from Crohn's disease patients. J. Med. Chem. 2013, 56, 5395–5406. [Google Scholar] [CrossRef] [PubMed]
- Chalopin, T.; Brissonnet, Y.; Sivignon, A.; Deniaud, D.; Cremet, L.; Barnich, N.; Bouckaert, J.; Gouin, S.G. Inhibition profiles of mono- and polyvalent FimH antagonists against 10 different Escherichia coli strains. Org. Biomol. Chem. 2015, 13, 11369–11375. [Google Scholar] [CrossRef] [PubMed]
- Sivignon, A.; Yan, X.; Alvarez Dorta, D.; Bonnet, R.; Bouckaert, J.; Fleury, E.; Bernard, J.; Gouin, S.G.; Darfeuille-Michaud, A.; Barnich, N. Development of heptylmannoside-based glycoconjugate antiadhesive compounds against adherent-invasive Escherichia coli bacteria associated with Crohn’s disease. MBio 2015. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, L.M.; Thomas, W.E.; Trintchina, E.; Vogel, V.; Sokurenko, E.V. Catch bond-mediated adhesion without a shear threshold—Trimannose versus monomannose interactions with the fimh adhesin of Escherichia coli. J. Biol. Chem. 2006, 281, 16656–16663. [Google Scholar] [CrossRef] [PubMed]
- Weissman, S.J.; Chattopadhyay, S.; Aprikian, P.; Obata-Yasuoka, M.; Yarova-Yarovaya, Y.; Stapleton, A.; Ba-Thein, W.; Dykhuizen, D.; HJohnson, J.R.; Sokurenko, E. Clonal analysis reveals high rate of structural mutations in fimbrial adhesins of extraintestinal pathogenic Escherichia coli. Mol. Microbiol. 2006, 59, 975–988. [Google Scholar] [CrossRef] [PubMed]
- Zakrisson, J.; Wiklund, K.; Axner, O.; Andersson, M. The shaft of the type-1 fimbriae regulates an external force to match the FimH catch bond. Biophys. J. 2013, 104, 2137–2148. [Google Scholar] [CrossRef] [PubMed]
- Cota, E.; Jones, C.; Simpson, P.; Altroff, H.; Anderson, K.L.; du Merle, L.; Guignot, J.; Servin, A.; Le Bougenec, C.; Mardon, H.; et al. The solution structure of the invasive tip complex from afa/dr fibrils. Mol. Microbiol. 2006, 62, 356–366. [Google Scholar] [CrossRef] [PubMed]
- Miller, E.; Garcia, T.; Hultgren, S.; Oberhauser, A.F. The mechanical properties of E. coli type-1 pili measured by atomic force microscopy techniques. Biophys. J. 2006, 91, 3848–3856. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, D.J.; Kalas, V.; Pinkner, J.S.; Chen, S.L.; Spaulding, C.N.; Dodson, K.W.; Hultgren, S.J. Positively selected FimH residues enhance virulence during urinary tract infection by altering FimH conformation. Proc. Natl. Acad. Sci. USA 2013, 110, 15530–15537. [Google Scholar] [CrossRef] [PubMed]
- Tchesnokova, V.; Aprikian, P.; Kisiela, D.; Gowey, S.; Korotkova, N.; Thomas, W.; Sokurenko, E. Type-1 fimbrial adhesin FimH elicits an immune response that enhances cell adhesion of Escherichia coli. Infect. Immun. 2011, 79, 3895–3904. [Google Scholar] [CrossRef] [PubMed]
- Kisiela, D.I.; Avagyan, H.; Friend, D.; Jalan, A.; Gupta, S.; Interlandi, G.; Liu, Y.; Tchesnokova, V.; Rodriguez, V.B.; Sumida, J.P. Inhibition and reversal of microbial attachment by an antibody with parasteric activity against the FimH adhesin of uropathogenic E. coli. PLoS Pathog. 2015, 11, e1004857. [Google Scholar] [CrossRef] [PubMed]
- Bouckaert, J.; Mackenzie, J.; de Paz, J.L.; Chipwaza, B.; Choudhury, D.; Zavialov, A.; Mannerstedt, K.; Anderson, J.; Pierard, D.; Wyns, L.; et al. The affinity of the FimH fimbrial adhesin is receptor-driven and quasi-independent of escherichia coli pathotypes. Mol. Microbiol. 2006, 61, 1556–1568. [Google Scholar] [CrossRef] [PubMed]
- Wellens, A.; Garofalo, C.; Nguyen, H.; van Gerven, N.; Slattegard, R.; Hernalsteens, J.P.; Wyns, L.; Oscarson, S.; de Greve, H.; Hultgren, S.; et al. Intervening with urinary tract infections using anti-adhesives based on the crystal structure of the FimH-oligomannose-3 complex. PLoS ONE 2008, 3, e2040. [Google Scholar] [CrossRef]
- Taganna, J.; de Boer, A.R.; Wuhrer, M.; Bouckaert, J. Glycosylation changes as important factors for the susceptibility to urinary tract infection. Biochem. Soc. Trans. 2011, 39, 349–354. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, L.M.; Thornas, W.E.; Sokurenko, E.V.; Vogel, V. Elevated shear stress protects Escherichia coli cells adhering to surfaces via catch bonds from detachment by soluble inhibitors. Appl. Environ. Microbiol. 2006, 72, 3005–3010. [Google Scholar] [CrossRef] [PubMed]
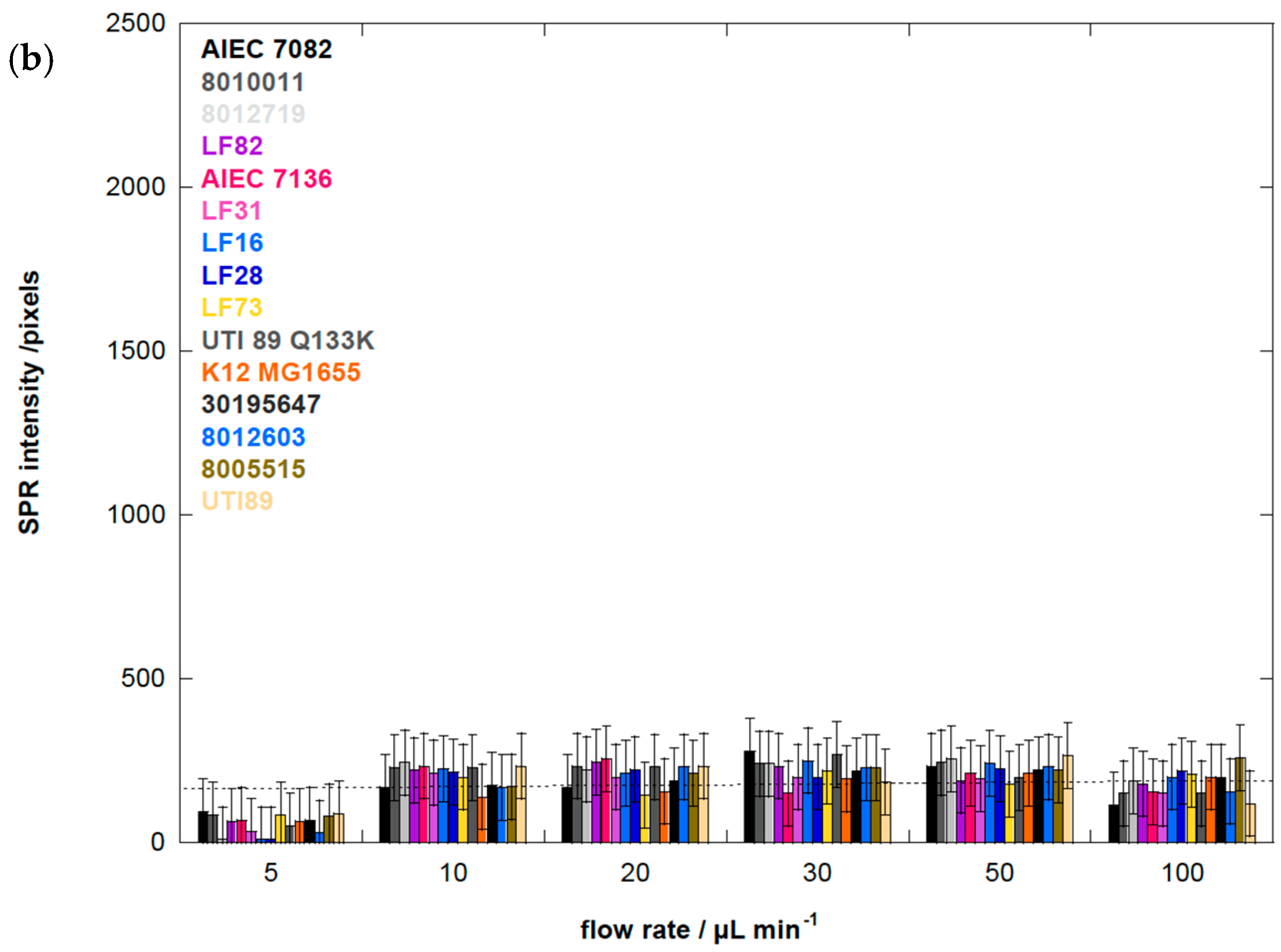
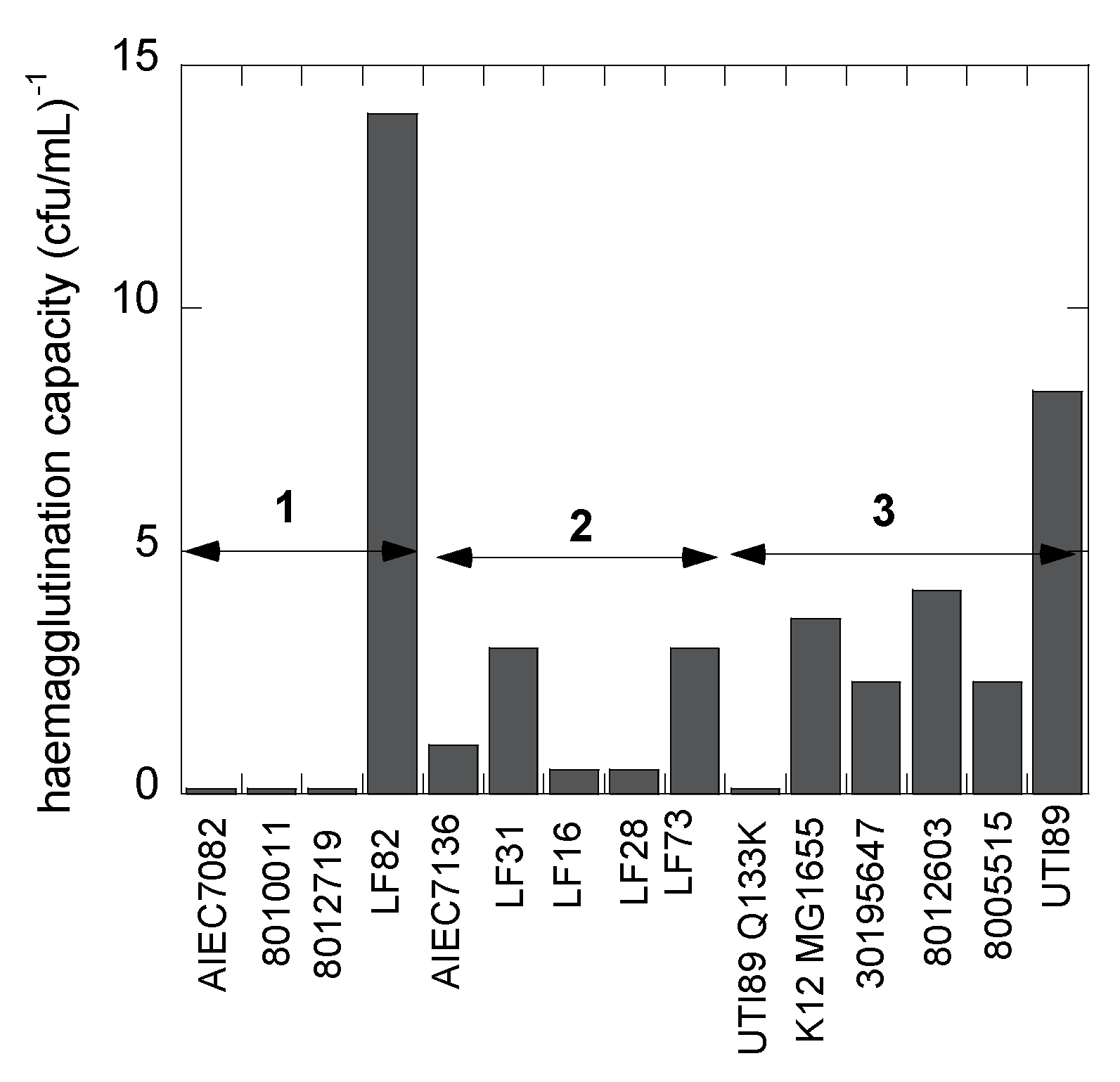
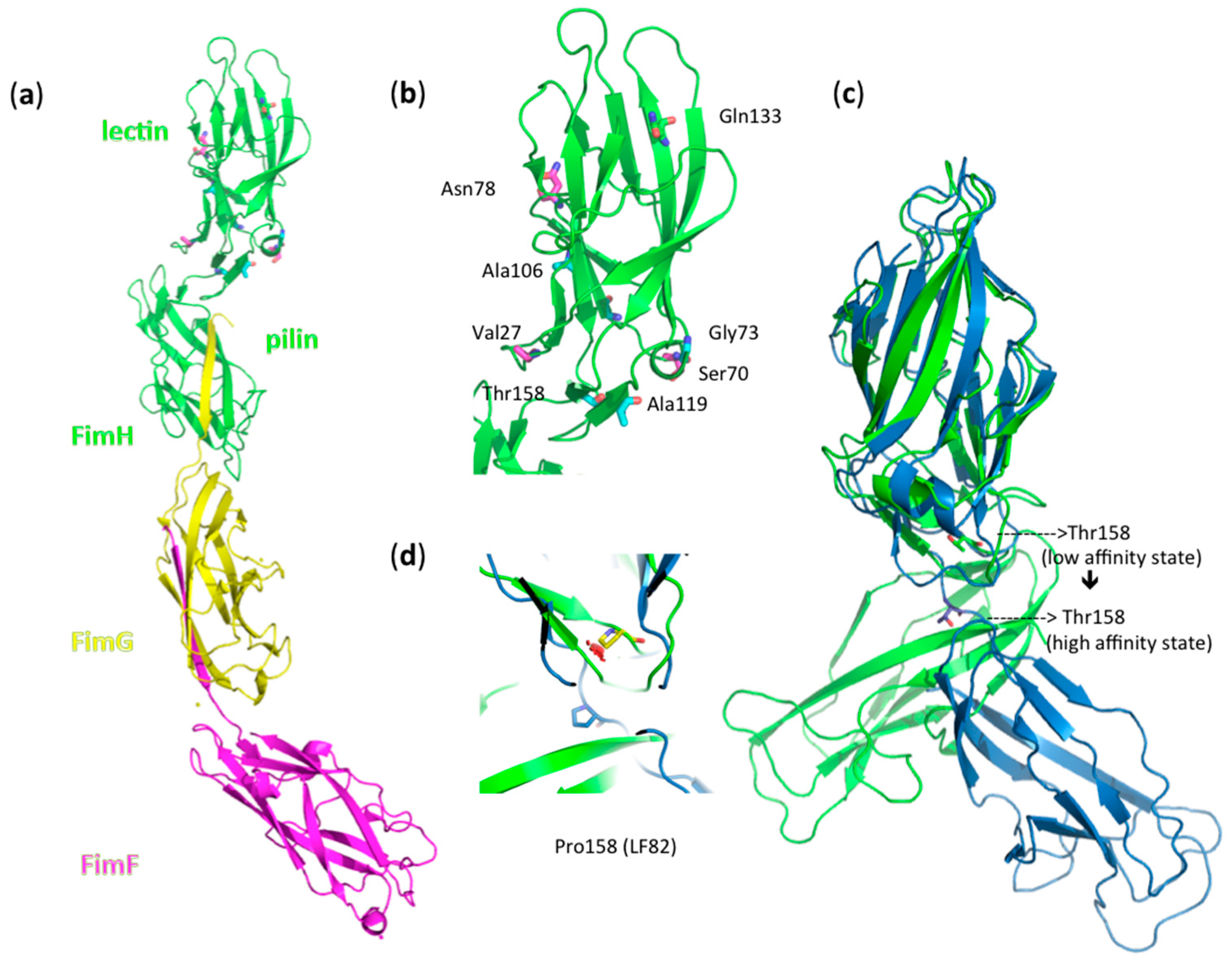
| E. coli Strain | fimH Sequence Amino Acid Differences with E. coli K12 MG1655 | Origin of E. coli Strain | Agglutination Titer (× s109 cfu·mL−1) | Group |
|---|---|---|---|---|
| 10 20 30 40 50 60 | ||||
| ACKTANGTA IPGGGSANV YVNLAPVNV GQNLVVDLST QIFCHPE TTYVTLQR | ||||
| 70 80 90 100 110 120 | ||||
| GSAYGVLS FSTVKYGS SYPFPTTSET PRVVYNSRTD KPWPVLYLT PVSSAGGVI | ||||
| 130 140 150 158 | ||||
| KAGSLIAVLI LRTNNSD DFQFVWNIYA NNDVVVP K12 MG1655 lectin domain | ||||
| AIEC7082 | ASN Thr158Ala | Crohn’s feces | neg. | 1 |
| 8010011 | ASN | knee prosthesis | neg. | 1 |
| 8012719 | ASN | hip prosthesis | neg. | 1 |
| LF82 | ASN Thr158Pro | Crohn’s biopsy | 0.07 | 1 |
| AIEC7136 | ASN Gly73Arg | Crohn’s feces | 0.97 | 2 |
| LF31 | ASN Gly66Ala Ala106Thr | Crohn’s biopsy | 0.30 | 2 |
| LF16 | ASN Gly73Trp | Crohn’s biopsy | 2.01 | 2 |
| LF28 | A Ala119Val | Crohn’s biopsy | 1.91 | 2 |
| LF73 | ASN | Crohn’s biopsy | 0.32 | 2 |
| UTI89 Q133K | ASN Gln133Lys | uropathogenic mutant FimH | neg. | 3 |
| K12 MG1655 | fecal | 0.28 | 3 | |
| 30195647 | fecal | 0.43 | 3 | |
| 8012603 | total hip replacement | 0.24 | 3 | |
| 8005515 | A Ala119Val | incomplete hip replacement | 0.42 | 3 |
| UTI89 | ASN | uropathogenic | 0.12 | 3 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szunerits, S.; Zagorodko, O.; Cogez, V.; Dumych, T.; Chalopin, T.; Alvarez Dorta, D.; Sivignon, A.; Barnich, N.; Harduin-Lepers, A.; Larroulet, I.; et al. Differentiation of Crohn’s Disease-Associated Isolates from Other Pathogenic Escherichia coli by Fimbrial Adhesion under Shear Force. Biology 2016, 5, 14. https://doi.org/10.3390/biology5020014
Szunerits S, Zagorodko O, Cogez V, Dumych T, Chalopin T, Alvarez Dorta D, Sivignon A, Barnich N, Harduin-Lepers A, Larroulet I, et al. Differentiation of Crohn’s Disease-Associated Isolates from Other Pathogenic Escherichia coli by Fimbrial Adhesion under Shear Force. Biology. 2016; 5(2):14. https://doi.org/10.3390/biology5020014
Chicago/Turabian StyleSzunerits, Sabine, Oleksandr Zagorodko, Virginie Cogez, Tetiana Dumych, Thibaut Chalopin, Dimitri Alvarez Dorta, Adeline Sivignon, Nicolas Barnich, Anne Harduin-Lepers, Iban Larroulet, and et al. 2016. "Differentiation of Crohn’s Disease-Associated Isolates from Other Pathogenic Escherichia coli by Fimbrial Adhesion under Shear Force" Biology 5, no. 2: 14. https://doi.org/10.3390/biology5020014
APA StyleSzunerits, S., Zagorodko, O., Cogez, V., Dumych, T., Chalopin, T., Alvarez Dorta, D., Sivignon, A., Barnich, N., Harduin-Lepers, A., Larroulet, I., Yanguas Serrano, A., Siriwardena, A., Pesquera, A., Zurutuza, A., Gouin, S. G., Boukherroub, R., & Bouckaert, J. (2016). Differentiation of Crohn’s Disease-Associated Isolates from Other Pathogenic Escherichia coli by Fimbrial Adhesion under Shear Force. Biology, 5(2), 14. https://doi.org/10.3390/biology5020014








