Biochemical and Functional Insights into the Integrated Regulation of Innate Immune Cell Responses by Teleost Leukocyte Immune-Type Receptors
Abstract
:1. Introduction
2. Examination of Stimulatory IpLITR-Mediated Responses
2.1. Stimulatory Immunoregulatory Receptors
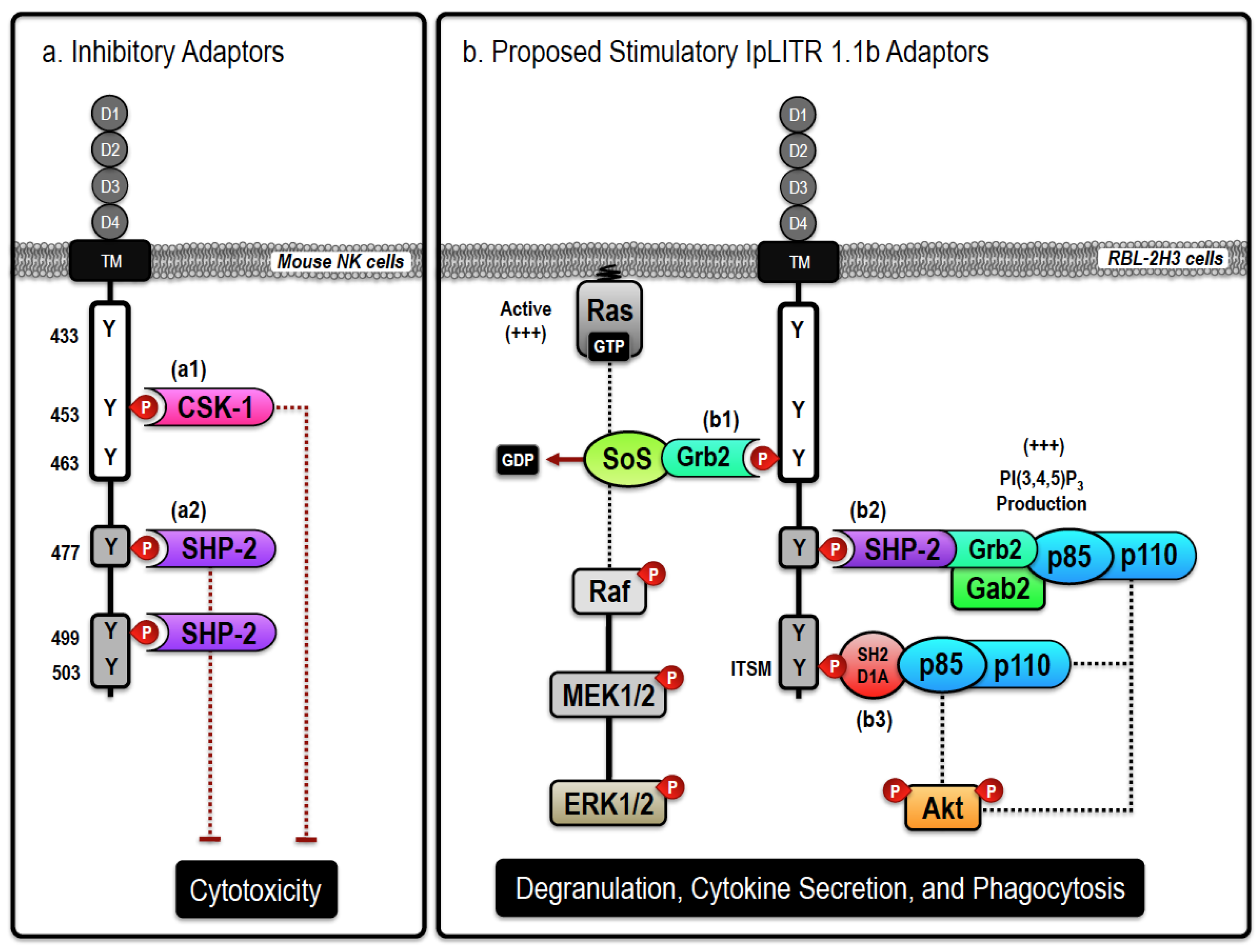
2.2. Stimulatory IpLITRs Demonstrate a Unique Ability to Associate with Intracellular Signaling Adaptors
2.3. Induction of Intracellular Signaling and Immune Cell Activation by a Stimulatory IpLITR-Adaptor Protein Complex
3. Examination of Inhibitory IpLITR-Mediated Responses
3.1. Inhibitory Immunoregulatory Receptors
3.2. Inhibitory IpLITRs Contain ITIMs and Recruit Protein Tyrosine Phosphatases
3.3. Inhibitory IpLITRs Abrogate the NK Cell Killing Response via Both SHP-1-Dependent and -Independent Signaling Pathways
4. Examination of IpLITR-Mediated Functional Plasticity
4.1. Functional Plasticity and Immunoregulatory Receptors
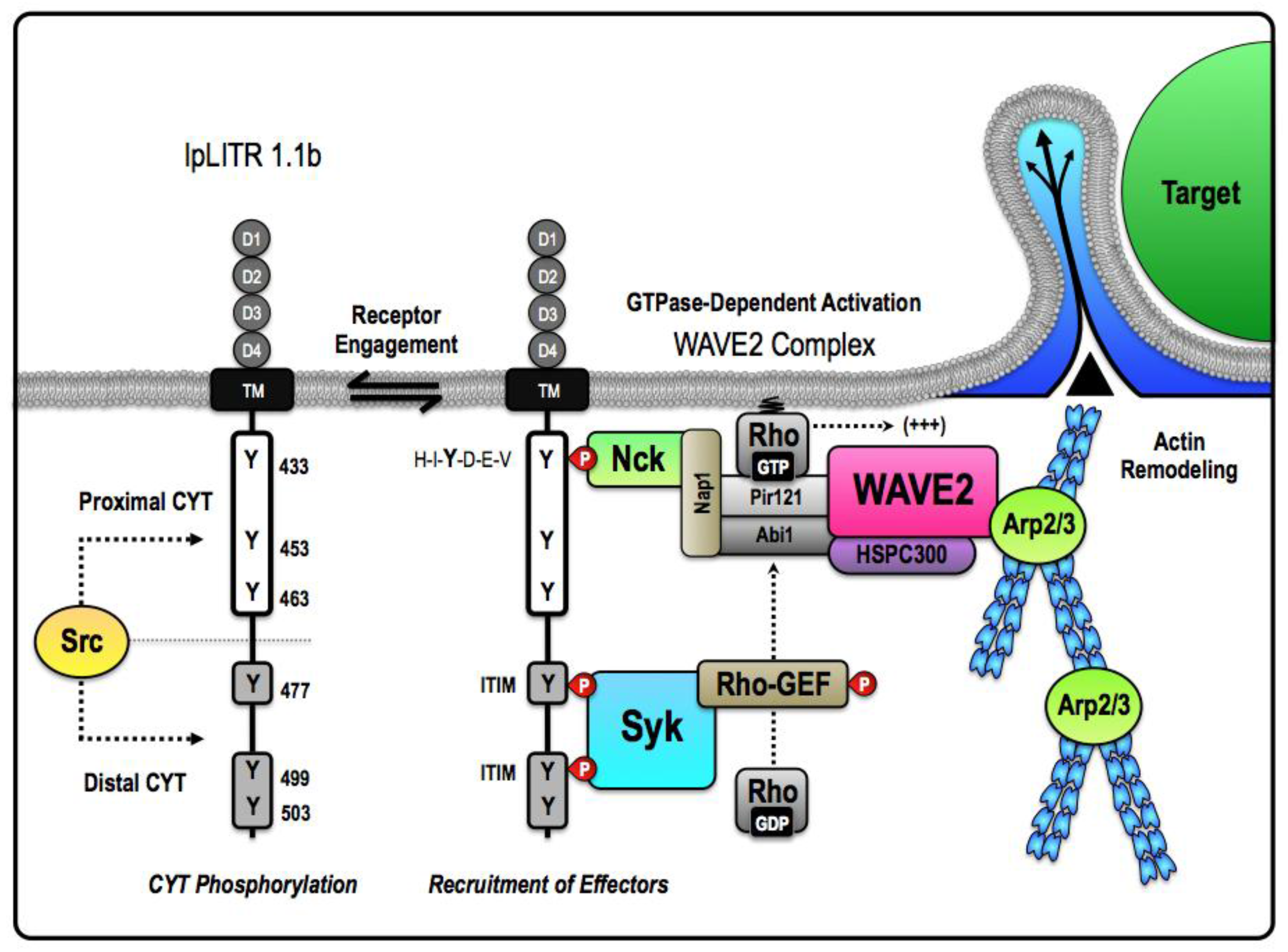
4.2. Induction of Phagocytosis and Stimulatory Signaling by an Inhibitory IpLITR
4.3. IpLITRs Activate Distinct Phagocytic Modes: Further Insights into IpLITR 1.1b-Mediated Functional Plasticity
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| Abs | antibodies |
| Akt | protein kinase B |
| CEACAM | carcinoembryonic antigen-related cell adhesion molecule |
| Csk | C-terminal Src kinase |
| CYT | cytoplasmic tail |
| EAT-2 | Ewing’s sarcoma (EWS)-activated transcript 2 |
| ERK | extracellular signal-regulated kinase |
| FcR | Fc receptor |
| FcRL | FcR-like protein |
| FcRg | FcR common gamma chain |
| Gab | Grb2-associated binder |
| GEF | guanine nucleotide exchange factors |
| Grb2 | growth factor receptor-bound 2 |
| HA | hemagglutinin |
| IgSF | immunoglobulin superfamily |
| ITAM | immunoreceptor tyrosine-based activation motif |
| ITIM | immunoreceptor tyrosine-based inhibition motif |
| ITSM | Immunoreceptor tyrosine-based switch motif |
| IL | interleukin |
| JNK | c-Jun N-terminal kinase |
| KIR | killer Ig-like receptor |
| LILR | leukocyte Ig-like receptor |
| LITR | leukocyte immune-type receptor |
| LRC | leukocyte receptor complex |
| mAb | monoclonal antibody |
| ERK | mitogen activated protein kinase |
| NITR | novel immune-type receptor |
| NK | natural killer |
| NKRs | NK receptors |
| PH | pleckstrin homology |
| PI3K | phosphatidylinositol 3-kinase |
| PI(3,4,5)P3 | phosphatidylinositol 3,4,5-trisphosphate |
| PTK | protein tyrosine kinases |
| PKC | protein kinase C |
| RBL | rat basophilic leukemia |
| SFK | Src family kinase |
| SHP | SH2 domain-containing cytoplasmic phosphatases |
| SH2D1A | SH2 domain protein 1A |
| SHIP | SH2-domain containing inositol 5-phosphatase |
| SoS | Son of Sevenless |
| Syk | spleen tyrosine kinase |
| TM | transmembrane |
| TNF-α | tumor necrosis factor-alpha |
| WAVE2 | WASp family verprolin-homologous protein-2 |
References
- Barrow, A.D.; Trowsdale, J. The extended leukocyte receptor complex: Diverse ways of modulating immune responses. Immunol. Rev. 2008, 224, 98–123. [Google Scholar] [CrossRef]
- Akula, S.; Mohammadamin, S.; Hellman, L. Fc Receptors and their appearance during vertebrate evolution. PLoS ONE 2014. [Google Scholar] [CrossRef] [PubMed]
- Barclay, A.N. Membrane proteins with immunoglobulin-like domains-a master superfamily of interaction molecules. Semin. Immunol. 2003, 15, 215–223. [Google Scholar] [CrossRef]
- Bezbradica, J.S.; Medzhitov, R. Role of ITAM signaling module in signal integration. Curr. Opin. Immunol. 2012, 24, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Engstad, R.E.; Robertsen, B. Recognition of yeast cell wall glucan by Atlantic salmon (Salmo salar L.) macrophages. Dev. Comp. Immunol. 1993, 17, 319–330. [Google Scholar] [CrossRef]
- Esteban, M.A.; Meseguer, J. Factors influencing phagocytic response of macrophages from the sea bass (Dicentrarchus labrax L.): An ultrastructural and quantitative study. Anat. Rec. 1997, 248, 533–541. [Google Scholar] [CrossRef]
- Frøystad, M.K.; Rode, M.; Berg, T.; Gjøen, T. A role for scavenger receptors in phagocytosis of protein-coated particles in rainbow trout head kidney macrophages. Dev. Comp. Immunol. 1998, 22, 533–549. [Google Scholar]
- Li, J.; Barreda, D.R.; Zhang, Y.A.; Boshra, H.; Gelman, A.E.; Lapatra, S.; Tort, L.; Sunyer, J.O. B lymphocytes from early vertebrates have potent phagocytic and microbicidal abilities. Nat. Immunol. 2006, 7, 1116–1124. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.A.; Salinas, I.; Li, J.; Parra, D.; Bjork, S.; Xu, Z.; LaPatra, S.E.; Bartholomew, J.; Sunyer, J.O. IgT, a primitive immunoglobulin class specialized in mucosal immunity. Nat. Immunol. 2010, 11, 827–835. [Google Scholar] [CrossRef] [PubMed]
- Stuge, T.B.; Wilson, M.R.; Zhou, H.; Barker, K.S.; Bengtén, E.; Chinchar, G.; Miller, N.W.; Clem, L.W. Development and analysis of various clonal alloantigen-dependent cytotoxic cell lines from channel catfish. J. Immunol. 2000, 15, 2971–2977. [Google Scholar] [CrossRef]
- Shen, L.; Stuge, T.B.; Bengtén, E.; Wilson, M.; Chinchar, V.G.; Naftel, J.P.; Bernanke, J.M.; Clem, L.W.; Miller, N.W. Identification and characterization of clonal NK-like cells from channel catfish (Ictalurus punctatus). Dev. Comp. Immunol. 2004, 28, 139–152. [Google Scholar] [CrossRef]
- Yoshida, S.H.; Stuge, T.B.; Miller, N.W.; Clem, L.W. Phylogeny of lymphocyte heterogeneity: Cytotoxic activity of channel catfish peripheral blood leukocytes directed against allogeneic targets. Dev. Comp. Immunol. 1995, 19, 71–77. [Google Scholar] [CrossRef]
- Vallejo, A.N.; Miller, N.W.; Clem, L.W. Phylogeny of immune recognition: Role of alloantigens in antigen presentation in channel catfish immune responses. Immunology 1991, 74, 165–168. [Google Scholar] [PubMed]
- Vallejo, A.N.; Miller, N.W.; Harvey, N.E.; Cuchens, M.A.; Warr, G.W.; Clem, L.W. Cellular pathway(s) of antigen processing and presentation in fish APC: Endosomal involvement and cell-free antigen presentation. Dev. Immunol. 1992, 3, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Dezfuli, B.S.; Giari, L. Mast cells in the gills and intestines of naturally infected fish: Evidence of migration and degranulation. J. Fish Dis. 2008, 31, 845–852. [Google Scholar] [CrossRef] [PubMed]
- Katzenback, B.A.; Belosevic, M. Isolation and functional characterization of neutrophil-like cells, from goldfish (Carassius auratus L.) kidney. Dev. Comp. Immunol. 2009, 33, 601–611. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Xu, W.; Wilson, M.; He, B.; Miller, N.W.; Bengtén, E.; Edholm, E-E.; Santini, P.A.; Rath, P.; Chiu, A.; et al. Immunoglobulin D enhances immune surveillance by activating antimicrobial, proinflammatory and B cell-stimulating programs in basophils. Nat. Immunol. 2009, 10, 889–898. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Nunez, I.; Wcisel, D.J.; Litman, G.W.; Yoder, J.A. Multigene families of immunoglobulin domain-containing innate immune receptors in zebrafish: Deciphering the differences. Dev. Comp. Immunol. 2014, 46, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Yoder, J.A.; Litman, G.W. The phylogenetic origins of natural killer receptors and recognition: Relationships, possibilities, and realities. Immunogenetics 2011, 63, 123–141. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, B.C.; Cortes, H.D.; Mewes-Ares, J.; Verheijen, K.; Stafford, J.L. Teleost IgSF immunoregulatory receptors. Dev. Comp. Immunol. 2011, 25, 1223–1237. [Google Scholar] [CrossRef] [PubMed]
- Strong, S.J.; Mueller, M.G.; Litman, R.T.; Hawke, N.A.; Haire, R.N.; Miracle, A.L.; Rast, J.P.; Amemiya, C.T.; Litman, G.W. A novel multigene family encodes diversified variable regions. Proc. Natl. Acad. Sci. USA 1999, 96, 15080–15085. [Google Scholar] [CrossRef] [PubMed]
- Yoder, J.A.; Mueller, M.G.; Wei, S.; Corliss, B.C.; Prather, D.M.; Willis, T.; Litman, R.T.; Djeu, J.Y.; Litman, G.W. Immune-type receptor genes in zebrafish share genetic and functional properties with genes encoded by mammalian leukocyte receptor cluster. Proc. Natl. Acad. Sci. USA 2001, 98, 6771–6776. [Google Scholar] [CrossRef] [PubMed]
- Cannon, J.P.; Haire, R.N.; Magis, A.T.; Eason, D.D.; Winfrey, K.N.; Hernandez Prada, J.A.; Bailey, K.M.; Jakoncic, J.; Litman, G.W.; Ostrov, D.A. A bony fish immunological receptor of the NITR multigene family mediates allogeneic recognition. Immunity 2008, 29, 228–237. [Google Scholar] [CrossRef] [PubMed]
- Yoder, J.A. Form, function and phylogenetics of NITRs in bony fish. Dev. Comp. Immunol. 2009, 33, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Litman, G.W.; Yoder, J.A.; Cannon, J.P.; Haire, R.N. Novel immune-type receptor genes and the origins of adaptive and innate immune recognition. Integr. Comp. Biol. 2003, 43, 331–337. [Google Scholar] [CrossRef] [PubMed]
- Litman, G.W.; Hawke, N.A.; Yoder, J.A. Novel immune-type receptor genes. Immunol. Rev. 2001, 181, 250–259. [Google Scholar] [CrossRef] [PubMed]
- Stafford, J.L.; Bengtén, E.; Du Pasquier, L.; McIntosh, R.D.; Quiniou, S.M.; Clem, L.W.; Miller, N.W.; Wilson, M. A novel family of diversified immunoregulatory receptors in teleosts is homologous to both mammalian Fc receptors and molecules encoded within the leukocyte receptor complex. Immunogenetics 2006, 58, 758–773. [Google Scholar] [CrossRef] [PubMed]
- Stafford, J.L.; Bengtén, E.; Du Pasquier, L.; Miller, N.W.; Wilson, M. Channel catfish leukocyte immune-type receptors contain a putative MHC class I binding site. Immunogenetics 2007, 59, 77–91. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Call, M.E.; Wucherpfennig, K.W. The assembly of diverse immune receptors is focused on a polar membrane-embedded interaction site. PLoS Biol. 2006. [Google Scholar] [CrossRef] [PubMed]
- Kuster, H.; Thompson, H.; Kinet, J.P. Characterization and expression of the gene for the human Fc receptor gamma subunit: Definition of a new gene family. J. Biol. Chem. 1999, 265, 6448–6452. [Google Scholar]
- Amoui, M.; Dráberová, L.; Tolar, P.; Dráber, P. Direct interaction of Syk and Lyn protein tyrosine kinases in rat basophilic leukemia cells activated via type I Fc epsilon receptors. Eur. J. Immunol. 1997, 27, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Mócsai, A.; Ruland, J.; Tybulewicz, V.L. The SYK tyrosine kinase: A crucial player in diverse biological functions. Nat. Rev. Immunol. 2010, 10, 387–402. [Google Scholar] [CrossRef] [PubMed]
- Underhill, D.M.; Goodridge, H.S. The many faces of ITAMs. Trends Immunol. 2007, 28, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Holowka, D.; Sil, D.; Torigoe, C.; Baird, B. Insights into immunoglobulin E receptor signaling from structurally defined ligands. Immunol. Rev. 2007, 217, 269–279. [Google Scholar] [CrossRef] [PubMed]
- Bakker, A.B.; Wu, J.; Phillips, J.H.; Lanier, L.L. NK cell activation: Distinct stimulatory pathways counterbalancing inhibitory signals. Hum. Immunol. 2000, 61, 18–27. [Google Scholar] [CrossRef]
- Lanier, L.L. Up on the tightrope: Natural killer cell activation and inhibition. Nat. Immunol. 2008, 9, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Van den Herik-Oudijk, I.E.; Ter Bekke, M.W.; Tempelman, M.J.; Capel, P.J.; van de Winkel, J.G. Functional differences between two Fc receptor ITAM signaling motifs. Blood 1995, 86, 3302–3307. [Google Scholar] [PubMed]
- Wu, J.; Cherwinski, H.; Spies, T.; Phillips, J.H.; Lanier, L.L. DAP10 and DAP12 form distinct, but functionally cooperative, receptor complexes in natural killer cells. J. Exp. Med. 2000, 192, 1059–1068. [Google Scholar] [CrossRef] [PubMed]
- Lanier, L.L.; Corliss, B.C.; Wu, J.; Leong, C.; Phillips, J.H. Immunoreceptor DAP12 bearing a tyrosine-based activation motif is involved in activating NK cells. Nature 1998, 391, 703–707. [Google Scholar] [CrossRef] [PubMed]
- Lanier, L.L.; Corliss, B.; Wu, J.; Phillips, J.H. Association of DAP12 with activating CD94/NKG2C NK cell receptors. Immunity 1998, 8, 693–701. [Google Scholar] [CrossRef]
- Yoder, J.A.; Orcutt, T.M.; Traver, D.; Litman, G.W. Structural characteristics of zebrafish orthologs of adaptor molecules that associate with transmembrane immune receptors. Gene 2007, 401, 154–164. [Google Scholar] [CrossRef] [PubMed]
- Mewes, J.; Verheijen, K.; Montgomery, B.C.; Stafford, J.L. Stimulatory catfish leukocyte immune-type receptors (IpLITRs) demonstrate a unique ability to associate with adaptor signaling proteins and participate in the formation of homo- and heterodimers. Mol. Immunol. 2009, 47, 318–331. [Google Scholar] [CrossRef] [PubMed]
- Takai, T.; Li, M.; Sylvestre, D.; Clynes, R.; Ravetch, J.V. FcR gamma chain deletion results in pleiotrophic effector cell defects. Cell 1994, 76, 519–529. [Google Scholar] [CrossRef]
- Kinet, J.P. The gamma-zeta dimers of Fc receptors as connectors to signal transduction. Curr. Opin. Immunol. 1992, 4, 43–48. [Google Scholar] [CrossRef]
- Cortes, H.D.; Montgomery, B.C.; Verheijen, K.; García-García, E.; Stafford, J.L. Examination of the stimulatory signaling potential of a channel catfish leukocyte immune-type receptor and associated adaptor. Dev. Comp. Immunol. 2012, 36, 62–73. [Google Scholar] [CrossRef] [PubMed]
- Cortes, H.D.; Lillico, D.M.; Zwozdesky, M.A.; Pemberton, J.G.; O'Brien, A.; Montgomery, B.C.; Wiersma, L.; Chang, J.P.; Stafford, J.L. Induction of phagocytosis and intracellular signaling by an inhibitory channel catfish leukocyte immune-type receptor, evidence for immunoregulatory functional plasticity in teleosts. J. Innate Immun. 2014, 6, 435–455. [Google Scholar] [CrossRef] [PubMed]
- Ghazizadeh, S.; Bolen, J.B.; Fleit, H.B. Tyrosine phosphorylation and association of Syk with FcγRII in monocytic THP-1 cells. Biochem. J. 1995, 305, 669–674. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.A.; Pleiman, C.M.; Pao, L.; Schneringer, J.; Hippen, K.; Cambier, J.C. Phosphorylated immunoreceptor signaling motifs (ITAMs) exhibit unique abilities to bind and activate Lyn and Syk tyrosine kinases. J. Immunol. 1995, 155, 4596–4603. [Google Scholar] [PubMed]
- Crowley, M.T.; Costello, P.S.; Fitzer-Attas, C.J.; Turner, M.; Meng, F.; Lowell, C.; Tybulewicz, V.L.; DeFranco, A.L. A critical role for Syk in signal transduction and phagocytosis mediated by Fcgamma receptors on macrophages. J. Exp. Med. 1997, 186, 1027–1039. [Google Scholar] [CrossRef] [PubMed]
- Kiefer, F.; Brumell, J.; Al-Alawi, N.; Latour, S.; Cheng, A.; Veillette, A.; Grinstein, S.; Pawson, T. The Syk protein tyrosine kinase is essential for Fcgamma receptor signaling in macrophages and neutrophils. Mol. Cell Biol. 1998, 18, 4209–4220. [Google Scholar] [CrossRef] [PubMed]
- Ninomiya, N.; Hazeki, K.; Fukui, Y.; Seya, T.; Okada, T.; Hazeki, O.; Ui, M. Involvement of phosphatidylinositol 3-kinase in Fcγ receptor signaling. J. Biol. Chem. 1994, 269, 22732–22737. [Google Scholar] [PubMed]
- Patel, J.C.; Hall, A.; Caron, E. Vav regulates activation of Rac but not Cdc42 during FcgammaR-mediated phagocytosis. Mol. Biol. Cell 2002, 13, 1215–1226. [Google Scholar] [CrossRef] [PubMed]
- Hoppe, A.D.; Swanson, J.A. Cdc42, Rac1, and Rac2 display distinct patterns of activation during phagocytosis. Mol. Biol. Cell 2004, 15, 3509–3519. [Google Scholar] [CrossRef] [PubMed]
- Hall, A.B.; Gakidis, M.A.M.; Glogauer, M.; Wilsbacher, J.L.; Gao, S.; Wojciech, S.; Brugge, J.S. Requirements for Vav guanine nucleotide exchange factors and Rho GTPases in FcgammaR- and complement-mediated phagocytosis. Immunity 2006, 24, 305–316. [Google Scholar] [CrossRef] [PubMed]
- May, R.C.; Caron, E.; Hall, A.; Machesky, L.M. Involvement of the Arp2/3 complex in phagocytosis mediated by FcgammaR or CR3. Nat. Cell Biol. 2000, 2, 246–248. [Google Scholar] [PubMed]
- Park, H.; Cox, D. Cdc42 regulates Fcgamma receptor-mediated phagocytosis through the activation and phosphorylation of Wiskott-Aldrich syndrome protein (WASP) and neural WASP. Mol. Biol. Cell 2009, 20, 4500–4508. [Google Scholar] [CrossRef] [PubMed]
- Lillico, D.M.; Zwozdesky, M.A.; Pemberton, J.G.; Deutscher, J.M.; Jones, L.O.; Chang, J.P.; Stafford, J.L. Teleost leukocyte immune-type receptors activate distinct phagocytic modes for target acquisition and engulfment. J. Leukoc. Biol. 2015, 98, 235–248. [Google Scholar] [CrossRef] [PubMed]
- Vivier, E.; Daeron, M. Immunoreceptor tyrosine-based inhibition motifs. Immunol. Today 1997, 18, 286–291. [Google Scholar] [CrossRef]
- Burshtyn, D.N.; Yang, W.; Yi, T.; Long, E.O. A novel phosphotyrosine motif with a critical amino acid at position-2 for the SH2 domain-mediated activation of the tyrosine phosphatase SHP-1. J. Biol. Chem. 1997, 272, 13066–13072. [Google Scholar] [CrossRef] [PubMed]
- Burshtyn, D.N.; Lam, A.S.; Weston, M.; Gupta, N.; Warmerdam, P.A.; Long, E.O. Conserved residues amino-terminal of cytoplasmic tyrosines contribute to the SHP-1-mediated inhibitory function of killer cell Ig-like receptors. J. Immunol. 1999, 162, 897–902. [Google Scholar] [PubMed]
- Long, E.O. Regulation of immune responses through inhibitory receptors. Annu. Rev. Immunol. 1999, 17, 875–904. [Google Scholar] [CrossRef] [PubMed]
- Ravetch, J.V.; Lanier, L.L. Immune Inhibitory Receptor. Science 2000, 290, 84–89. [Google Scholar] [CrossRef] [PubMed]
- Sidorenko, S.P.; Clark, E.A. The dual-function CD150 receptor subfamily: The viral attraction. Nat. Immunol. 2003, 4, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, B.C.; Mewes, J.; Davidson, C.; Burshtyn, D.N.; Stafford, J.L. Cell surface expression of channel catfish leukocyte immune-type receptors (IpLITRs) and recruitment of both Src homology 2 domain-containing protein tyrosine phosphatase (SHP)-1 and SHP-2. Dev. Comp. Immunol. 2009, 33, 570–582. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, B.C.; Cortes, H.D.; Burshtyn, D.N.; Stafford, J.L. Channel catfish leukocyte immune-type receptor mediated inhibition of cellular cytotoxicity is facilitated by SHP-1-dependent and -independent mechanisms. Dev. Comp. Immunol. 2012, 37, 151–163. [Google Scholar] [CrossRef] [PubMed]
- Songyang, Z.; Shoelson, S.E.; McGlade, J.; Olivier, P.; Pawson, T.; Bustelo, X.R. Specific motifs recognized by the SH2 domains of Csk, 3BP2, fps/fes, GRB-2, HCP, SHC, Syk, and Vav. Mol. Cell. Biol. 1994, 14, 2777–2785. [Google Scholar] [CrossRef] [PubMed]
- Okada, M.; Nada, S.; Yamanashi, Y.; Yamamoto, T.; Nakagawa, H. Csk: A protein-tyrosine kinase involved in regulation of Src family kinases. J. Biol. Chem. 1991, 266, 24249–24252. [Google Scholar] [PubMed]
- Chong, Y-P.; Mulhern, T.D.; Cheng, H.-C. C-terminal kinase (Csk) and CSK-homologous kinase (CHK)-endogenous negative regulators of Src-family protein kinases. Growth Factors 2005, 23, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Verbrugge, A.; Rijkers, E.S.K.; de Ruiter, T.; Meyaard, L. Leukocyte associated Ig-like receptor-1 has SH-2 domain-containing phosphatase independent function and recruits C-terminal Src kinase. Eur. J. Immmunol. 2006, 36, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Sayos, J.; Martinez-Barriocanal, A.; Kitzig, F.; Bellon, T.; Lopez-Botet, M. Recruitment of C-terminal Src kinase by the leukocyte inhibitory receptor CD85j. Biochem. Biophys. Res. Commun. 2004, 324, 640–647. [Google Scholar] [CrossRef] [PubMed]
- Àlvarez-Errico, D.; Sayós, J.; López-Botet, N. The IREM-1 (CD300f) inhibitory receptor associates with the p85 α subunit of phosphoinositide 3-kinase. J. Immunol. 2007, 178, 808–816. [Google Scholar] [CrossRef] [PubMed]
- Izawa, K.; Kitaura, J.; Yamanishi, Y.; Matsuoka, T.; Kaitani, A.; Sugiuchi, M.; Takahashi, M.; Maehara, A.; Enomoto, Y.; Oki, T.; et al. An activating and inhibitory signal from an inhibitory receptor LMIR3/CLM-1: LMIR3 augments lipopolysaccharide response through association with FcRγ in mast cells. J. Immunol. 2009, 183, 925–936. [Google Scholar] [CrossRef] [PubMed]
- Newman, P.J.; Newman, D.K. Signal transduction pathways mediated by PECAM-1: New roles for an old molecule in platelet and vascular cell biology. Arterioscler. Thromb. Vasc. Biol. 2003, 23, 953–964. [Google Scholar] [CrossRef] [PubMed]
- Faure, M.; Long, E.O. KIR2DL4 (CD158d), an NK cell-activating receptor with inhibitory potential. J. Immunol. 2002, 168, 6208–6214. [Google Scholar] [CrossRef] [PubMed]
- Rajagopalan, S.; Fu, J.; Long, E.O. Cutting edge: Induction of IFN-gamma production but not cytotoxicity by the killer cell Ig-like receptor KIR2DL4 (CD185d) in resting NK cells. J. Immunol. 2001, 167, 1877–1881. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi-Maki, A.; Yusa, S.; Catina, T.L.; Campbell, K.S. KIR2DL4 is an IL-2 regulated NK cell receptor that exhibits limited expression in humans but triggers strong IFN gamma production. J. Immunol. 2003, 171, 3415–3425. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi-Maki, A.; Catina, T.L.; Campbell, K.S. Cutting edge: KIR2DL4 transduces signals into human NK cells through association with the Fc receptor gamma protein. J. Immunol. 2005, 174, 3859–3863. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhou, Y.; Yang, Q.; Mu, C.; Duan, E.; Chen, J.; Yang, M.; Xia, P.; Cui, B. Ligation of Fc gamma receptor IIB enhances levels of antiviral cytokine in response to PRRSV infection in vitro. Vet. Microbiol. 2012, 160, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Blank, U.; Launay, P.; Benhamou, M.; Monteiro, R.C. Inhibitory ITAMs as novel regulators of immunity. Immunol. Rev. 2009, 232, 59–71. [Google Scholar] [CrossRef] [PubMed]
- Ivashkiv, L.B. How ITAMs inhibit signaling. Sci. Signal. 2011. [Google Scholar] [CrossRef] [PubMed]
- Pfirsch-Maisonnas, S.; Aloulou, M.; Xu, T.; Claver, J.; Kanamaru, T.; Tiwari, M.; Launay, P.; Monteiro, R.C.; Blank, U. Inhibitory ITAM signaling traps activating receptors with the phosphatase SHP-1 to form polarized “inhibisome” clusters. Sci. Signal. 2011. [Google Scholar] [CrossRef] [PubMed]
- Chemnitz, J.M.; Parry, R.V.; Nichols, K.E.; June, C.H.; Riley, J.L. SHP-1 and SHP-2 associate with immunoreceptor tyrosine-based switch motif of programmed death 1 upon primary human T cell stimulation, but only receptor ligation prevents T cell activation. J. Immunol. 2004, 173, 945–954. [Google Scholar] [CrossRef] [PubMed]
- Shlapatska, L.M.; Mikhalap, S.V.; Berdova, A.G.; Zelensky, O.M.; Yun, T.J.; Nichols, K.E.; Clark, E.A.; Sidorenko, S.P. CD150 association with either the SH2 containing inositol phosphatase or the SH2-containing protein tyrosine phosphatase is regulated by the adaptor protein SH2D1A. J. Immunol. 2001, 166, 5480–5487. [Google Scholar] [CrossRef] [PubMed]
- Mikhalap, S.V.; Shlapatska, L.M.; Berdova, A.G.; Law, C.L.; Clark, E.A.; Sidorenko, S.P. CDw150 associates with src-homology 2-containing inositol phosphatase and modulates CD96-mediated apoptosis. J. Immunol. 1999, 15, 5719–5727. [Google Scholar]
- Aoukaty, A.; Tan, R. Association of the X-linked lymphoproliferative disease gene product SAP/SH2D1A with 2B4, a natural killer cell-activating molecule, is dependent on phosphoinositide 3-kinase. J. Biol. Chem. 2002, 277, 13331–13337. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Iosef, C.; Jia, C.Y.; Han, V.K.; Li, S.S. Dual functional roles for the X-linked lymphoproliferative syndrome gene product SAP/SH2D1A in signaling through the signaling lymphocyte activation molecule (SLAM) family of immune receptors. J. Biol. Chem. 1993, 278, 3852–3859. [Google Scholar] [CrossRef] [PubMed]
- Brummer, T.; Schmitz-Peiffer, C.; Daly, R.J. Docking proteins. FEBS J. 2010, 277, 4356–4369. [Google Scholar] [CrossRef] [PubMed]
- Belov, A.A.; Mohammadi, N. Grb2, a double-edged sword of receptor tyrosine kinase signaling. Sci. Signal. 2012. [Google Scholar] [CrossRef] [PubMed]
- Gotoh, N. Regulation of growth factor signaling by FRS2 family docking/scaffold adaptor proteins. Cancer Sci. 2008, 99, 1319–1325. [Google Scholar] [CrossRef] [PubMed]
- Gu, H.; Saito, K.; Klaman, L.D.; Shen, J.; Fleming, T.; Wang, Y.; Pratt, J.C.; Lin, G.; Lim, B.; Kinet, J.P.; et al. Essential role for Gab2 in the allergic response. Nature 2001, 412, 186–190. [Google Scholar] [CrossRef] [PubMed]
- Bohdanowicz, M.; Cosío, G.; Backer, J.M.; Grinstein, S. Class I and class III phosphoinositide 3-kinases are required for actin polymerization that propels phagosomes. J. Cell Biol. 2010, 191, 999–1012. [Google Scholar] [CrossRef] [PubMed]
- Gu, H.; Botelho, R.J.; Yu, M.; Grinstein, S.; Neel, B.G. Critical role for scaffolding adapter Gab2 in Fc gamma R-mediated phagocytosis. J. Cell Biol. 2003, 161, 1151–1161. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Nishimura, R.; Kashishian, A.; Batzer, A.G.; Kim, W.J.; Cooper, J.A.; Schlessinger, J. A new function for a phosphotyrosine phosphatase: Linking GRB2-Sos to a receptor tyrosine kinase. Mol. Cell. Biol. 1994, 12, 509–517. [Google Scholar] [CrossRef]
- Bennett, A.M.; Tang, T.L.; Sugimoto, S.; Walsh, C.T.; Neel, B.G. Protein-tyrosine phosphatase SHPTP2 couples platelet-derived growth factor receptor β to Ras. Proc. Natl. Acad. Sci. USA 1991, 91, 7335–7339. [Google Scholar] [CrossRef]
- Gu, H.; Botelho, R.J.; Yu, M.; Grinstein, S.; Neel, B.G. Critical role for scaffolding adapter Gab2 in FcγR-mediated phagocytosis. J. Cell. Biol. 2003, 161, 1151–1161. [Google Scholar] [CrossRef] [PubMed]
- Lowenstein, E.J.; Daly, R.J.; Batzer, A.G.; Li, W.; Margolis, B.; Lammers, R.; Ullrich, A.; Skolnik, E.Y.; Bar-Sagi, D.; Schlessinger, J. The SH2 and SH3 domain containing protein GRB2 links receptor tyrosine kinases to ras signaling. Cell 1992, 70, 431–442. [Google Scholar] [CrossRef]
- Simon, M.A.; Bowtell, D.D.; Dodson, G.S.; Laverty, T.R.; Rubin, G.M. Ras1 and a putative guanine nucleotide exchange factor perform crucial steps in signaling by the sevenless protein tyrosine kinase. Cell 1991, 67, 701–716. [Google Scholar] [CrossRef]
- Li, N.; Batzer, A.; Daly, R.; Yajnik, V.; Skolnik, E.; Chardin, P.; Bar-Sagi, D.; Margolis, B.; Schlessinger, J. Guanine-nucleotide-releasing factor hSos1 binds to Grb2 and links receptor tyrosine kinases to Ras signalling. Nature 1993, 363, 85–88. [Google Scholar] [CrossRef] [PubMed]
- Araki, N.; Johnson, M.T.; Swanson, J.A. A role for phosphoinositide 3-kinase of macropinocytosis and phagocytosis by macrophages. J. Cell. Biol. 1996, 135, 1249–1260. [Google Scholar] [CrossRef] [PubMed]
- Cox, D.; Tseng, C.C.; Bjekic, G.; Greenberg, S. A requirement for phosphatidylinositol 3-kinase in pseudopod extension. J. Biol. Chem. 1999, 274, 1240–1247. [Google Scholar] [CrossRef] [PubMed]
- Katzav, S. Vav1: A hematopoietic signal transduction molecule involved in human malignancies. Int. J. Biochem. Cell. Biol. 2009, 41, 1245–1248. [Google Scholar] [CrossRef] [PubMed]
- Schmitter, T.; Pils, S.; Sakk, V.; Frank, R.; Fischer, K-D.; Hauck, C.R. The granulocyte receptor carcinoembryonic antigen-related cell adhesion molecule 3 (CEACAM3) directly associates with Vav to promote phagocytosis of human pathogens. J. Immunol. 2007, 178, 3797–3805. [Google Scholar] [CrossRef] [PubMed]
- Pils, S.; Kopp, K.; Peterson, L.; Delgado Tascón, J.; Nyffenegger-Jann, N.J.; Hauck, C.R. The adaptor molecule Nck localizes the WAVE complex to promote actin polymerization during CEACAM3-mediated phagocytosis of bacteria. PLoS ONE 2012, 7, e32808. [Google Scholar] [CrossRef] [PubMed]
- Takenawa, T.; Suetsugu, S. The WASP-WAVE protein network, connecting the membrane to the cytoskeleton. Nat. Rev. Nol. Cell Biol. 2007, 8, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Frese, S.; Schubert, W.D.; Findeis, A.C.; Marquardt, T.; Roske, Y.S.; Stradal, T.E.; Heinz, D.W. The phosphotyrosine peptide binding specificity of Nck1 and Nck2 Src homology 2 domains. J. Biol. Chem. 2006, 281, 18236–18245. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, M.C. Phosphoregulation of the WAVE regulatory complex and signal integration. Semin. Cell Dev. Biol. 2013, 24, 272–279. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wu, Y.; Hu, H.; Wang, W.; Lu, Y.; Mao, H.; Liu, X.; Liu, Z.; Chen, B.G. Syk protein tyrosine kinase involves PECAM-1 signaling through tandem immunotyrosine inhibitory motifs in human THP-1 macrophages. Cell. Immunol. 2011, 279, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Rougerie, P.; Miskolci, V.; Cox, D. Generation of membrane structures during phagocytosis and chemotaxis of macrophages: Role and regulation of the actin cytoskeleton. Immunol. Rev. 2013, 256, 222–239. [Google Scholar] [CrossRef] [PubMed]
- Beemiller, P.; Zhang, Y.; Mohan, S.; Levinsohn, E.; Gaeta, I.; Hoppe, A.D.; Swanson, J.A. A Cdc42 activation cycle coordinated by PI 3-kinase during Fc receptor-mediated phagocytosis. Mol. Biol. Cell. 2010, 21, 470–480. [Google Scholar] [CrossRef] [PubMed]
- Niedergang, F.; Colucci-Guyon, E.; Dubois, T.; Raposo, G.; Chavrier, P. ADP ribosylation factor 6 is activated and controls membrane delivery during phagocytosis in macrophages. J. Cell Biol. 2003, 161, 1143–1150. [Google Scholar] [CrossRef] [PubMed]
- Cox, D.; Lee, D.J.; Dale, B.M.; Calafat, J.; Greenberg, S. A Rab11-containing rapidly recycling compartment in macrophages that promotes phagocytosis. Proc. Natl. Acad. Sci. USA 2000, 97, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Braun, V.; Fraisier, V.; Raposo, G.; Hurbain, I.; Sibarita, J.B.; Chavrier, P.; Galli, T.; Niedergang, F. TI-VAMP/VAMP7 is required for optimal phagocytosis of opsonised particles in macrophages. EMBO J. 2004, 23, 4166–4176. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.L.; Mason, D.; Schreiber, A.D.; Grinstein, S. Quantitative analysis of membrane remodeling at the phagocytic cup. Mol. Biol. Cell 2007, 18, 2883–2892. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, R.P.; Reményi, A.; Yeh, B.J.; Lim, W.A. Domains, motifs, and scaffolds: The role of modular interactions in the evolution and wiring of cell signaling circuits. Annu. Rev. Biochem. 2006, 75, 655–680. [Google Scholar] [CrossRef] [PubMed]
| Human FcRLs | Top Matching Catfish Sequences | E value | Score | ID | Pos. | Coverage |
|---|---|---|---|---|---|---|
| FcRL6 (XP_005245185) | FcRI (NP_001187150) | 4e-09 | 55.8 | 26% | 37% | 89% |
| FcRL5 (AAK31327) | LITR TS32.15 L2.1a (ABI23564) | 1e-19 | 165 | 26% | 40% | 90% |
| FcRL4 (NP_112572) | LITR TS32.15 L2.2c (ABI23567) | 3e-11 | 63.2 | 25% | 38% | 89% |
| FcRL3 (NP_443171) | LITR TS32.15 L1.1a (ABI16036) | 1e-18 | 88.2 | 25% | 39% | 90% |
| FcRL2 (NP_110391) | LITR TS32.17 L2.1a (ABI23578) | 6e-06 | 46.2 | 24% | 39% | 79% |
| FcRL1 (NP_443170) | Hepacam 2 (NP_00118775) | 2e-04 | 40.8 | 23% | 46% | 38% |
| Human FcRs | Top Matching Catfish Sequences | E value | Score | ID | Pos. | Coverage |
| FcyRI (NP_000557) | FcRI (NP_001187150) | 2e-17 | 79.0 | 26% | 40% | 96% |
| LITR TS32.15 L1.1a (ABI16036) | 2e-11 | 62.4 | 26% | 40% | 50% | |
| FcεRIα (XP_003821022) | LITR TS32.17 L1.1a (ABI16049) | 2e-07 | 48.5 | 29% | 43% | 96% |
| FcγRIIA (NP_001129691) | −no matches− | − | − | − | − | − |
| FcγRIIIA (NP_001121068) | LITR TS32.15 L2.1a (ABI23564) | 1e-07 | 49.7 | 25% | 44% | 77% |
| FcγRIIC (NP_963857) | −no matches− | − | − | − | − | − |
| FcγRIIIB (NP_001231682) | LITR TS32.15 L2.1a (ABI23564) | 1e-05 | 44.3 | 25% | 44% | 66% |
| FcγRIIB (NP_001002274) | −no matches− | − | − | − | − | − |
| FcµR (AAP36498) | −no matches− | − | − | − | − | − |
| PIGR (NP_002635) | −no matches− | − | − | − | − | − |
| FcαµR (NP_114418) | −no matches− | − | − | − | − | − |
| Human LRC Proteins | Top Matching Catfish Sequences | E value | Score | ID | Pos. | Coverage |
| FcαR (NP_001991) | −no matches− | − | − | − | − | − |
| NKp46 (O76036) | −no matches− | − | − | − | − | − |
| LAIR1 (NP_001275954) | −no matches− | − | − | − | − | − |
| LILRB1 (AAC51179) | LITR1 (AAW82352) | 6e-10 | 98.2 | 25% | 39% | 66% |
| LILRB2 (XP_011546934) | LITR TS32.15 L2.2c (ABI23567) | 9e-13 | 68.6 | 23% | 38% | 85% |
| LILRB3 (O75022) | LITR1 (AAW82352) | 4e-07 | 50.4 | 23% | 39% | 73% |
| LILRB4 (NP_001265355) | −no matches− | − | − | − | − | − |
| LILRB5 (NP_006831) | LITR1 (AAW82352) | 2e-10 | 60.8 | 24% | 38% | 66% |
| LILRA1 (NP_006854) | LITR TS32.15 L2.2c (ABI23567) | 2e-10 | 61.2 | 24% | 38% | 89% |
| LILRA2 (XP_011545392) | LITR1 (AAW82352) | 2e-13 | 70.1 | 24% | 38% | 66% |
| LILRA3 (Q8N6C8) | LITR TS32.17 L1.2b (ABII6052) | 4e-09 | 55.5 | 24% | 40% | 53% |
| LILRA4 (P59901) | LITR1 (AAW82352) | 2e-08 | 54.7 | 26% | 40% | 65% |
| LILRA5 (A6NI73) | −no matches− | − | − | − | − | − |
| LILRA6 (AGZ61988) | LITR1 (AAW82352) | 1e-09 | 51.2 | 23% | 38% | 68% |
| KIR3DL1 (ADM64608) | LITR3 (NP_001187136) | 7e-08 | 52.4 | 27% | 41% | 65% |
| KIR3DS1 (ABX88987) | LITR3 (NP_001187136) | 8e-07 | 48.9 | 27% | 41% | 65% |
| KIR2DL1 (AAC50335) | −no matches− | − | − | − | − | − |
| KIR2DS1 (XP_011546300) | −no matches− | − | − | − | − | − |
| KIR2DL4 (ABW73959) | −no matches− | − | − | − | − | − |
| Human Siglecs | Top Matching Catfish Sequences | E value | Score | ID | Pos. | Coverage |
| Siglec-1 (Q9BZZ2) | LITR3 (NP_001187136) | 2e-16 | 137 | 25% | 39% | 51% |
| Siglec-2 (NP_001762) | LITR TS32.15 L1.1a (ABI16036) | 7e-18 | 170 | 26% | 40% | 74% |
| Siglec-3 (P20138) | −no matches− | − | − | − | − | − |
| Siglec-4 (NP_002352) | −no matches− | − | − | − | − | − |
| Siglec-15 (Q6ZMC9) | −no matches− | − | − | − | − | − |
| Human CEACAMs | Top Matching Catfish Sequences | E value | Score | ID | Pos. | Coverage |
| Ceacam-1 (NP_001703) | Hepacam 2 (NP_00118775) | 4e-14 | 144 | 31% | 44% | 64% |
| LITR3 (NP_001187136) | 1e-09 | 58.5 | 24% | 43% | 65% | |
| Ceacam-3 (NP_291021) | Hepacam 2 (NP_00118775) | 9e-11 | 59.3 | 31% | 50% | 56% |
| Ceacam-4 (NP_001808) | −no matches− | − | − | − | − | − |
| Ceacam-5 (NP_001295327) | LITR3 (NP_001187136) | 1e-22 | 166 | 25% | 41% | 75% |
| Ceacam-6 (P40199) | Hepacam 2 (NP_00118775) | 4e-14 | 71.2 | 31% | 47% | 48% |
| LITR TS32.17 L2.2b (ABI23581) | 3e-09 | 56.2 | 27% | 41% | 56% | |
| Ceacam-7 (Q14002) | Hepacam 2 (NP_00118775) | 2e-07 | 49.7 | 36% | 52% | 32% |
| Ceacam-8 (NP_001807) | Hepacam 2 (NP_00118775) | 6e-10 | 58.5 | 27% | 42% | 51% |
| LITR TS32.15 L2.3b (ABI23569) | 2e-08 | 53.1 | 28% | 42% | 56% | |
| Mouse PIRs | Top Matching Catfish Sequences | E value | Score | ID | Pos. | Coverage |
| PIRA (NP_035217) | LITR TS32.15 L1.1a (ABI16036) | 1e-12 | 61.6 | 24% | 39% | 88% |
| PIRB (AAC53219) | LITR TS32.15 L1.1a (ABI16036) | 1e-11 | 105 | 24% | 39% | 92% |
| Chicken Receptors | Top Matching Catfish Sequences | E value | Score | ID | Pos. | Coverage |
| FcRL1-like (XP_015135539) | LITR TS32.17 L2.1a (ABI23578) | 2e-06 | 46.6 | 26% | 43% | 73% |
| FcRL2-like (NP_001090998) | IpFcRI (NP_001187150) | 4e-21 | 91.3 | 24% | 43% | 65% |
| LITR3 (NP_001187136) | 3e-11 | 63.2 | 25% | 41% | 69% | |
| CHIR-A (AAG37067) | LITR TS32.17 L2.1a (ABI23578) | 5e-09 | 53.9 | 28% | 45% | 75% |
| CHIR-B (AAG37068) | LITR1 (AAW82352) | 6e-10 | 101 | 27% | 42% | 83% |
| CHIR-AB1 (NP_001139613) | LITR TS32.15 L2.2c (ABI23567) | 3e-07 | 79.7 | 30% | 46% | 78% |
| Xenopus Receptors | Top Matching Catfish Sequences | E value | Score | ID | Pos. | Coverage |
| FcγRI-L (XP_012809801) | FcRI (NP_001187150) | 1e-18 | 84.0 | 34% | 45% | 63% |
| LITR TS32.15 L2.2c (ABI23567) | 1e-12 | 67.4 | 28% | 45% | 62% | |
| FcR3-L (XP_012824905) | FcRI (NP_001187150) | 6e-17 | 80.1 | 22% | 46% | 43% |
| LITR TS32.15 L2.3c (ABI23570) | 2e-15 | 76.6 | 25% | 43% | 61% | |
| FcR4-L (XP_012809008) | FcRI (NP_001187150) | 4e-18 | 83.2 | 21% | 44% | 58% |
| LITR TS32.15 L2.2c (ABI23567) | 6e-15 | 75.5 | 23% | 40% | 92% | |
| FcR5-L (XP_012825305) | LITR TS32.17 L2.1a (ABI23578) | 8e-19 | 89.7 | 25% | 44% | 41% |
| ILR-1 (XP_002938564) | LITR TS32.17 L2.2b (ABI23581) | 2e-12 | 102 | 28% | 44% | 67% |
| ILR-2 (NP_001121201) | LITR TS32.17 L2.1a (ABI23578) | 3e-12 | 65.1 | 20% | 45% | 69% |
| Human Signaling Proteins | Top Matching Catfish Sequences | E value | Score | ID | Pos. | Coverage |
| c-Src (P12931) | Lymphocyte PTP (AJW77401) | 0.0 | 560 | 59% | 77% | 82% |
| SYK (P43405) | SYK (NP_998008) * | 0.0 | 863 | 67% | 77% | 98% |
| PI3K p85α (NP_852664) | PI3K p85α (AHH41763) | 0.0 | 864 | 62% | 76% | 99% |
| Csk (NP_004374) | Csk-like (AJW77401) | 3e-105 | 318 | 40% | 58% | 95% |
| GRB2 (CAG467401) | GRB2 (NP_001187313) | 3e-151 | 421 | 91% | 95% | 100% |
| Gab2 (BAA76737) | Gab2 (XP_692935) | 0.0 | 793 | 63% | 75% | 99% |
| Nck1 (NP_001278928) | Nck1 (NP_001278928) * | 0.0 | 615 | 77% | 87% | 100% |
| SHP-1 (AAA36610) | SHP-1-like (XP_009290704) * | 0.0 | 783 | 66% | 78% | 95% |
| SHP-2 (BAA02740) | SHP-2-like (CBX19678) * | 0.0 | 1076 | 91% | 94% | 100% |
| SHIP-1 (AAB49680) | SHIP-1 (AJK26904) | 0.0 | 1164 | 56% | 67% | 99% |
| SH2D1A (NP_001108409) | SH2D1A (NP_001187495) | 2e-49 | 155 | 64% | 76% | 89% |
| Vav3 (EAW51251) | Vav3 (XP_009296581) * | 0.0 | 1173 | 68% | 80% | 100% |
| Rac1 (NP_008839) | Rac1 (AD027935) | 4e-136 | 381 | 94% | 98% | 100% |
| Cdc42 (AAM21109) | Cdc42 (NP_001188177) | 2e-134 | 377 | 96% | 97% | 100% |
| RhoA (P61586) | RhoA-like (NP_001187623) | 2e-138 | 387 | 95% | 98% | 100% |
| Wave2 (P61586) | Wave3 (NP_001074059) * | 1e-104 | 322 | 44% | 53% | 100% |
| Wave2 (NP_957375) * | 2e-104 | 313 | 76% | 85% | 38% | |
| WASp (NP_000368) | WASp (NP_956232) * | 1e-106 | 327 | 54% | 69% | 59% |
| N-WASp (BAA20128) | N-WASp (NP_001076475) * | 3e-145 | 428 | 75% | 85% | 53% |
| Human Receptors | Teleost Matches | E value | Score | ID | Pos. | Coverage |
|---|---|---|---|---|---|---|
| FcRL5 (AAK31327) | Zebrafish CD22 (XP_009293714) | 3e-46 | 184 | 27% | 43% | 94% |
| Salmon Sialoadhesin-like (XP_009293614) | 3e-46 | 184 | 26% | 45% | 97% | |
| Herring Sialoadhesin-like (XP_012675080) | 8e-33 | 316 | 30% | 45% | 96% | |
| Cichlid CD22-like (XP_014269289) | 1e-32 | 276 | 28% | 47% | 76% | |
| Herring FcRL5 (XP_012678524) | 4e-31 | 261 | 25% | 40% | 94% | |
| FcγRI (NP_000557) | Asian arowana FcRL5 (KPP56756) | 6e-21 | 98.2 | 29% | 44% | 91% |
| Trout unamed protien (CDQ78931) | 2e-18 | 88.6 | 27% | 44% | 81% | |
| Salmon FcγRI-like (ACN10126) | 4e-18 | 88.2 | 28% | 42% | 81% | |
| Catfish FcRI (NP_001187150) | 3e-15 | 79.0 | 26% | 40% | 96% | |
| Catfish TS32.15 L1.1a (ABI16036) | 4e-09 | 62.4 | 29% | 44% | 50% | |
| FcγRIIA (NP_001129691) * | Herring FcRL5 (XP_012678524) | 7e-10 | 208 | 31% | 41% | 79% |
| Yellow Croaker FcγRII-like (XP_010752023) | 9e-10 | 60.5 | 25% | 45% | 78% | |
| Tilapia FcγRIB-like (XP_013119926) | 2e-09 | 59.7 | 37% | 54% | 41% | |
| Mexican tetra CD22 (XP_015459483) | 3e-09 | 60.8 | 31% | 47% | 70% | |
| Herring FcRL3 (XP_012675232) | 2e-08 | 58.9 | 27% | 50% | 70% | |
| FcγRIIB (NP_001002274) * | Yellow Croaker FcγRII-like (XP_010752023) | 1e-11 | 65.9 | 28% | 48% | 67% |
| Killifish unknown protein (XP_013889184) | 2e-10 | 65.1 | 27% | 43% | 82% | |
| Mexican tetra CD22 (XP_015459483) | 4e-10 | 63.9 | 31% | 46% | 61% | |
| Herring FcRL3 (XP_012675232) | 2e-09 | 62.0 | 28% | 51% | 70% | |
| Cichlid FcγRII-like (XP_006808904) | 3e-08 | 58.2 | 28% | 42% | 76% | |
| FcµR (AAP36498) * | Damselfish unknown protein (XP_008286766) | 4e-06 | 52.8 | 34% | 47% | 42% |
| Salmon CMRF35-like (XP_014058627) | 8e-06 | 51.2 | 34% | 47% | 42% | |
| Killifish PIGR-like (012722396) | 2e-05 | 50.1 | 29% | 41% | 40% | |
| PIGR (NP_002635) * | Tilapia PIGR-like (XP_013123119) | 4e-25 | 115 | 27% | 44% | 67% |
| Yellow Croaker PIGR (KKF27361) | 1e-23 | 111 | 25% | 39% | 81% | |
| Medaka PIGR (XP_011484914) | 3e-23 | 262 | 28% | 44% | 72% | |
| Carp PIGR (ADB97624) | 1e-18 | 92.8 | 31% | 47% | 73% | |
| Zebrafish PIGR (NP_001289179) | 7e-18 | 90.5 | 30% | 47% | 68% | |
| FcαR (NP_001991) * | Cichlid unknown protein (XP_014265321) | 7e-05 | 48.5 | 29% | 46% | 51% |
| NKp46 (O76036) * | Atlantic molly CD276-like (XP_014823336) | 4e-05 | 49.7 | 24% | 37% | 65% |
| Sailfin molly IgSF 1-like Rc (XP_014882122) | 4e-04 | 46.6 | 24% | 38% | 69% | |
| LAIR1 (NP_001275954) * | −no matches− | − | − | − | − | − |
| LILRB1 (AAC51179) | Pike DSCAM-like (XP_12987993) | 1e-15 | 84.3 | 28% | 41% | 85% |
| Pike FcRL5 (XP_010867787) | 6e-14 | 79.7 | 26% | 43% | 82% | |
| Salmon IgSF 1-like Rc (XP_01401166) | 2e-11 | 71.6 | 27% | 41% | 83% | |
| Pike LILRA4-like (XP_012988338) | 4e-10 | 64.3 | 28% | 44% | 42% | |
| Cafish LITR1 (AAW82352) | 2e-09 | 65.1 | 26% | 40% | 86% | |
| LILRB4 (NP_001265355) * | Cichlid IgSF 1-like Rc (XP_014267965) | 2e-11 | 68.9 | 27% | 47% | 78% |
| Amazon molly PEACAM-like (XP_007571377) | 4e-07 | 56.2 | 28% | 42% | 77% | |
| Atlantic molly IgSF 1-like Rc (XP_014827576) | 1e-05 | 51.6 | 29% | 45% | 78% | |
| Yellow Croaker LILRA2-like (KKF09113) | 2e-05 | 51.2 | 31% | 45% | 64% | |
| LILRA2 (XP_011545392) | Trout unamed protien (CDQ78931) | 1e-12 | 75.9 | 27% | 42% | 90% |
| Pike DSCAM-like (XP_12987993) | 5e-12 | 73.6 | 27% | 42% | 82% | |
| Catfish LITR1 (AAW82352) | 4e-11 | 70.1 | 24% | 38% | 66% | |
| Mexican tetra FcRL5 (XP_015464057) | 8e-11 | 69.7 | 28% | 41% | 72% | |
| Pike FcRL5 (XP_010867787) | 7e-10 | 66.7 | 27% | 42% | 77% | |
| LILRA5 (A6NI73) * | Pllatyfish IgSF 1-like Rc (XP_005816340) | 4e-06 | 53.1 | 28% | 40% | 72% |
| Cichlid KIR2DL4-like (XP_005951820) | 4e-05 | 48.9 | 35% | 48% | 40% | |
| Cichlid FcγRI-like (XP_014267764) | 6e-05 | 48.5 | 34% | 49% | 40% | |
| Cichlid IgSF 1-like Rc (XP_014267965) | 3e-04 | 47.0 | 32% | 45% | 44% | |
| KIR3DL1 (ADM64608) | Asian arowana FcRL5 (KPP56756) | 1e-11 | 70.5 | 25% | 42% | 63% |
| Salmon KIR3DL1-like (XP_014042396) | 2e-11 | 69.7 | 30% | 43% | 85% | |
| Cichlid IgSF 1-like Rc (XP_014267965) | 9e-08 | 70.5 | 25% | 43% | 85% | |
| Salmon KIR3DS1-like (XP_014043811) | 2e-07 | 55.5 | 31% | 44% | 56% | |
| Catfish LITR3 (NP_001187136) | 1e-05 | 52.0 | 27% | 41% | 65% | |
| KIR2DL1 (AAC50335) * | Salmon KIR3DS1-like (XP_014043811) | 1e-05 | 49.7 | 28% | 40% | 78% |
| Salmon KIR3DL1-like (XP_014042396) | 4e-05 | 49.3 | 28% | 40% | 78% | |
| Salmon LILRA4-like (XP_014037968) | 8e-05 | 48.5 | 29% | 41% | 75% | |
| Pike OSCAR-like (XP_012988534) | 1e-04 | 47.8 | 29% | 44% | 81% | |
| Pike DSCAM-like (XP_12987993) | 1e-04 | 48.1 | 26% | 43% | 81% | |
| Siglec-2 (NP_001762) | Zebrafish CD22 (XP_009293714) | 8e-71 | 738 | 30% | 45% | 99% |
| Cichlid CD22-like (XP_005755784) | 4e-64 | 221 | 33% | 52% | 67% | |
| Tilapia CD22-like (XP_013132285) | 6e-61 | 218 | 30% | 46% | 85% | |
| Trout unamed protien (CDQ84455) | 2e-58 | 212 | 31% | 47% | 86% | |
| Siglec-4 (NP_002352) * | Salmon Siglec-4 (MAG) (XP_014021122) | 4e-154 | 459 | 44% | 64% | 96% |
| Pike Siglec-4 (MAG) (XP_014021122) | 7e-152 | 454 | 43% | 63% | 96% | |
| Black cod Siglec-4 (MAG) (XP_010788688) | 4e-151 | 452 | 44% | 63% | 96% | |
| Tiger puffer Siglec-4 (MAG) (NP_001027876) | 2e-149 | 449 | 43% | 63% | 96% | |
| Ceacam-3 (NP_291021) | Herring Hemicentin-1-like (XP_012676907) | 1e-18 | 90.5 | 34% | 52% | 65% |
| Cichlid CEACAM 5-like (XP_014266060) | 4e-16 | 82.8 | 33% | 53% | 82% | |
| Salmon CEACAM 1-like (XP_014038441) | 1e-15 | 79.3 | 33% | 52% | 55% | |
| Medaka CEACAM 1-like (XP_011483887) | 5e-15 | 77.8 | 28% | 42% | 94% | |
| Ceacam-4 (NP_001808) * | −no matches− | − | − | − | − | − |
| Ceacam-5 (NP_001295327) | Tongue sole CEACAM 5-like (XP_008322222) | 4e-80 | 277 | 30% | 50% | 85% |
| Yellow Croaker CEACAM 5 (KKF27703) | 4e-75 | 918 | 32% | 53% | 98% | |
| Pike CEACAM 5 (XP_012994902) | 4e-69 | 570 | 32% | 48% | 86% | |
| Cichlid Hemicentin-1-like (XP_004550953) | 1e-65 | 892 | 30% | 47% | 83% | |
| Salmon Hemicentin-1-like (XP_014056825) | 3e-65 | 1041 | 30% | 46% | 86% |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fei, C.; Pemberton, J.G.; Lillico, D.M.E.; Zwozdesky, M.A.; Stafford, J.L. Biochemical and Functional Insights into the Integrated Regulation of Innate Immune Cell Responses by Teleost Leukocyte Immune-Type Receptors. Biology 2016, 5, 13. https://doi.org/10.3390/biology5010013
Fei C, Pemberton JG, Lillico DME, Zwozdesky MA, Stafford JL. Biochemical and Functional Insights into the Integrated Regulation of Innate Immune Cell Responses by Teleost Leukocyte Immune-Type Receptors. Biology. 2016; 5(1):13. https://doi.org/10.3390/biology5010013
Chicago/Turabian StyleFei, Chenjie, Joshua G. Pemberton, Dustin M. E. Lillico, Myron A. Zwozdesky, and James L. Stafford. 2016. "Biochemical and Functional Insights into the Integrated Regulation of Innate Immune Cell Responses by Teleost Leukocyte Immune-Type Receptors" Biology 5, no. 1: 13. https://doi.org/10.3390/biology5010013
APA StyleFei, C., Pemberton, J. G., Lillico, D. M. E., Zwozdesky, M. A., & Stafford, J. L. (2016). Biochemical and Functional Insights into the Integrated Regulation of Innate Immune Cell Responses by Teleost Leukocyte Immune-Type Receptors. Biology, 5(1), 13. https://doi.org/10.3390/biology5010013





