Patterns and Functional Insights of DNA Methylation Variation in a South American Mayfly Across an Agriculturally Impacted Semi-Arid Watershed
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Study System
2.2. Biological Sampling and MethylRAD Production
2.3. Reference Genome
2.4. Analysis of Methylome Structure Based on Geographical Locations
2.5. Analysis of Main Discriminant Methylome Structure Patterns
2.6. Functional Enrichment Analysis
3. Results
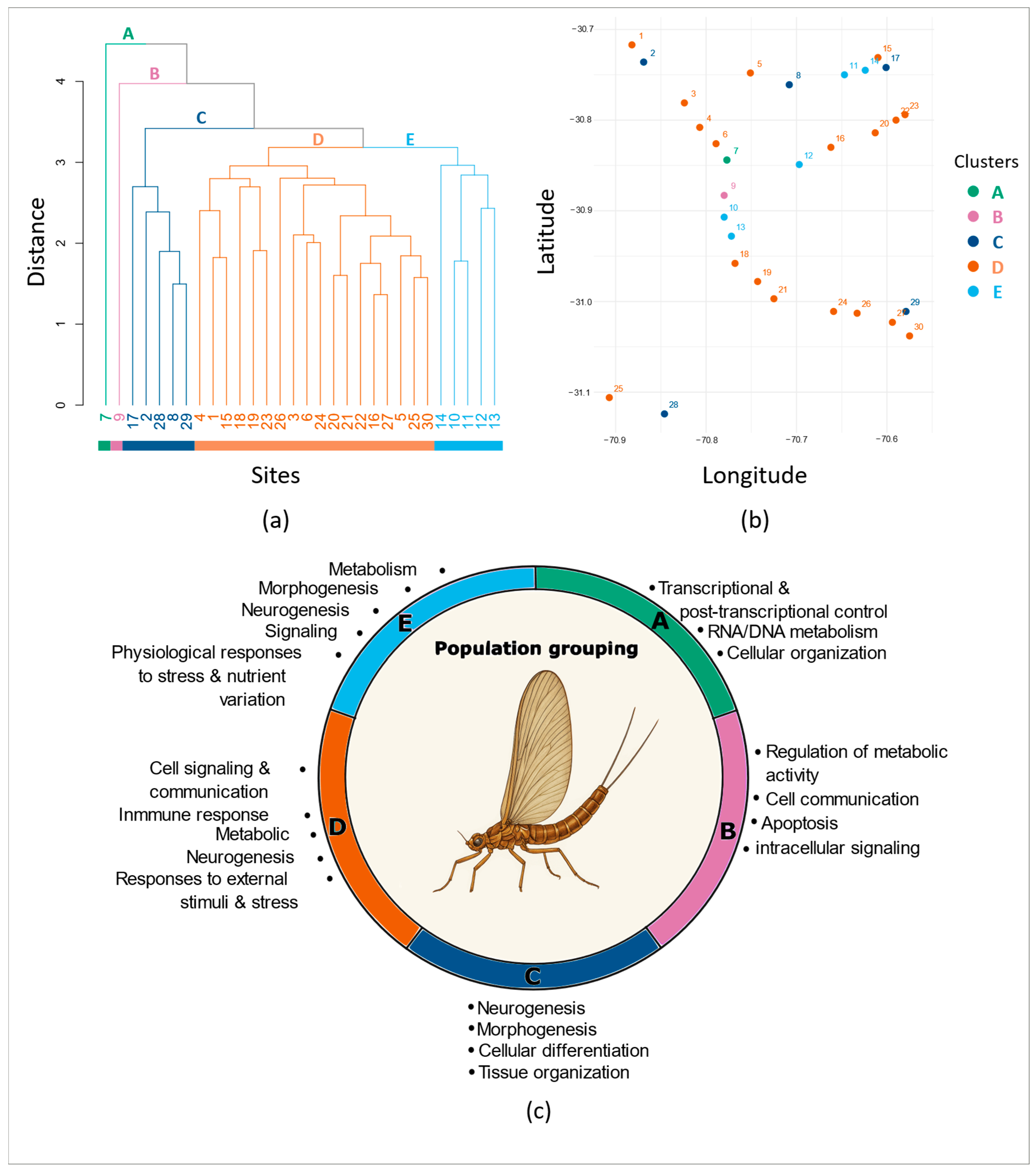
3.1. Methylome Patterns Across Geographical Locations
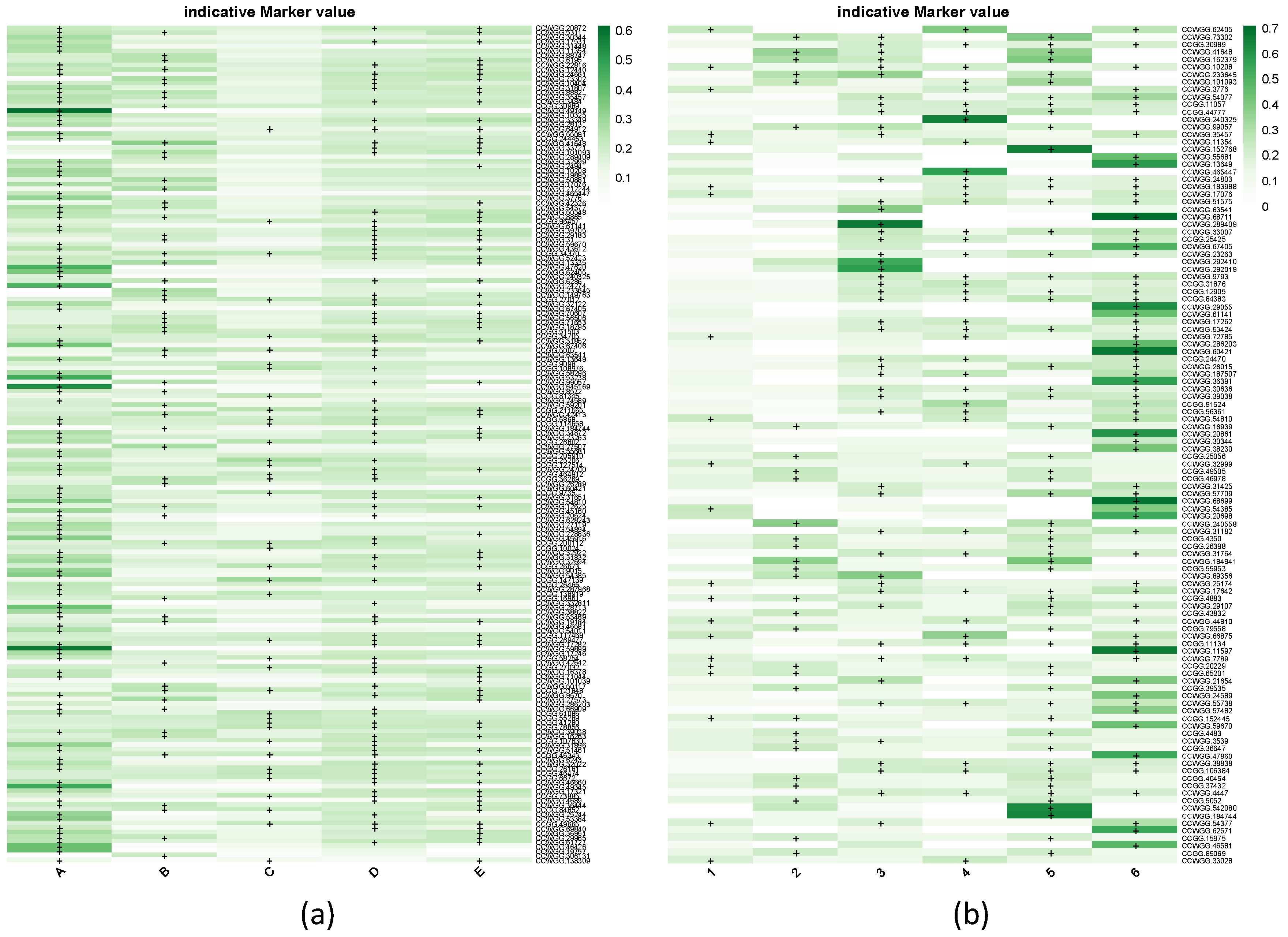
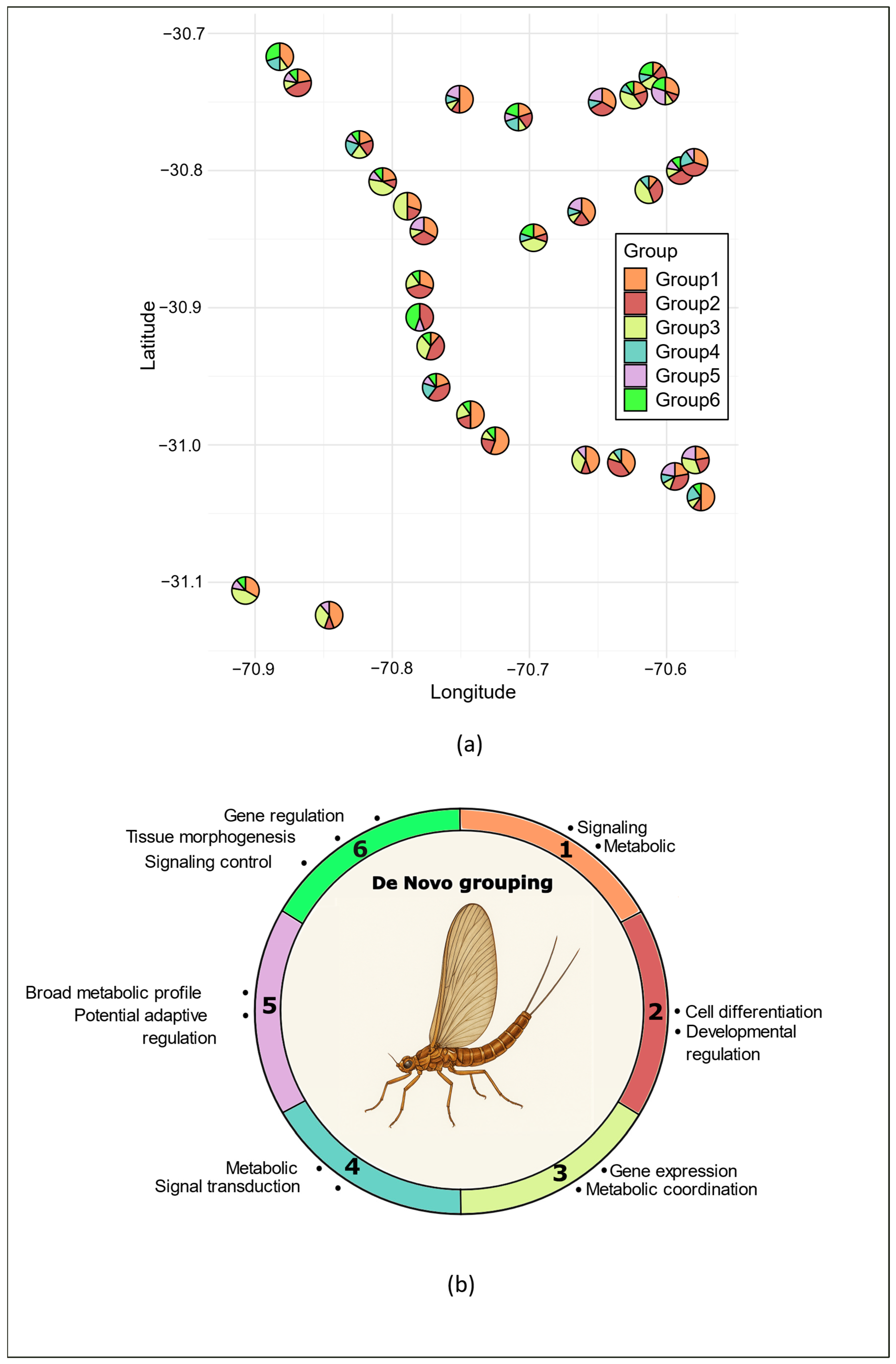
3.2. Main Methylome Structure Patterns
3.3. Genome Sequencing and Enrichment Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baltazar-Soares, M.; Balard, A.; Heckwolf, M.J. Epigenetic diversity and the evolutionary potential of wild populations. Evol. Appl. 2024, 17, e70011. [Google Scholar] [CrossRef] [PubMed]
- Brennan, R.S.; deMayo, J.A.; Finiguerra, M.; Baumann, H.; Dam, H.G.; Pespeni, M.H. Complementary genetic and epigenetic changes facilitate rapid adaptation to multiple global change stressors. Proc. Natl. Acad. Sci. USA 2025, 122, e2422782122. [Google Scholar] [CrossRef]
- Carneiro, V.C.; Lyko, F. Rapid epigenetic adaptation in animals and its role in invasiveness. Integr. Comp. Biol. 2020, 60, 267–274. [Google Scholar] [CrossRef]
- Hendry, A.P.; Farrugia, T.J.; Kinnison, M.T. Human influences on rates of phenotypic change in wild animal populations. Mol. Ecol. 2008, 17, 20–29. [Google Scholar] [CrossRef]
- Petitjean, Q.; Jean, S.; Gandar, A.; Côte, J.; Laffaille, P.; Jacquin, L. Stress responses in fish: From molecular to evolutionary processes. Sci. Total Environ. 2019, 684, 371–380. [Google Scholar] [CrossRef]
- Ashe, A.; Colot, V.; Oldroyd, B.P. How does epigenetics influence the course of evolution? Philos. Trans. R. Soc. B Biol. Sci. 2021, 376, 20200111. [Google Scholar] [CrossRef]
- Rey, O.; Danchin, E.; Mirouze, M.; Loot, C.; Blanchet, S. Adaptation to global change: A transposable element-epigenetics perspective. Trends Ecol. Evol. 2016, 31, 514–526. [Google Scholar] [CrossRef]
- Baduel, P.; Sammarco, I.; Barrett, R.; Coronado-Zamora, M.; Crespel, A.; Díez-Rodríguez, B.; Fox, J.; Galanti, D.; González, J.; Jueterbock, A.; et al. The evolutionary consequences of interactions between the epigenome, the genome and the environment. Evol. Appl. 2024, 17, e13730. [Google Scholar] [CrossRef]
- Husby, A. Wild epigenetics: Insights from epigenetic studies on natural populations. Proc. R. Soc. B Biol. Sci. 2022, 289, 20211633. [Google Scholar] [CrossRef] [PubMed]
- Guil, S.; Esteller, M. DNA methylomes, histone codes and miRNAs: Tying it all together. Int. J. Biochem. Cell Biol. 2009, 41, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Richards, E.J. Inherited epigenetic variation—revisiting soft inheritance. Nat. Rev. Genet. 2006, 7, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Becker, C.; Hagmann, J.; Müller, J.; Koenig, D.; Stegle, O.; Borgwardt, K.; Weigel, D. Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature 2011, 480, 245–249. [Google Scholar] [CrossRef] [PubMed]
- Johannes, F.; Schmitz, R.J. Spontaneous epimutations in plants. New Phytol. 2019, 221, 1253–1259. [Google Scholar] [CrossRef] [PubMed]
- Kronholm, I.; Collins, S. Epigenetic mutations can both help and hinder adaptive evolution. Mol. Ecol. 2016, 25, 1856–1868. [Google Scholar] [CrossRef]
- Lynch, M.; Burger, R.; Butcher, D.; Gabriel, W. The mutational meltdown in asexual populations. J. Hered. 1993, 84, 339–344. [Google Scholar] [CrossRef]
- Sammarco, I.; Rodríguez, B.D.; Galanti, D.; Nunn, A.; Becker, C.; Bossdorf, O.; Münzbergová, Z.; Latzel, V. DNA methylation in the wild: Epigenetic transgenerational inheritance can mediate adaptation in clones of wild strawberry (Fragaria vesca). New Phytol. 2024, 241, 1621–1635. [Google Scholar] [CrossRef]
- Latzel, V.; Allan, E.; Silveira, A.B.; Colot, V.; Fischer, M.; Bossdorf, O. Epigenetic diversity increases the productivity and stability of plant populations. Nat. Commun. 2013, 4, 2875. [Google Scholar] [CrossRef]
- Bogan, S.N.; Yi, S. Potential role of DNA methylation as a driver of plastic responses to the environment across cells, organisms, and populations. Genome Biol. Evol. 2024, 16, evae022. [Google Scholar] [CrossRef]
- Schrey, A.W.; Coon, C.A.C.; Grispo, M.T.; Awad, M.; Imboma, T.; McCoy, E.D.; Mushinsky, H.R.; Richards, C.L.; Martin, L.B. Epigenetic variation may compensate for decreased genetic variation with introductions: A case study using house sparrows (Passer domesticus) on two continents. Genet. Res. Int. 2012, 2012, 979751. [Google Scholar] [CrossRef]
- Schmitz, R.J.; Lewis, Z.A.; Galli, M.G. DNA methylation: Shared and divergent features across eukaryotes. Trends Genet. 2019, 35, 818–827. [Google Scholar] [CrossRef]
- van der Graaf, A.; Wardenaar, R.; Neumann, D.A.; Taudt, A.; Shaw, R.G.; Jansen, R.C.; Schmitz, R.J.; Colomé-Tatché, M.; Johannes, F. Rate, spectrum, and evolutionary dynamics of spontaneous epimutations. Proc. Natl. Acad. Sci. USA 2015, 112, 6676–6681. [Google Scholar] [CrossRef] [PubMed]
- Tourancheau, A.; Mead, E.A.; Zhang, X.S.; Fang, G. Discovering multiple types of DNA methylation from bacteria and microbiome using nanopore sequencing. Nat. Methods 2021, 18, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Bewick, A.J.; Vogel, K.J.; Moore, A.J.; Schmitz, R.J. Evolution of DNA Methylation across Insects. Mol. Biol. Evol. 2017, 34, 654–665. [Google Scholar] [CrossRef] [PubMed]
- Lewis, S.H.; Ross, L.; Bain, S.A.; Pahita, E.; Smith, S.A.; Cordaux, R.; Miska, E.A.; Lenhard, B.; Jiggins, F.M.; Sarkies, P. Widespread conservation and lineage-specific diversification of genome-wide DNA methylation patterns across arthropods. PLoS Genet. 2020, 16, e1008864. [Google Scholar] [CrossRef]
- Feng, S.H.; Jacobsen, S.E.; Reik, W. Epigenetic reprogramming in plant and animal development. Science 2010, 330, 622–627. [Google Scholar] [CrossRef]
- Rey, O.; Eizaguirre, C.; Angers, B.; Baltazar-Soares, M.; Sagonas, K.; Prunier, J.G.; Blanchet, S. Linking epigenetics and biological conservation: Towards a conservation epigenetics perspective. Funct. Ecol. 2020, 34, 414–427. [Google Scholar] [CrossRef]
- Jeremias, G.; Veloso, T.; Gonçalves, F.J.M.; Van Nieuwerburgh, F.; Pereira, J.L.; Asselman, J. Multigenerational DNA methylation responses to copper exposure in Daphnia: Potential targets for epigenetic biomarkers? Chemosphere 2022, 308, 136231. [Google Scholar] [CrossRef]
- Wang, S.; Lv, J.; Zhang, L.L.; Dou, J.Z.; Sun, Y.; Li, X.; Fu, X.T.; Dou, H.Q.; Mao, J.X.; Hu, X.L.; et al. MethylRAD: A simple and scalable method for genome-wide DNA methylation profiling using methylation-dependent restriction enzymes. Open Biol. 2015, 5, 150130. [Google Scholar] [CrossRef]
- Gouin, N.; Bertin, A.; Snow, D.D.; Lozada, A.; Grandjean, F.; Kolok, A.S. Occurrence and environmental risk assessment of pesticides reveal important threats to aquatic organisms in precordilleran rivers of north-central Chile. Sci. Total Environ. 2025, 984, 179701. [Google Scholar] [CrossRef]
- Holmstrup, M.; Bindesbol, A.M.; Oostingh, G.J.; Duschl, A.; Scheil, V.; Köhler, H.R.; Loureiro, S.; Soares, A.; Ferreira, A.L.G.; Kienle, C.; et al. Interactions between effects of environmental chemicals and natural stressors: A review. Sci. Total Environ. 2010, 408, 3746–3762. [Google Scholar] [CrossRef]
- Liess, M.; Foit, K.; Knillmann, S.; Schäfer, R.B.; Liess, H.D. Predicting the synergy of multiple stress effects. Sci. Rep. 2016, 6, 32965. [Google Scholar] [CrossRef]
- Russo, R.; Becker, J.M.; Liess, M. Sequential exposure to low levels of pesticides and temperature stress increase toxicological sensitivity of crustaceans. Sci. Total Environ. 2018, 610, 563–569. [Google Scholar] [CrossRef] [PubMed]
- Gouin, N.; Bertin, A.; Espinosa, M.I.; Snow, D.D.; Ali, J.M.; Kolok, A.S. Pesticide contamination drives adaptive genetic variation in the endemic mayfly Andesiops torrens within a semi-arid agricultural watershed of Chile. Environ. Pollut. 2019, 255, 113099. [Google Scholar] [CrossRef] [PubMed]
- Gouin, N.; Notte, A.M.; Kolok, A.S.; Bertin, A. Pesticide exposure affects DNA methylation patterns in natural populations of a mayfly. Sci. Total Environ. 2023, 864, 161096. [Google Scholar] [CrossRef] [PubMed]
- Epele, L.B.; Miserendino, M.L.; Pessacq, P. Life history, seasonal variation and production of Andesiops torrens (Lugo-Ortiz and McCafferty) and Andesiops peruvianus (Ulmer) (Ephemeroptera: Baetidae) in a headwater Patagonian stream. Limnologica 2011, 41, 57–62. [Google Scholar] [CrossRef]
- Fierro, P.; Barrientos, P.; Montiel, S.; Valdovinos, C. Distribution and life cycles of mayflies Andesiops torrens (Lugo-Ortiz and McCafferty, 1999) and Andesiops peruvianus (Ulmer, 1920) (Ephemeroptera: Baetidae): Importance for environmental monitoring. Aquat. Insects 2025, 46, 38–54. [Google Scholar] [CrossRef]
- Camousseight, A. Estado de conocimiento de los Ephemeroptera de Chile. Gayana 2006, 70, 50–56. [Google Scholar] [CrossRef]
- Sabando, M.C.; Vila, I.; Peñaloza, R.; Véliz, D. Contrasting population genetic structure of two widespread aquatic insects in the Chilean high-slope rivers. Mar. Freshw. Res. 2011, 62, 1–10. [Google Scholar] [CrossRef]
- Falvey, M.; Garreaud, R. Wintertime precipitation episodes in central Chile: Associated meteorological conditions and orographic influences. J. Hydrometeorol. 2007, 8, 171–193. [Google Scholar] [CrossRef]
- Ali, J.M.; Montecinos, A.; Schulze, T.T.; Allmon, L.G.; Kallenbach, A.T.; Watson, G.F.; Davis, P.H.; Snow, D.D.; Bertin, A.; Gouin, N. Assessment of gene expression biomarkers in the Chilean pencil catfish, Trichomycterus areolatus, from the Choapa river basin, Coquimbo Chile. Arch. Environ. Contam. Toxicol. 2020, 78, 137–148. [Google Scholar] [CrossRef]
- Álamos, N.; Alvarez-Garreton, C.; Muñoz, A.; González-Reyes, A. The influence of human activities on streamflow reductions during the megadrought in central Chile. Hydrol. Earth Syst. Sci. 2024, 28, 2483–2503. [Google Scholar] [CrossRef]
- Provataris, P.; Meusemann, K.; Niehuis, O.; Grath, S.; Misof, B. Signatures of DNA methylation across insects suggest reduced DNA methylation levels in holometabola. Genome Biol. Evol. 2018, 10, 1185–1197. [Google Scholar] [CrossRef] [PubMed]
- Fatemi, M.; Hermann, A.; Gowher, H.; Jeltsch, A. Dnmt3a and Dnmt1 functionally cooperate during de novo methylation of DNA. Eur. J. Biochem. 2002, 269, 4981–4984. [Google Scholar] [CrossRef]
- Zhang, J.J.; Kobert, K.; Flouri, T.; Stamatakis, A. PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 2014, 30, 614–620. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.M.; Niu, B.F.; Zhu, Z.W.; Wu, S.T.; Li, W.Z. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Li, R.Q.; Yu, C.; Li, Y.R.; Lam, T.W.; Yiu, S.M.; Kristiansen, K.; Wang, J. SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics 2009, 25, 1966–1967. [Google Scholar] [CrossRef]
- Ruan, J.; Li, H. Fast and accurate long-read assembly with wtdbg2. Nat. Methods 2020, 17, 155–158. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.D.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Korf, I. Gene finding in novel genomes. BMC Bioinform. 2004, 5, 59. [Google Scholar] [CrossRef]
- Holt, C.; Yandell, M. MAKER2: An annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinform. 2011, 12, 491. [Google Scholar] [CrossRef]
- Jones, P.; Binns, D.; Chang, H.Y.; Fraser, M.; Li, W.Z.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST plus: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Huerta-Cepas, J.; Szklarczyk, D.; Forslund, K.; Cook, H.; Heller, D.; Walter, M.C.; Rattei, T.; Mende, D.R.; Sunagawa, S.; Kuhn, M.; et al. eggNOG 4.5: A hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016, 44, D286–D293. [Google Scholar] [CrossRef] [PubMed]
- Marçais, G.; Kingsford, C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 2011, 27, 764–770. [Google Scholar] [CrossRef]
- Vurture, G.W.; Sedlazeck, F.J.; Nattestad, M.; Underwood, C.J.; Fang, H.; Gurtowski, J.; Schatz, M.C. GenomeScope: Fast reference-free genome profiling from short reads. Bioinformatics 2017, 33, 2202–2204. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The genome analysis toolkit: A map reduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Broad Institute. Picard Toolkit. Available online: https://broadinstitute.github.io/picard/ (accessed on 8 December 2025).
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Jombart, T.; Devillard, S.; Balloux, F. Discriminant analysis of principal components: A new method for the analysis of genetically structured populations. BMC Genet. 2010, 11, 94. [Google Scholar] [CrossRef]
- Thia, J.A. Guidelines for standardizing the application of discriminant analysis of principal components to genotype data. Mol. Ecol. Resour. 2023, 23, 523–538. [Google Scholar] [CrossRef]
- Legendre, P.; Gallagher, E.D. Ecologically meaningful transformations for ordination of species data. Oecologia 2001, 129, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Muggeo, V.M.R. Estimating regression models with unknown break-points. Stat. Med. 2003, 22, 3055–3071. [Google Scholar] [CrossRef]
- Muggeo, V.M.R. Segmented: An R package to fit regression models with broken-line relationships. R News 2008, 8, 20–25. [Google Scholar]
- Borcard, D.; Gillet, F.; Legendre, P. Numerical Ecology with R.; Springer: Berlin, German, 2011. [Google Scholar]
- Dûfrene, M.; Legendre, P. Species assemblages and indicator species: The need for a flexible asymmetrical approach. Ecol. Monogr. 1997, 67, 345–366. [Google Scholar] [CrossRef]
- Kriventseva, E.V.; Kuznetsov, D.; Tegenfeldt, F.; Manni, M.; Dias, R.; Simao, F.A.; Zdobnov, E.M. OrthoDB v10: Sampling the diversity of animal, plant, fungal, protist, bacterial and viral genomes for evolutionary and functional annotations of orthologs. Nucleic Acids Res. 2019, 47, D807–D811. [Google Scholar] [CrossRef]
- Chapelle, V.; Silvestre, F. Population epigenetics: The extent of DNA methylation variation in wild animal populations. Epigenomes 2022, 6, 31. [Google Scholar] [CrossRef]
- Hu, J.; Barrett, R.D.H. Epigenetics in natural animal populations. J. Evol. Biol. 2017, 30, 1612–1632. [Google Scholar] [CrossRef]
- Liu, S.; Sun, K.P.; Jiang, T.L.; Feng, J. Natural epigenetic variation in bats and its role in evolution. J. Exp. Biol. 2015, 218, 100–106. [Google Scholar] [CrossRef]
- Schield, D.R.; Walsh, M.R.; Card, D.C.; Andrew, A.L.; Adams, R.H.; Castoe, T.A. EpiRADseq: Scalable analysis of genomewide patterns of methylation using next-generation sequencing. Methods Ecol. Evol. 2016, 7, 60–69. [Google Scholar] [CrossRef]
- Richards, C.L.; Alonso, C.; Becker, C.; Bossdorf, O.; Bucher, E.; Colomé-Tatché, M.; Durka, W.; Engelhardt, J.; Gaspar, B.; Gogol-Döring, A.; et al. Ecological plant epigenetics: Evidence from model and non-model species, and the way forward. Ecol. Lett. 2017, 20, 1576–1590. [Google Scholar] [CrossRef]
- Gáspár, B.; Bossdorf, O.; Durka, W. Structure, stability and ecological significance of natural epigenetic variation: A large-scale survey in Plantago lanceolata. N. Phytol. 2019, 221, 1585–1596. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.Z.; He, Z.H. Small DNA methylation, big player in plant abiotic stress responses and memory. Front. Plant Sci. 2020, 11, 595603. [Google Scholar] [CrossRef] [PubMed]
- Galanti, D.; Ramos-Cruz, D.; Nunn, A.; Rodríguez-Arévalo, I.; Scheepens, J.F.; Becker, C.; Bossdorf, O. Genetic and environmental drivers of large-scale epigenetic variation in Thlaspi arvense. PLoS Genet. 2022, 18, e1010452. [Google Scholar] [CrossRef] [PubMed]
- Vannote, R.L.; Minshall, G.W.; Cummins, K.W.; Sedell, J.R.; Cushing, C.E. The River Continuum Concept. Can. J. Fish. Aquat. Sci. 1980, 37, 130–137. [Google Scholar] [CrossRef]
- Gupta, A.; Nair, S. Epigenetic processes in insect adaptation to environmental stress. Curr. Opin. Insect Sci. 2025, 67, 101294. [Google Scholar] [CrossRef]
- Gallegos-Sánchez, S.; Encalada, A.C.; Ríos-Touma, B.; Dominguez, E. An experimental approach to test the effect of temperature increase and nutrient enrichment on Andean aquatic insects. Aquat. Sci. 2024, 86, 43. [Google Scholar] [CrossRef]
- Li, B.; Hu, P.; Zhu, L.B.; You, L.L.; Cao, H.H.; Wang, J.; Zhang, S.Z.; Liu, M.H.; Toufeeq, S.; Huang, S.J.; et al. DNA methylation is correlated with gene expression during diapause termination of early embryonic development in the silkworm (Bombyx mori). Int. J. Mol. Sci. 2020, 21, 671. [Google Scholar] [CrossRef]
- Hunt, B.; Pegoraro, M.; Marshall, H.; Mallon, E.B. A role for DNA methylation in bumblebee morphogenesis hints at female-specific developmental erasure. Insect Mol. Biol. 2024, 33, 481–492. [Google Scholar] [CrossRef]
- Chen, P.; Xiao, W.F.; Pan, M.H.; Xiao, J.S.; Feng, Y.J.; Dong, Z.Q.; Zou, B.X.; Zhou, L.; Zhang, Y.H.; Lu, C. Comparative genome-wide DNA methylation analysis reveals epigenomic differences in response to heat-humidity stress in Bombyx mori. Int. J. Biol. Macromol. 2020, 164, 3771–3779. [Google Scholar] [CrossRef]
- Bogdanovic, O.; Lister, R. DNA methylation and the preservation of cell identity. Curr. Opin. Genet. Dev. 2017, 46, 9–14. [Google Scholar] [CrossRef]
- Wood, J.G.; Schwer, B.; Wickremesinghe, P.C.; Hartnett, D.A.; Burhenn, L.; Garcia, M.; Li, M.; Verdin, E.; Helfand, S.L. Sirt4 is a mitochondrial regulator of metabolism and lifespan in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2018, 115, 1564–1569. [Google Scholar] [CrossRef] [PubMed]
- Gresková, A.; Petrivalsky, M. Thioredoxin system in insects: Uncovering the roles of thioredoxins and thioredoxin reductase beyond the antioxidant defences. Insects 2024, 15, 797. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.L.; Ding, J.H.; Liu, Z.X.; Shao, Z.M.; Liang, X.H.; Wang, J.; Wu, F.A.; Sheng, S. A role of peptidoglycan recognition protein in mediating insecticide detoxification in Glyphodes pyloalis. Arch. Insect Biochem. Physiol. 2021, 108, e21842. [Google Scholar] [CrossRef] [PubMed]
- Mariani, A.; Comolli, D.; Fanelli, R.; Forloni, G.; De Paola, M. Neonicotinoid Pesticides Affect Developing Neurons in Experimental Mouse Models and in Human Induced Pluripotent Stem Cell (iPSC)-Derived Neural Cultures and Organoids. Cells 2024, 13, 1295. [Google Scholar] [CrossRef]
- Rodríguez, A.; Castrejón-Godínez, M.L.; Monterrosas-Brisson, N. Pesticides: Environmental stressors implicated in the development of central nervous system disorders and neurodegeneration. Stresses 2025, 5, 31. [Google Scholar] [CrossRef]
- El Kenawy, A.M. Chapter 1—Hydroclimatic extremes in arid and semi-arid regions: Status, challenges, and future outlook. Assess. Attrib. Socioecon. Impacts 2024, 1–22. [Google Scholar] [CrossRef]
- Buckley, L.B.; Arakaki, A.J.; Cannistra, A.F.; Kharouba, H.M.; Kingsolver, J.G. Insect Development, Thermal Plasticity and Fitness Implications in Changing, Seasonal Environments. Integr. Comp. Biol. 2017, 57, 988–998. [Google Scholar] [CrossRef]
- Rodrigues, Y.K.; Beldade, P. Thermal plasticity in insects’ response to climate change and to multifactorial environments. Front. Ecol. Evol. 2020, 8, 271. [Google Scholar] [CrossRef]
- Nufio, C.R.; Sheffer, M.M.; Smith, J.M.; Troutman, M.T.; Bawa, S.J.; Taylor, E.D.; Schoville, S.D.; Williams, C.M.; Buckley, L.B. Insect size responses to climate change vary across elevations according to seasonal timing. PLoS Biol. 2025, 23, e3002805. [Google Scholar] [CrossRef]
- Johnson, M.F.; Albertson, L.K.; Algar, A.C.; Dugdale, S.J.; Edwards, P.; England, J.; Gibbins, C.; Kazama, S.; Komori, D.; MacColl, A.D.C.; et al. Rising water temperature in rivers: Ecological impacts and future resilience. Wiley Interdiscip. Rev. Water 2024, 11, e1724. [Google Scholar] [CrossRef]
- Dimond, J.L.; Gamblewood, S.K.; Roberts, S.B. Genetic and epigenetic insight into morphospecies in a reef coral. Mol. Ecol. 2017, 26, 5031–5042. [Google Scholar] [CrossRef]
- Dixon, G.; Liao, Y.; Bay, L.K.; Matz, M.V. Role of gene body methylation in acclimatization and adaptation in a basal metazoan. Proc. Natl. Acad. Sci. USA 2018, 115, 13342–13346. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.L.; Chen, J.H.; Tsang, M.H.; Chung, J.D.; Chang, C.T.; Hwang, S.Y. Influences of environmental and spatial factors on genetic and epigenetic variations in Rhododendron oldhamii (Ericaceae). Tree Genet. Genomes 2015, 11, 823. [Google Scholar] [CrossRef]
- Rodríguez-Casariego, J.A.; Mercado-Molina, A.E.; Garcia-Souto, D.; Ortiz-Rivera, I.M.; Lopes, C.; Baums, I.B.; Sabat, A.M.; Eirin-Lopez, J.M. Genome-wide DNA methylation analysis reveals a conserved epigenetic response to seasonal environmental variation in the staghorn coral Acropora cervicornis. Front. Mar. Sci. 2020, 7, 560424. [Google Scholar] [CrossRef]
- Wang, X.X.; Li, A.; Wang, W.; Que, H.Y.; Zhang, G.F.; Li, L. DNA methylation mediates differentiation in thermal responses of Pacific oyster (Crassostrea gigas) derived from different tidal levels. Heredity 2021, 126, 10–22. [Google Scholar] [CrossRef]
- Xie, H.H.; Konate, M.; Sai, N.; Tesfamicael, K.G.; Cavagnaro, T.; Gilliham, M.; Breen, J.; Metcalfe, A.; Stephen, J.R.; De Bei, R.; et al. Global DNA methylation patterns can play a role in defining terroir in grapevine (Vitis vinifera cv. Shiraz). Front. Plant Sci. 2017, 8, 1860. [Google Scholar] [CrossRef]
- Jeremias, G.; Barbosa, J.; Marques, S.M.; Asselman, J.; Gonçalves, F.J.M.; Pereira, J.L. Synthesizing the role of epigenetics in the response and adaptation of species to climate change in freshwater ecosystems. Mol. Ecol. 2018, 27, 2790–2806. [Google Scholar] [CrossRef]
- Mathers, T.C.; Mugford, S.T.; Percival-Alwyn, L.; Chen, Y.Z.; Kaithakottil, G.; Swarbreck, D.; Hogenhout, S.A.; van Oosterhout, C. Sex-specific changes in the aphid DNA methylation landscape. Mol. Ecol. 2019, 28, 4228–4241. [Google Scholar] [CrossRef]
- McCaw, B.A.; Leonard, A.M.; Stevenson, T.J.; Lancaster, L.T. A role of epigenetic mechanisms in regulating female reproductive responses to temperature in a pest beetle. Insect Mol. Biol. 2024, 33, 516–533. [Google Scholar] [CrossRef]
- Boman, J.; Zhu, Y.S.; Höök, L.; Vila, R.; Talavera, G.; Backström, N. Environmental stress during larval development induces DNA methylation shifts in the migratory painted lady butterfly (Vanessa cardui). Mol. Ecol. 2023, 32, 3513–3523. [Google Scholar] [CrossRef]
- Royle, J.W.; Hurwood, D.; Sadowski, P.; Dudley, K.J. Non-CG DNA methylation marks the transition from pupa to adult in Helicoverpa armigera. Insect Mol. Biol. 2024, 33, 493–502. [Google Scholar] [CrossRef]
- Song, X.W.; Huang, F.; Liu, J.J.; Li, C.J.; Gao, S.S.; Wu, W.; Zhai, M.F.; Yu, X.J.; Xiong, W.F.; Xie, J.; et al. Genome-wide DNA methylomes from discrete developmental stages reveal the predominance of non-CpG methylation in Tribolium castaneum. DNA Res. 2017, 24, 445–458. [Google Scholar] [CrossRef]
- Gavery, M.R.; Roberts, S.B. DNA methylation patterns provide insight into epigenetic regulation in the Pacific oyster (Crassostrea gigas). BMC Genom. 2010, 11, 483. [Google Scholar] [CrossRef]
- Almudi, I.; Vizueta, J.; Wyatt, C.D.R.; de Mendoza, A.; Marlétaz, F.; Firbas, P.N.; Feuda, R.; Masiero, G.; Medina, P.; Alcaina-Caro, A.; et al. Genomic adaptations to aquatic and aerial life in mayflies and the origin of insect wings. Nat. Commun. 2020, 11, 2631. [Google Scholar] [CrossRef]
- de la Peña, M.V.; Piskobulu, V.; Murgatroyd, C.; Hager, R. DNA methylation patterns respond to thermal stress in the viviparous cockroach Diploptera punctata. Epigenetics 2021, 16, 313–326. [Google Scholar] [CrossRef]
- Wu, J.; Zhang, W.W.; Li, C.H. Heat and hypoxia exposure mediates circadian rhythms response via methylation modification in Apostichopus japonicas. Front. Mar. Sci. 2021, 8, 721465. [Google Scholar] [CrossRef]
- Vaiserman, A.M. Hormesis and epigenetics: Is there a link? Ageing Res. Rev. 2011, 10, 413–421. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Bertin, A.; Notte, A.M.; Moumen, B.; Coral-Santacruz, D.; Grandjean, F.; Gouin, N. Patterns and Functional Insights of DNA Methylation Variation in a South American Mayfly Across an Agriculturally Impacted Semi-Arid Watershed. Biology 2026, 15, 90. https://doi.org/10.3390/biology15010090
Bertin A, Notte AM, Moumen B, Coral-Santacruz D, Grandjean F, Gouin N. Patterns and Functional Insights of DNA Methylation Variation in a South American Mayfly Across an Agriculturally Impacted Semi-Arid Watershed. Biology. 2026; 15(1):90. https://doi.org/10.3390/biology15010090
Chicago/Turabian StyleBertin, Angéline, Ana María Notte, Bouziane Moumen, Diana Coral-Santacruz, Frédéric Grandjean, and Nicolas Gouin. 2026. "Patterns and Functional Insights of DNA Methylation Variation in a South American Mayfly Across an Agriculturally Impacted Semi-Arid Watershed" Biology 15, no. 1: 90. https://doi.org/10.3390/biology15010090
APA StyleBertin, A., Notte, A. M., Moumen, B., Coral-Santacruz, D., Grandjean, F., & Gouin, N. (2026). Patterns and Functional Insights of DNA Methylation Variation in a South American Mayfly Across an Agriculturally Impacted Semi-Arid Watershed. Biology, 15(1), 90. https://doi.org/10.3390/biology15010090





