CRISPR-Cas9 Gene Editing in Aspergillus: From Pathogenesis to Metabolic Engineering
Simple Summary
Abstract
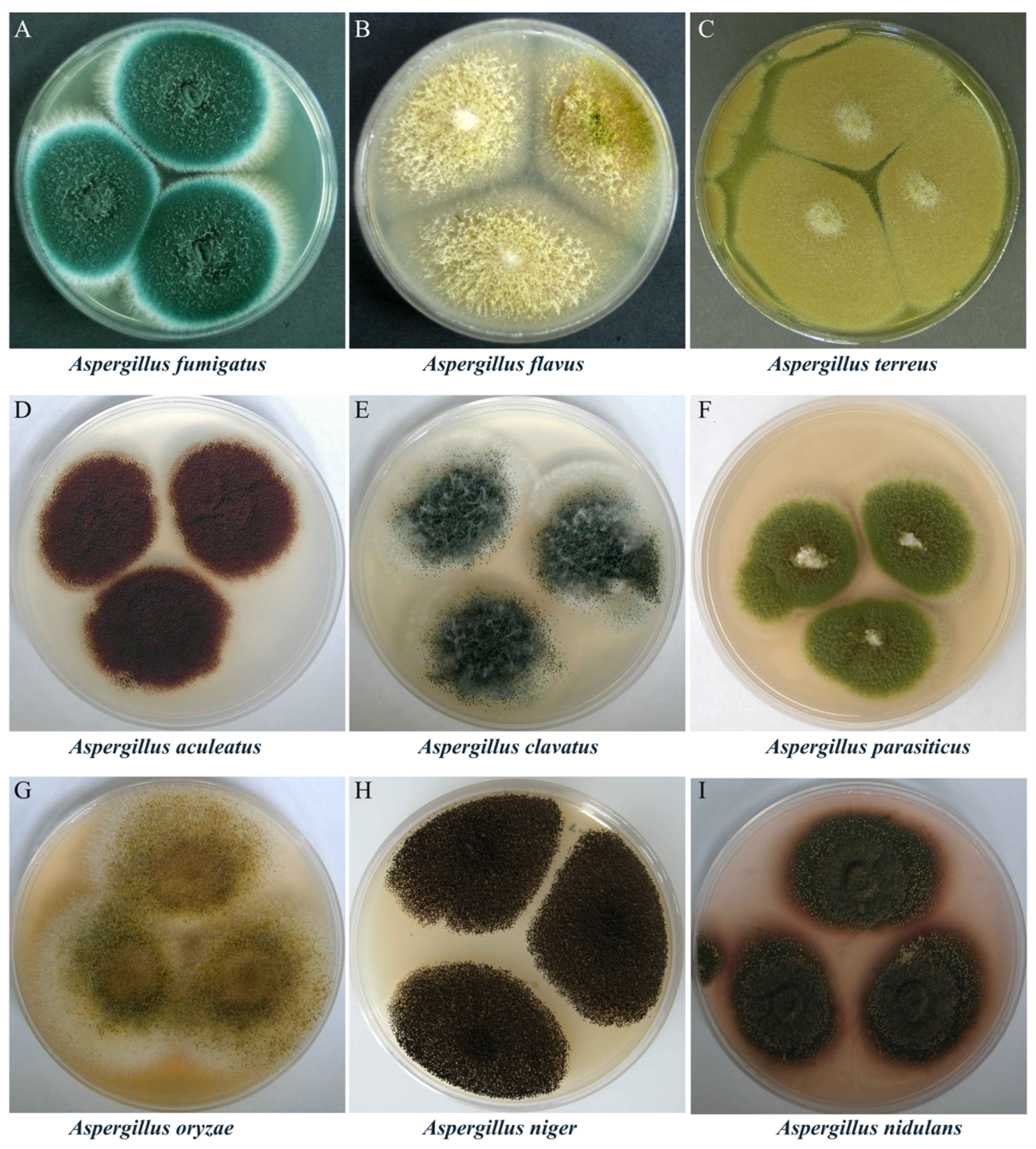
1. Introduction
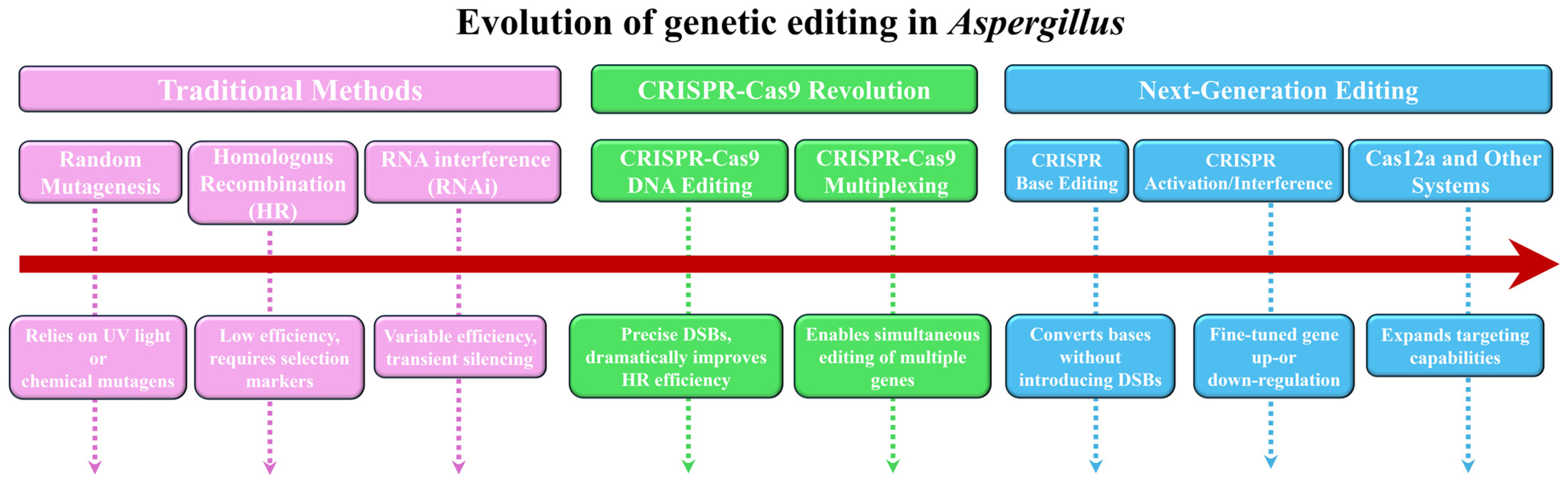
2. Genetic Editing Techniques in Aspergillus Research
2.1. Limitations of Traditional Genetic Tools in Aspergillus
2.2. Efficient Editing of CRISPR-Cas9 Technology in Aspergillus
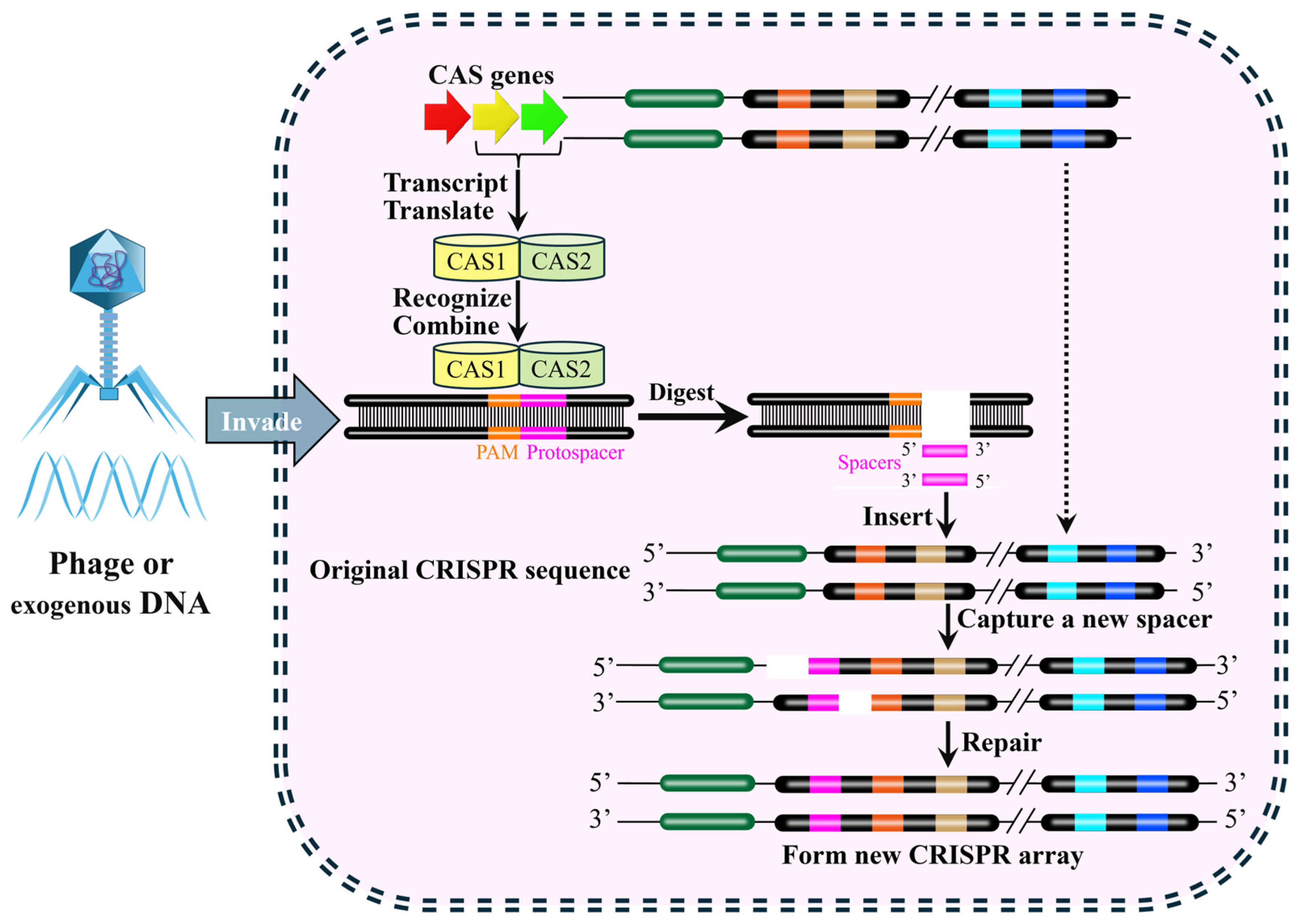
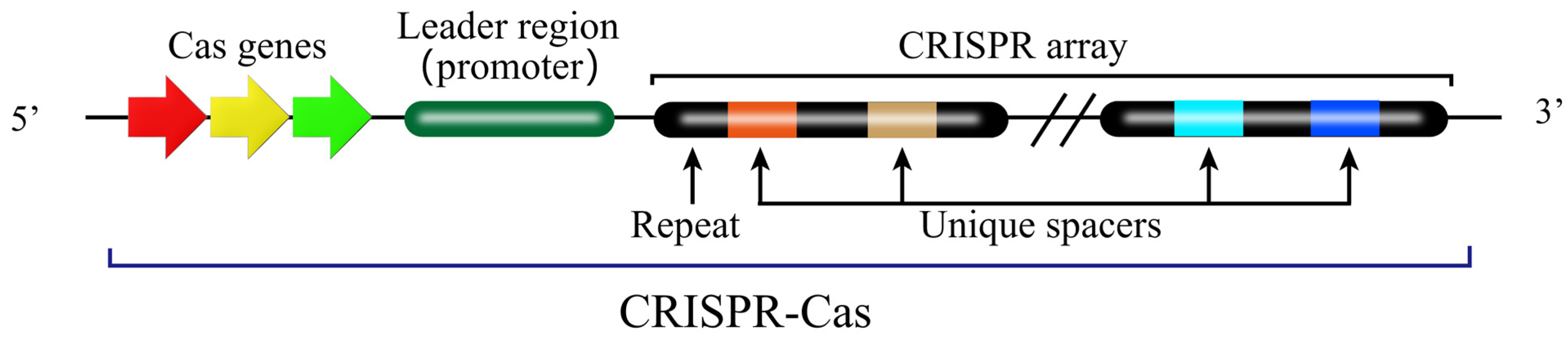
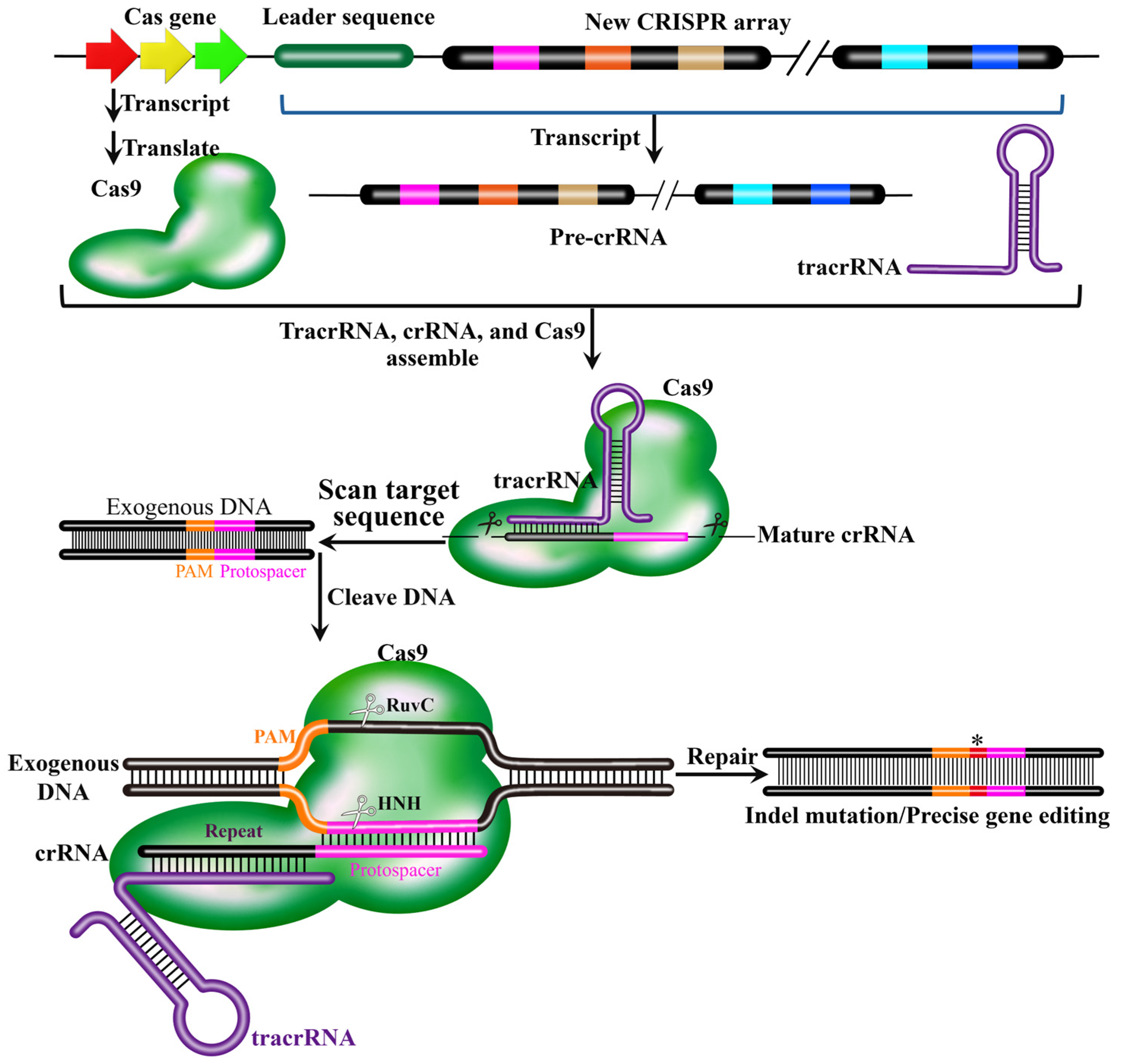
3. Working Mechanism of CRISPR-Cas9 System
3.1. Type of CRISPR-Cas System
3.2. Composition and Working Mechanism of CRISPR-Cas9 System
3.3. Functional Features of Cas9 and PAM in CRISPR-Cas9 System
4. Key Applications of CRISPR-Cas9 Technology in Aspergillus
4.1. Elucidating Pathogenicity and Drug Resistance
4.2. Disruption of Toxin Biosynthesis in Aspergillus
4.3. Metabolic Engineering for Bioproduction
4.3.1. Enhancing Industrial Enzyme and Protein Production
4.3.2. Engineering for Organic Acid Production
4.3.3. Reprogramming Natural Product Biosynthetic Pathways
5. Discussion and Future Perspectives
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Baniya, S. Aspergillus: Morphology, Clinical Features, and Lab Diagnosis. Mycology. Available online: https://microbeonline.com/aspergillus-morphology-clinical-features-and-lab-diagnosis/#Aspergillus_glaucus (accessed on 20 June 2025).
- Samson, R.A.; Visagie, C.M.; Houbraken, J.; Hong, S.B.; Hubka, V.; Klaassen, C.H.; Perrone, G.; Seifert, K.A.; Susca, A.; Tanney, J.B.; et al. Phylogeny, identification and nomenclature of the genus Aspergillus. Stud. Mycol. 2014, 78, 141–173. [Google Scholar] [CrossRef]
- Amaike, S.; Keller, N.P. Aspergillus flavus. Annu. Rev. Phytopathol. 2011, 49, 107–133. [Google Scholar] [CrossRef]
- Reijula, K.; Tuomi, T. Mycotoxins of aspergilli: Exposure and health effects. Front. Biosci. 2003, 8, s232–s235. [Google Scholar] [CrossRef] [PubMed]
- Lass-Florl, C.; Dietl, A.M.; Kontoyiannis, D.P.; Brock, M. Aspergillus terreus species complex. Clin. Microbiol. Rev. 2021, 34, e00311-20. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, J.C. Aspergillus fumigatus: Growth and virulence. Med. Mycol. 2006, 44, S77–S81. [Google Scholar] [CrossRef] [PubMed]
- Hedayati, M.T.; Pasqualotto, A.C.; Warn, P.A.; Bowyer, P.; Denning, D.W. Aspergillus flavus: Human pathogen, allergen and mycotoxin producer. Microbiology 2007, 153, 1677–1692. [Google Scholar] [CrossRef]
- Allwood, J.G.; Wakeling, L.T.; Bean, D.C. Fermentation and the microbial community of Japanese koji and miso: A review. J. Food Sci. 2021, 86, 2194–2207. [Google Scholar] [CrossRef]
- Cairns, T.C.; Nai, C.; Meyer, V. How a fungus shapes biotechnology: 100 years of Aspergillus niger research. Fungal Biol. Biotechnol. 2018, 5, 13. [Google Scholar] [CrossRef]
- Oikawa, H. Heterologous production of fungal natural products: Reconstitution of biosynthetic gene clusters in model host Aspergillus oryzae. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2020, 96, 420–430. [Google Scholar] [CrossRef]
- Frandsen, R.J.N.; Khorsand-Jamal, P.; Kongstad, K.T.; Nafisi, M.; Kannangara, R.M.; Staerk, D.; Okkels, F.T.; Binderup, K.; Madsen, B.; Møller, B.L.; et al. Heterologous production of the widely used natural food colorant carminic acid in Aspergillus nidulans. Sci. Rep. 2018, 8, 12853. [Google Scholar] [CrossRef]
- Huynh, H.H.; Morita, N.; Sakamoto, T.; Katayama, T.; Miyakawa, T.; Tanokura, M.; Chiba, Y.; Shinkura, R.; Maruyama, J.I. Functional production of human antibody by the filamentous fungus Aspergillus oryzae. Fungal Biol. Biotechnol. 2020, 7, 7. [Google Scholar] [CrossRef]
- Nishimura, I.; Shinohara, Y.; Oguma, T.; Koyama, Y. Survival strategy of the salt-tolerant lactic acid bacterium Tetragenococcus halophilus to counteract koji mold, Aspergillus oryzae, in soy sauce brewing. Biosci. Biotechnol. Biochem. 2018, 82, 1437–1443. [Google Scholar] [CrossRef]
- Dao, T.T.; de Mattos-Shipley, K.M.J.; Prosser, I.M.; Williams, K.; Zacharova, M.K.; Lazarus, C.M.; Willis, C.L.; Bailey, A.M. Cleaning the cellular factory—Deletion of McrA in Aspergillus oryzae NSAR1 and the generation of a novel kojic acid deficient strain for cleaner heterologous production of secondary metabolites. Front. Fungal Biol. 2021, 2, 632542. [Google Scholar] [CrossRef] [PubMed]
- Daba, G.M.; Mostafa, F.A.; Elkhateeb, W.A. The ancient koji mold (Aspergillus oryzae) as a modern biotechnological tool. Bioresour. Bioprocess. 2021, 8, 52. [Google Scholar] [CrossRef]
- Hama, S.; Tamalampudi, S.; Suzuki, Y.; Yoshida, A.; Fukuda, H.; Kondo, A. Preparation and comparative characterization of immobilized Aspergillus oryzae expressing Fusarium heterosporum lipase for enzymatic biodiesel production. Appl. Microbiol. Biotechnol. 2008, 81, 637–645. [Google Scholar] [CrossRef] [PubMed]
- Papagianni, M. Advances in citric acid fermentation by Aspergillus niger: Biochemical aspects, membrane transport and modeling. Biotechnol. Adv. 2007, 25, 244–263. [Google Scholar] [CrossRef]
- Idris, A.D.; Abalaka, M.E.; Oyewole, O.A.; Egwim, E.C.; Maddela, N.R. Purification and characterization of β-galactosidase from Aspergillus niger PQ570689 for lactose hydrolysis and prebiotic synthesis. Folia Microbiol. 2025, 70, 01293. [Google Scholar] [CrossRef] [PubMed]
- Park, H.S.; Jun, S.C.; Han, K.H.; Hong, S.B.; Yu, J.H. Diversity, application, and synthetic biology of industrially important Aspergillus fungi. In Advances in Applied Microbiology; Sariaslani, S., Gadd, G.M., Eds.; Academic Press: Cambridge, MA, USA, 2017; Volume 100, pp. 161–202. [Google Scholar]
- Książek, E. Citric acid: Properties, microbial production, and applications in industries. Molecules 2024, 29, 22. [Google Scholar] [CrossRef]
- Shi, T.; Liu, G.; Ji, R.; Shi, K.; Song, P.; Ren, L.; Huang, H.; Ji, X. CRISPR/Cas9-based genome editing of the filamentous fungi: The state of the art. Appl. Microbiol. Biotechnol. 2017, 101, 7435–7443. [Google Scholar] [CrossRef]
- Wang, S.; Chen, H.; Tang, X.; Zhang, H.; Chen, W.; Chen, Y.Q. Molecular tools for gene manipulation in filamentous fungi. Appl. Microbiol. Biotechnol. 2017, 101, 8063–8075. [Google Scholar] [CrossRef]
- Chang, P.K. Creating large chromosomal segment deletions in Aspergillus flavus by a dual CRISPR/Cas9 system: Deletion of gene clusters for production of aflatoxin, cyclopiazonic acid, and ustiloxin B. Fungal Genet. Biol. 2024, 170, 103863. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, J. Genome editing technology and its application potentials in the industrial filamentous fungus Aspergillus oryzae. J. Fungi 2021, 7, 638. [Google Scholar] [CrossRef]
- Maruyama, J.; Nakajima, H.; Kitamoto, K. Visualization of nuclei in Aspergillus oryzae with EGFP and analysis of the number of nuclei in each conidium by FACS. Biosci. Biotechnol. Biochem. 2001, 65, 1504–1510. [Google Scholar] [CrossRef] [PubMed]
- McCarty, N.S.; Graham, A.E.; Studená, L.; Ledesma-Amaro, R. Multiplexed CRISPR technologies for gene editing and transcriptional regulation. Nat. Commun. 2020, 11, 1281. [Google Scholar] [CrossRef]
- Nodvig, C.S.; Nielsen, J.B.; Kogle, M.E.; Mortensen, U.H. A CRISPR-Cas9 system for genetic engineering of filamentous fungi. PLoS ONE 2015, 10, e0133085. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Chen, L.; Jiang, Y.; Zhou, Z.; Zou, G. Efficient genome editing in filamentous fungus Trichoderma reesei using the CRISPR/Cas9 system. Cell Discov. 2015, 1, 15007. [Google Scholar] [CrossRef]
- Pohl, C.; Kiel, J.A.; Driessen, A.J.; Bovenberg, R.A.; Nygård, Y. CRISPR/Cas9 based genome editing of Penicillium chrysogenum. ACS Synth. Biol. 2016, 5, 754–764. [Google Scholar] [CrossRef]
- Matsu-Ura, T.; Baek, M.; Kwon, J.; Hong, C. Efficient gene editing in Neurospora crassa with CRISPR technology. Fungal Biol. Biotechnol. 2015, 2, 4. [Google Scholar] [CrossRef]
- Gruttner, S.; Kempken, F. A user-friendly CRISPR/Cas9 system for mutagenesis of Neurospora crassa. Sci. Rep. 2024, 14, 20469. [Google Scholar] [CrossRef]
- Schuster, M.; Kahmann, R. CRISPR-Cas9 genome editing approaches in filamentous fungi and oomycetes. Fungal Genet. Biol. 2019, 130, 43–53. [Google Scholar] [CrossRef]
- Yuan, G.; Deng, S.; Czajka, J.J.; Dai, Z.; Hofstad, B.A.; Kim, J.; Pomraning, K.R. CRISPR-Cas9/Cas12a systems for efficient genome editing and large genomic fragment deletions in Aspergillus niger. Front. Bioeng. Biotechnol. 2024, 12, 1452496. [Google Scholar] [CrossRef]
- Mayorga, M.E.; Timberlake, W.E. Isolation and molecular characterization of the Aspergillus nidulans wA gene. Genetics 1990, 126, 73–79. [Google Scholar] [CrossRef]
- Kuivanen, J.; Korja, V.; Holmstrom, S.; Richard, P. Development of microtiter plate scale CRISPR/Cas9 transformation method for Aspergillus niger based on in vitro assembled ribonucleoprotein complexes. Fungal Biol. Biotechnol. 2019, 6, 3. [Google Scholar] [CrossRef] [PubMed]
- Nan, Y.; Ouyang, L.; Chu, J. In vitro CRISPR/Cas9 system for genome editing of Aspergillus niger based on removable bidirectional selection marker AmdS. Biotechnol. Appl. Biochem. 2021, 68, 964–970. [Google Scholar] [CrossRef]
- Chang, P.K. A simple CRISPR/Cas9 system for efficiently targeting genes of Aspergillus section Flavi species, Aspergillus nidulans, Aspergillus fumigatus, Aspergillus terreus, and Aspergillus niger. Microbiol. Spectr. 2023, 11, e0464822. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Lu, J.; Zhang, G.; Zhou, J.; Li, J.; Du, G.; Chen, J. CRISPR/Cas9-mediated multiplexed genome editing in Aspergillus oryzae. J. Fungi 2023, 9, 109. [Google Scholar] [CrossRef] [PubMed]
- Todokoro, T.; Hata, Y.; Ishida, H. CRISPR/Cas9 improves targeted knock-in efficiency in Aspergillus oryzae. Biotechnol. Notes 2024, 5, 58–63. [Google Scholar] [CrossRef]
- Mojica, F.J.M.; Rodriguez-Valera, F. The discovery of CRISPR in archaea and bacteria. FEBS J. 2016, 283, 3162–3169. [Google Scholar] [CrossRef]
- Amitai, G.; Sorek, R. CRISPR-Cas adaptation: Insights into the mechanism of action. Nat. Rev. Microbiol. 2016, 14, 67–76. [Google Scholar] [CrossRef]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef]
- Makarova, K.S.; Grishin, N.V.; Shabalina, S.A.; Wolf, Y.I.; Koonin, E.V. A putative RNA-interference based immune system in prokaryotes: Computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct 2006, 1, 7. [Google Scholar] [CrossRef]
- Makarova, K.S.; Wolf, Y.I.; Alkhnbashi, O.S.; Costa, F.; Shah, S.A.; Saunders, S.J.; Barrangou, R.; Brouns, S.J.J.; Charpentier, E.; Haft, D.H.; et al. An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 2015, 13, 722. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, E.; Sharma, S.; Tiwari, V.; Garg, M. Different Classes of CRISPR-Cas Systems. In Gene Editing in Plants; Kumar, A., Arora, S., Ogita, S., Yau, Y.Y., Mukherjee, K., Eds.; Springer: Singapore, 2024; pp. 73–94. [Google Scholar]
- Jackson, S.A.; McKenzie, R.E.; Fagerlund, R.D.; Kieper, S.N.; Fineran, P.C.; Brouns, S.J.J. CRISPR-Cas: Adapting to change. Science 2017, 356, eaal5056. [Google Scholar] [CrossRef]
- Brouns, S.J.; Jore, M.M.; Lundgren, M.; Westra, E.R.; Slijkhuis, R.J.; Snijders, A.P.; Dickman, M.J.; Makarova, K.S.; Koonin, E.V.; van der Oost, J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 2008, 321, 960–964. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Samai, P.; Pyenson, N.; Jiang, W.; Goldberg, G.W.; Hatoum-Aslan, A.; Marraffini, L.A. Co-transcriptional DNA and RNA Cleavage during Type III CRISPR-Cas Immunity. Cell 2015, 161, 1164–1174. [Google Scholar] [CrossRef]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; van der Oost, J.; Regev, A.; et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef]
- Abudayyeh, O.O.; Gootenberg, J.S.; Konermann, S.; Joung, J.; Slaymaker, I.M.; Cox, D.B.; Shmakov, S.; Makarova, K.S.; Semenova, E.; Minakhin, L.; et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 2016, 353, aaf5573. [Google Scholar] [CrossRef]
- Gaj, T.; Gersbach, C.A.; Barbas, C.F., 3rd. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013, 31, 397–405. [Google Scholar] [CrossRef]
- Gasiunas, G.; Barrangou, R.; Horvath, P.; Siksnys, V. Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. USA 2012, 109, E2579–E2586. [Google Scholar] [CrossRef]
- Deltcheva, E.; Chylinski, K.; Sharma, C.M.; Gonzales, K.; Chao, Y.; Pirzada, Z.A.; Eckert, M.R.; Vogel, J.; Charpentier, E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 2011, 471, 602–607. [Google Scholar] [CrossRef]
- Hale, C.R.; Zhao, P.; Olson, S.; Duff, M.O.; Graveley, B.R.; Wells, L.; Terns, R.M.; Terns, M.P. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 2009, 139, 945–956. [Google Scholar] [CrossRef]
- Sander, J.D.; Joung, J.K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 2014, 32, 347–355. [Google Scholar] [CrossRef]
- Hsu, P.D.; Scott, D.A.; Weinstein, J.A.; Ran, F.A.; Konermann, S.; Agarwala, V.; Li, Y.; Fine, E.J.; Wu, X.; Shalem, O.; et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013, 31, 827–832. [Google Scholar] [CrossRef]
- Hu, J.H.; Miller, S.M.; Geurts, M.H.; Tang, W.; Chen, L.; Sun, N.; Zeina, C.M.; Gao, X.; Rees, H.A.; Lin, Z.; et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 2018, 556, 57–63. [Google Scholar] [CrossRef]
- Teng, Y.; Wang, J.; Jiang, T.; Zou, Y.; Yan, Y. Engineering a Streptococcus Cas9 ortholog with an RxQ PAM-binding motif for PAM-free gene control in bacteria. ACS Synth. Biol. 2023, 12, 2764–2772. [Google Scholar] [CrossRef]
- Kleinstiver, B.P.; Pattanayak, V.; Prew, M.S.; Tsai, S.Q.; Nguyen, N.T.; Zheng, Z.; Joung, J.K. High-fidelity CRISPR-Cas9 nucleases with no detectable genomewide off-target effects. Nature 2016, 529, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Kleinstiver, B.P.; Prew, M.S.; Tsai, S.Q.; Topkar, V.V.; Nguyen, N.T.; Zheng, Z.; Gonzales, A.P.W.; Li, Z.; Peterson, R.T.; Yeh, J.R.J.; et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 2015, 523, 481–485. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Hsu, P.D.; Lin, C.Y.; Gootenberg, J.S.; Konermann, S.; Trevino, A.E.; Scott, D.A.; Inoue, A.; Matoba, S.; Zhang, Y.; et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 2013, 154, 1380–1389. [Google Scholar] [CrossRef] [PubMed]
- Walton, R.T.; Christie, K.A.; Whittaker, M.N.; Kleinstiver, B.P. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science 2020, 368, 290–296. [Google Scholar] [CrossRef]
- Wang, S.; Tao, C.; Mao, H.; Hou, L.; Wang, Y.; Qi, T.; Yang, Y.; Ong, S.G.; Hu, S.; Chai, R.; et al. Correction: Identification of SaCas9 orthologs containing a conserved serine residue that determines simple NNGG PAM recognition. PLoS Biol. 2025, 23, e3003036. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Cai, Y.; Sun, S.; Han, T.; Chen, L.; Hou, W. Using Staphylococcus aureus Cas9 to expand the scope of potential gene targets for genome editing in soybean. Int. J. Mol. Sci. 2022, 23, 12789. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Koo, T.; Park, S.W.; Kim, D.; Kim, K.; Cho, H.Y.; Song, D.W.; Lee, K.J.; Jung, M.H.; Kim, S.; et al. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat. Commun. 2017, 8, 14500. [Google Scholar] [CrossRef]
- Gao, S.; Wang, Y.; Qi, T.; Wei, J.; Hu, Z.; Liu, J.; Sun, S.; Liu, H.; Wang, Y. Genome editing with natural and engineered CjCas9 orthologs. Mol. Ther. 2023, 31, 1177–1187. [Google Scholar] [CrossRef]
- Miller, S.M.; Wang, T.; Randolph, P.B.; Arbab, M.; Shen, M.W.; Huang, T.P.; Matuszek, Z.; Newby, G.A.; Rees, H.A.; Liu, D.R. Continuous evolution of SpCas9 variants compatible with non-G PAMs. Nat. Biotechnol. 2020, 38, 471–481. [Google Scholar] [CrossRef]
- Chatterjee, P.; Jakimo, N.; Lee, J.; Amrani, N.; Rodríguez, T.; Koseki, S.R.T.; Tysinger, E.; Qing, R.; Hao, S.; Sontheimer, E.J.; et al. An engineered ScCas9 with broad PAM range and high specificity and activity. Nat. Biotechnol. 2020, 38, 1154–1158. [Google Scholar] [CrossRef] [PubMed]
- Upton, A.; Kirby, K.A.; Carpenter, P.; Boeckh, M.; Marr, K.A. Invasive aspergillosis following hematopoietic cell transplantation: Outcomes and prognostic factors associated with mortality. Clin. Infect. Dis. 2007, 44, 531–540. [Google Scholar] [CrossRef]
- Krappmann, S.; Bayram, O.; Braus, G.H. Deletion and allelic exchange of the Aspergillus fumigatus veA locus via a novel recyclable marker module. Eukaryot. Cell 2005, 4, 1298–1307. [Google Scholar] [CrossRef]
- Hassane, A.M.A.; Obiedallah, M.; Karimi, J.; Khattab, S.M.R.; Hussein, H.R.; Abo-Dahab, Y.; Eltoukhy, A.; Abo-Dahab, N.F.; Abouelela, M.E. Unravelling fungal genome editing revolution: Pathological and biotechnological application aspects. Arch. Microbiol. 2025, 207, 150. [Google Scholar] [CrossRef]
- Fuller, K.K.; Chen, S.; Loros, J.J.; Dunlap, J.C. Development of the CRISPR/Cas9 system for targeted gene disruption in Aspergillus fumigatus. Eukaryot. Cell 2015, 14, 1073–1080. [Google Scholar] [CrossRef]
- Zhang, C.; Meng, X.; Wei, X.; Lu, L. Highly efficient CRISPR mutagenesis by microhomology-mediated end joining in Aspergillus fumigatus. Fungal Genet. Biol. 2016, 86, 47–57. [Google Scholar] [CrossRef]
- Sastre-Velasquez, L.E.; Mach, N.; Mertens, B.; Kuhbacher, A.; Merschak, P.; Dallemulle, A.; Lechner, L.; Baldin, C.; Diallinas, G.; Gsaller, F. Simultaneous multigene integration in Aspergillus fumigatus using CRISPR/Cas9 and endogenous counter-selectable markers. J. Biol. Eng. 2025, 19, 69. [Google Scholar] [CrossRef] [PubMed]
- Nair, A.; Rao, A.S.; Surabhi, M.A.; Gnanika, M.; More, S.S. Unravelling fungal pathogenesis: Advances in CRISPR-Cas9 for understanding virulence and adaptation. Fungal Genet. Biol. 2025, 179, 104006. [Google Scholar] [CrossRef]
- Dudakova, A.; Spiess, B.; Tangwattanachuleeporn, M.; Sasse, C.; Buchheidt, D.; Weig, M.; Groß, U.; Bader, O. Molecular tools for the detection and deduction of azole antifungal drug resistance phenotypes in Aspergillus species. Clin. Microbiol. Rev. 2017, 30, 1065–1091. [Google Scholar] [CrossRef]
- Meis, J.F.; Chowdhary, A.; Rhodes, J.L.; Fisher, M.C.; Verweij, P.E. Clinical implications of globally emerging azole resistance in Aspergillus fumigatus. Philos. Trans. R. Soc. B 2016, 371, 20150460. [Google Scholar] [CrossRef]
- Rybak, J.M.; Ge, W.; Wiederhold, N.P.; Parker, J.E.; Kelly, S.L.; Rogers, P.D.; Fortwendel, J.R. Mutations in hmg1, challenging the paradigm of clinical triazole resistance in Aspergillus fumigatus. mBio 2019, 10, e00437-19. [Google Scholar] [CrossRef]
- Arai, T.; Umeyama, T.; Majima, H.; Inukai, T.; Watanabe, A.; Miyazaki, Y.; Kamei, K. hmg1 mutations in Aspergillus fumigatus and their contribution to triazole susceptibility. Med. Mycol. 2021, 59, 980–984. [Google Scholar] [CrossRef]
- Handelman, M.; Osherov, N. Efficient generation of multiple seamless point mutations conferring triazole resistance in Aspergillus fumigatus. J. Fungi 2023, 9, 644. [Google Scholar] [CrossRef]
- Souza, A.C.O.; Martin-Vicente, A.; Nywening, A.V.; Ge, W.; Lowes, D.J.; Peters, B.M.; Fortwendel, J.R. Loss of septation initiation network (SIN) kinases blocks tissue invasion and unlocks echinocandin cidal activity against Aspergillus fumigatus. PLoS Pathog. 2021, 17, e1009806. [Google Scholar] [CrossRef] [PubMed]
- Bandyopadhyay, R.; Atehnkeng, J.; Ortega Beltran, A.; Akande, A.; Falade, T.D.O.; Cotty, P.J. ‘Ground-Truthing’ Efficacy of Biological Control for Aflatoxin Mitigation in Farmers’ Fields in Nigeria: From Field Trials to Commercial Usage, a 10-Year Study. Front. Microbiol. 2019, 10, 2528. [Google Scholar] [CrossRef] [PubMed]
- Zhi, Q.Q.; Li, J.Y.; Liu, Q.Y.; He, Z.M. A cytosine methyltransferase ortholog dmtA is involved in the sensitivity of Aspergillus flavus to environmental stresses. Fungal Biol. 2017, 121, 501–514. [Google Scholar] [CrossRef]
- Lehmbeck, J.; Andersen, B.; Sáez-Sáez, J.; Frisvad, J.C.; Arnau, J. Mycotoxin-Free Aspergillus oryzae Strain Lineage for Alternative and Novel Protein Production at Industrial Scale. Appl. Microbiol. Biotechnol. 2025, 109, 94. [Google Scholar] [CrossRef]
- Frisvad, J.C.; Møller, L.L.; Larsen, T.O.; Kumar, R.; Arnau, J. Safety of the Fungal Workhorses of Industrial Biotechnology: Update on the Mycotoxin and Secondary Metabolite Potential of Aspergillus niger, Aspergillus oryzae, and Trichoderma reesei. Appl. Microbiol. Biotechnol. 2018, 102, 9481–9515. [Google Scholar] [CrossRef]
- Jin, F.J.; Hu, S.; Wang, B.T.; Jin, L. Advances in Genetic Engineering Technology and Its Application in the Industrial Fungus Aspergillus oryzae. Front. Microbiol. 2021, 12, 644404. [Google Scholar] [CrossRef]
- Anantayanon, J.; Jeennor, S.; Panchanawaporn, S.; Chutrakul, C.; Laoteng, K. Significance of Two Intracellular Triacylglycerol Lipases of Aspergillus oryzae in Lipid Mobilization: A Perspective in Industrial Implication for Microbial Lipid Production. Gene 2021, 793, 145745. [Google Scholar] [CrossRef]
- Chutrakul, C.; Panchanawaporn, S.; Jeennor, S.; Anantayanon, J.; Laoteng, K. Promoter Exchange of the Cryptic Nonribosomal Peptide Synthetase Gene for Oligopeptide Production in Aspergillus oryzae. J. Microbiol. 2022, 60, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Zheng, J.; Yu, D.; Wang, B.; Pan, L. Efficient Genome Editing in Aspergillus niger with an Improved Recyclable CRISPR-HDR Toolbox and Its Application in Introducing Multiple Copies of Heterologous Genes. J. Microbiol. Methods 2019, 163, 105655. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, C.; Fu, Y.; Zhang, Q.; Ma, J.; Zhou, J.; Li, J.; Du, G.; Liu, S. A CRISPR/Cas9-Based Visual Toolkit Enabling Multiplex Integration at Specific Genomic Loci in Aspergillus niger. Synth. Syst. Biotechnol. 2024, 9, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Nie, H.; Jin, M.; Wang, Z.; Zheng, J.; Zhang, Y.; Li, X. Heterologous Expression of Candida antarctica Lipase B in Aspergillus niger Using CRISPR/Cas9-Mediated Multi-Gene Editing. Biotechnol. Bioeng. 2025, 122, 1770–1779. [Google Scholar] [CrossRef]
- Benites-Pariente, J.S.; Samolski, I.; Ludeña, Y.; Villena, G.K. CRISPR/Cas9 Mediated Targeted Knock-In of eglA Gene to Improve Endoglucanase Activity of Aspergillus fumigatus LMB-35Aa. Sci. Rep. 2024, 14, 19661. [Google Scholar] [CrossRef]
- Cairns, T.C.; Feurstein, C.; Zheng, X.; Zhang, L.H.; Zheng, P.; Sun, J.; Meyer, V. Functional Exploration of Co-Expression Networks Identifies a Nexus for Modulating Protein and Citric Acid Titres in Aspergillus niger Submerged Culture. Fungal Biol. Biotechnol. 2019, 6, 18. [Google Scholar] [CrossRef]
- Dong, L.; Lin, X.; Yu, D.; Huang, L.; Wang, B.; Pan, L. High-Level Expression of Highly Active and Thermostable Trehalase from Myceliophthora thermophila in Aspergillus niger by Using the CRISPR/Cas9 Tool and Its Application in Ethanol Fermentation. J. Ind. Microbiol. Biotechnol. 2020, 47, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Yu, D.; Lin, X.; Wang, B.; Pan, L. Improving Expression of Thermostable Trehalase from Myceliophthora sepedonium in Aspergillus niger Mediated by the CRISPR/Cas9 Tool and Its Purification, Characterization. Protein Expr. Purif. 2020, 165, 105482. [Google Scholar] [CrossRef]
- Gerhardt, J.A.; Rubio, M.V.; Terrasan, C.R.F.; Wassano, N.S.; Rodrigues, A.; Figueiredo, F.L.; Antoniel, E.P.; Contesini, F.J.; Dias, A.H.S.; Mortensen, U.H.; et al. Improving recombinant protein secretion in Aspergillus nidulans by targeting the N-glycosylation machinery. Metab. Eng. Commun. 2025, 20, e00264. [Google Scholar] [CrossRef]
- Zheng, X.; Zheng, P.; Zhang, K.; Cairns, T.C.; Meyer, V.; Sun, J.; Ma, Y. 5S rRNA Promoter for Guide RNA Expression Enabled Highly Efficient CRISPR/Cas9 Genome Editing in Aspergillus niger. ACS Synth. Biol. 2019, 8, 1568–1574. [Google Scholar] [CrossRef]
- Tong, Z.; Zheng, X.; Tong, Y.; Shi, Y.C.; Sun, J. Systems metabolic engineering for citric acid production by Aspergillus niger in the post-genomic era. Microb. Cell Fact. 2019, 18, 28. [Google Scholar] [CrossRef]
- Chen, M.; Zheng, X.; Du, P.; Ni, X.; Li, Z.; Zheng, P.; Sun, J. Genetic context-dependent regulation of FlbE enhances morphology modulation and citric acid production in Aspergillus niger. Bioresour. Technol. 2025, 439, 133350. [Google Scholar] [CrossRef] [PubMed]
- Kuivanen, J.; Wang, Y.M.J.; Richard, P. Engineering Aspergillus niger for Galactaric Acid Production: Elimination of Galactaric Acid Catabolism by Using RNA Sequencing and CRISPR/Cas9. Microb. Cell Fact. 2016, 15, 210. [Google Scholar] [CrossRef] [PubMed]
- Kuivanen, J.; Arvas, M.; Richard, P. Clustered Genes Encoding 2-Keto-L-Gulonate Reductase and L-Idonate 5-Dehydrogenase in the Novel Fungal D-Glucuronic Acid Pathway. Front. Microbiol. 2017, 8, 225. [Google Scholar] [CrossRef]
- Yang, L.; Henriksen, M.M.; Hansen, R.S.; Lubeck, M.; Vang, J.; Andersen, J.E.; Bille, S.; Lubeck, P.S. Metabolic engineering of Aspergillus niger via ribonucleoprotein-based CRISPR-Cas9 system for succinic acid production from renewable biomass. Biotechnol. Biofuels 2020, 13, 206. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, C.; Pei, L.; Qian, Q.; Lu, L. Production of L-Malic Acid by Metabolically Engineered Aspergillus nidulans Based on Efficient CRISPR-Cas9 and Cre-loxP Systems. J. Fungi 2023, 9, 719. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Shi, M.; Xu, Y.; Yang, D.; Lu, L.; Xue, F.; Xu, Q. Conditional Expression of FumA in Aspergillus niger Enhances Synthesis of L-Malic Acid. Appl. Environ. Microbiol. 2024, 90, e0000824. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Zhang, Z.; Long, C.; He, B.; Hu, Z.; Jiang, C.; Zeng, B. Identification and Functional Characterization of Glycerol Dehydrogenase Reveal the Role in Kojic Acid Synthesis in Aspergillus oryzae. World J. Microbiol. Biotechnol. 2020, 36, 136. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, H.; Chen, Z.; Fan, J.; Chen, T.; Zeng, B.; Zhang, Z. Construction of single, double, or triple mutants within kojic acid synthesis genes kojA, kojR, and kojT by the CRISPR/Cas9 tool in Aspergillus oryzae. Folia Microbiol. 2022, 67, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Qiu, T.; Chen, Z.; Zhang, H.; Deng, B.; Yao, L.; Zhang, Z. The Zn(II)2-Cys6-type zinc finger protein AoKap7 is involved in the growth, oxidative stress and kojic acid synthesis in Aspergillus oryzae. IMA Fungus 2025, 16, e153994. [Google Scholar] [CrossRef]
- Qiu, T.; Zhang, H.; Li, Y.; Yao, L.; Zhang, Z. Aokap9 gene knockout contributes to kojic acid synthesis in Aspergillus oryzae. Microb. Cell Factories 2025, 24, 221. [Google Scholar] [CrossRef]
- Weber, J.; Valiante, V.; Nodvig, C.S.; Mattern, D.J.; Slotkowski, R.A.; Mortensen, U.H.; Brakhage, A.A. Functional Reconstitution of a Fungal Natural Product Gene Cluster by Advanced Genome Editing. ACS Synth. Biol. 2017, 6, 62–68. [Google Scholar] [CrossRef]
- Shih, S.Y.; Mortensen, U.H.; Chang, F.R.; Tsai, H. Editing Aspergillus terreus Using the CRISPR-Cas9 System. Synth. Biol. 2022, 7, ysac031. [Google Scholar] [CrossRef]
- Zeng, X.; Zheng, J.; Lu, F.; Pan, L.; Wang, B. Heterologous Synthesis of Monacolin J by Reconstructing Its Biosynthetic Gene Cluster in Aspergillus niger. J. Fungi 2022, 8, 407. [Google Scholar] [CrossRef]
- Saito, N.; Katayama, T.; Minami, A.; Oikawa, H.; Maruyama, J.I. Versatile Filamentous Fungal Host Highly-Producing Heterologous Natural Products Developed by Genome Editing-Mediated Engineering of Multiple Metabolic Pathways. Commun. Biol. 2024, 7, 1263. [Google Scholar] [CrossRef]
- Roux, I.; Woodcraft, C.; Hu, J.; Wolters, R.; Gilchrist, C.L.M.; Chooi, Y.H. CRISPR-Mediated Activation of Biosynthetic Gene Clusters for Bioactive Molecule Discovery in Filamentous Fungi. ACS Synth. Biol. 2020, 9, 1843–1854. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Xu, Q.; Pang, M.; Ma, Y.; Zhang, Z.; Zhang, D.; Guo, D.; Wang, L.; Li, Q.; Li, Y.; et al. CRISPR-Cas9 Cytidine-Base-Editor Mediated Continuous In Vivo Evolution in Aspergillus nidulans. ACS Synth. Biol. 2025, 14, 621–628. [Google Scholar] [CrossRef]
- Li, X.; Huang, L.; Pan, L.; Wang, B.; Pan, L. CRISPR/dCas9-mediated epigenetic modification reveals differential regulation of histone acetylation on Aspergillus niger secondary metabolite. Microbiol. Res. 2021, 245, 126694. [Google Scholar] [CrossRef]
- Li, X. Study on Epigenetic Regulation Mechanisms of Secondary Metabolism in Aspergillus niger. Ph.D. Thesis, South China University of Technology, Guangdong, China, 2022. [Google Scholar]
- Maini Rekdal, V.; van der Luijt, C.R.B.; Chen, Y.; Kakumanu, R.; Baidoo, E.E.K.; Petzold, C.J.; Cruz-Morales, P.; Keasling, J.D. Edible mycelium bioengineered for enhanced nutritional value and sensory appeal using a modular synthetic biology toolkit. Nat. Commun. 2024, 5, 2099. [Google Scholar] [CrossRef]
- Li, L.; Liu, X.; Wei, K.; Lu, Y.; Jiang, W. Synthetic biology approaches for chromosomal integration of genes and pathways in industrial microbial systems. Biotechnol. Adv. 2019, 37, 730–745. [Google Scholar] [CrossRef]
- Wang, Q.; Cao, R.; Xie, Y.; Zhang, Z.; Li, X.; Zhang, Y.; Luo, H.; Yao, H.; Xue, P.; Ni, S. Unlocking the potential of engineered microbes in immunotoxin-based cancer therapy. Front. Microbiol. 2025, 16, 1603671. [Google Scholar] [CrossRef]
- Jeennor, S.; Anantayanon, J.; Panchanawaporn, S.; Chutrakul, C.; Vongsangnak, W.; Laoteng, K. Efficient de novo production of bioactive cordycepin by Aspergillus oryzae using a food-grade expression platform. Microb. Cell Fact. 2023, 22, 253. [Google Scholar] [CrossRef]
- Song, R.; Zhai, Q.; Sun, L.; Huang, E.; Zhang, Y.; Zhu, Y.; Guo, Q.; Tian, Y.; Zhao, B.; Lu, H. CRISPR/Cas9 genome editing technology in filamentous fungi: Progress and perspective. Appl. Microbiol. Biotechnol. 2019, 103, 6919–6932. [Google Scholar] [CrossRef] [PubMed]
- Cai, G.; Lin, Z.; Shi, S. Development and expansion of the CRISPR/Cas9 toolboxes for powerful genome engineering in yeast. Enzym. Microb. Technol. 2022, 159, 110056. [Google Scholar] [CrossRef]
- Gong, G.; Zhang, Y.; Wang, Z.; Liu, L.; Shi, S.; Siewers, V.; Yuan, Q.; Nielsen, J.; Zhang, X.; Liu, Z. GTR 2.0: gRNA-tRNA Array and Cas9-NG Based Genome Disruption and Single-Nucleotide Conversion in Saccharomyces cerevisiae. ACS Synth. Biol. 2021, 10, 1328–1337. [Google Scholar] [CrossRef] [PubMed]

| System Type | Signature Protein(s) | Target Molecule | Key Features | PAM Requirement | Cleavage Mechanism | References |
|---|---|---|---|---|---|---|
| Type I | Cas3 (helicase-nuclease) | DNA | Features a multi-subunit Cascade complex for surveillance and the Cas3 protein for processive DNA degradation. | Yes | Cas3 makes long-range deletions. | [45,47] |
| Type II | Cas9 | DNA | The most well-studied system. Uses a single Cas9 protein and a sgRNA. Cuts DNA, creating double-strand breaks. Diverse subtypes (II-A, B, C, D) with varying sizes and efficiencies exist. | Yes (e.g., SpCas9: NGG) | HNH and RuvC domains cut complementary and non-complementary strands, often resulting in blunt ends. | [45,48] |
| Type III | Cas10 | RNA/DNA | Complex systems that can target RNA. Some subtypes exhibit collateral, non-specific cleavage of RNA or DNA upon target recognition. | - | Involves the Cas10 protein. | [45,49] |
| Type IV | Unknown | DNA | Functionally less characterized. Believed to play a role in plasmid interference. | - | - | [45] |
| Type V | Cas12 (e.g., Cas12a/Cpf1) | DNA | Effectors like Cas12 are often smaller than Cas9. They originate from TnpB transposons, an evolutionary path revealed by the discovery of “TranC” intermediates. Cas12a. | Yes | A single RuvC domain cuts both DNA strands, creating staggered ends with overhangs. | [45,50] |
| Type VI | Cas13 | RNA | Targets and cleaves RNA molecules. Upon activation, some Cas13 proteins exhibit non-specific “collateral” cleavage of nearby RNAs, which has been leveraged for sensitive diagnostic tools like SHERLOCK. | - | - | [45,51] |
| Cas9 Source | Cas9 Type | Cas9 Name | PAM Sequence | Characteristics | References |
|---|---|---|---|---|---|
| Streptococcus pyogenes | Natural Cas9 orthologs | SpCas9 | NGG | Widely used; 3′-NGG on the non-target strand. | [58,59] |
| Staphylococcus aureus | Natural Cas9 orthologs | SaCas9 | NNGRRT (R=A or G) | Original PAM, restrictive. | [64,65] |
| Campylobacter jejuni | Natural Cas9 orthologs | CjCas9 | NNNNACAC | Ultra-compact; useful for tight spaces. | [66] |
| Streptococcus equinus HC5 | Natural Cas9 orthologs | SeHCas9 | NAG, NKG (K=G or T), NAW (W=A or T) | Similar to SpCas9 but harbored a different RxQ PAM-binding motif. | [59] |
| Streptococcus pyogenes | Engineered variants | xCas9 | NG, GAA, GAT | Broadened PAM recognition via directed evolution. | [58] |
| Campylobacter jejuni | Chimeric variants | Hsp1-Hsp2Cas9 | N4CY (Y=C or T) | Few off-targets compared to SpCas9. | [67] |
| Streptococcus equinus HC5 | Engineered variants | SeHdCas9-RR | NNG, NNN | Achieved PAM-free editing in bacterial systems. | [68] |
| Streptococcus canis | engineered variants | ScCas9++ | NNG | With high specificity and minimal off-target effects. | [69] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Hu, D.; Zhao, R.; Lin, Y.; Jiang, C. CRISPR-Cas9 Gene Editing in Aspergillus: From Pathogenesis to Metabolic Engineering. Biology 2026, 15, 53. https://doi.org/10.3390/biology15010053
Hu D, Zhao R, Lin Y, Jiang C. CRISPR-Cas9 Gene Editing in Aspergillus: From Pathogenesis to Metabolic Engineering. Biology. 2026; 15(1):53. https://doi.org/10.3390/biology15010053
Chicago/Turabian StyleHu, Danni, Ruoyu Zhao, Yingxu Lin, and Chunmiao Jiang. 2026. "CRISPR-Cas9 Gene Editing in Aspergillus: From Pathogenesis to Metabolic Engineering" Biology 15, no. 1: 53. https://doi.org/10.3390/biology15010053
APA StyleHu, D., Zhao, R., Lin, Y., & Jiang, C. (2026). CRISPR-Cas9 Gene Editing in Aspergillus: From Pathogenesis to Metabolic Engineering. Biology, 15(1), 53. https://doi.org/10.3390/biology15010053







