Study of FOXL2 Regulation on Ovarian Function in Chlamys farreri Through Comparative ChIP-Seq and Transcriptome Analysis Using RNA Interference
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Samples
2.2. RNA-Seq and Transcriptome Differential Expression Analysis
2.3. Real-Time Quantitative PCR Validation
2.4. ChIP-Seq
2.5. ChIP-qPCR Validation
3. Results
3.1. RNA-Seq Analysis
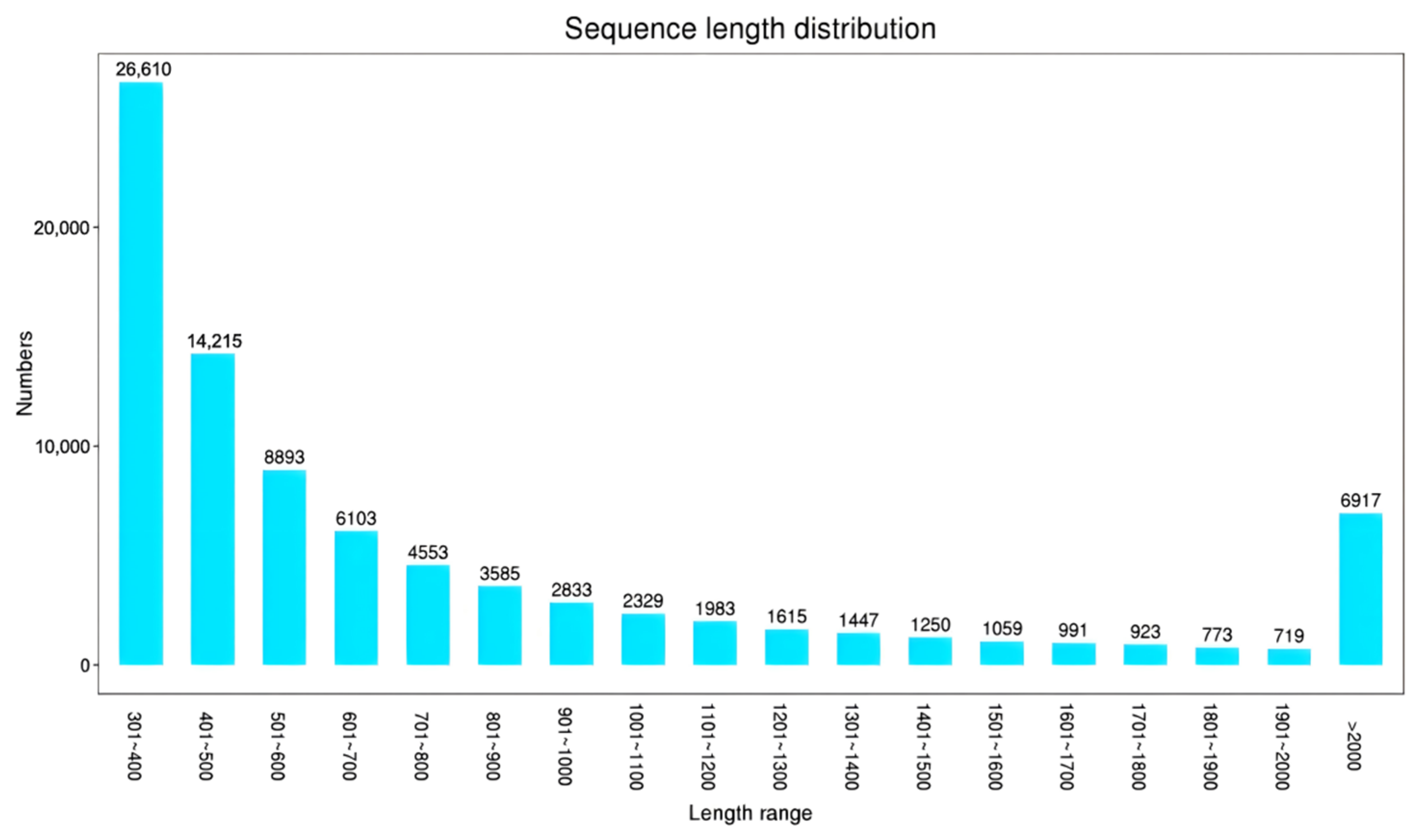
3.1.1. De Novo Assembly and Functional Gene Annotation
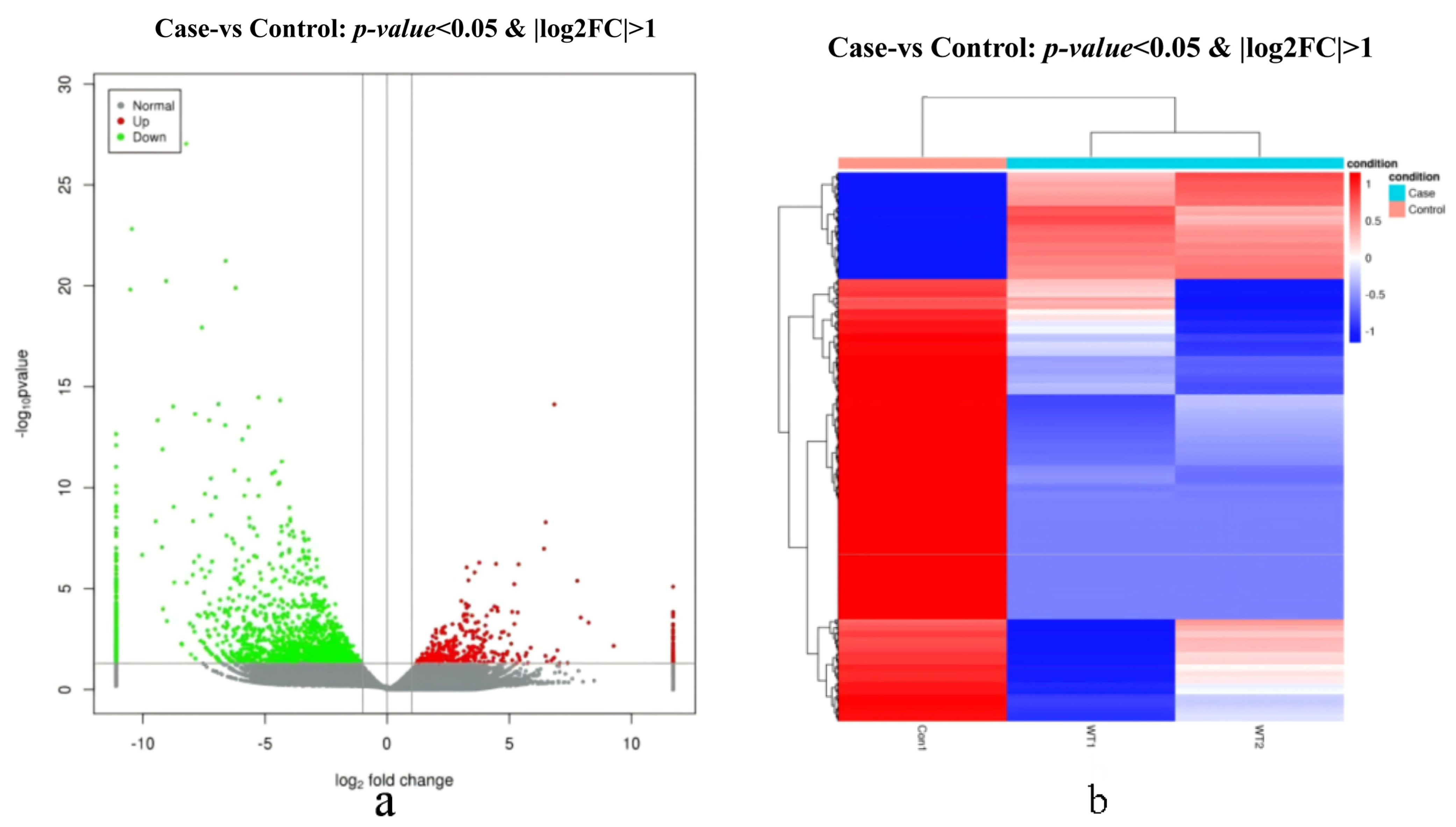
3.1.2. DEGs Analysis
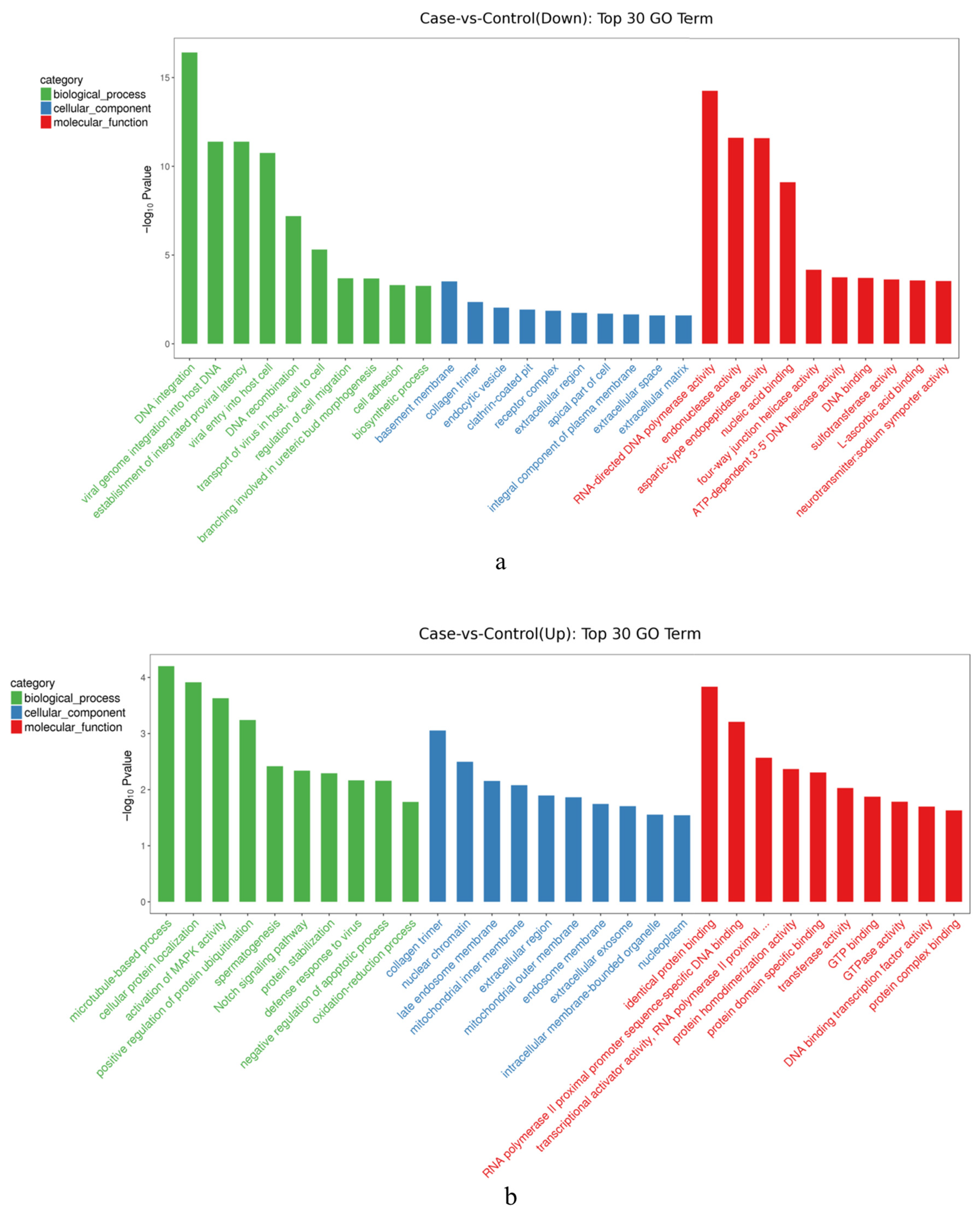
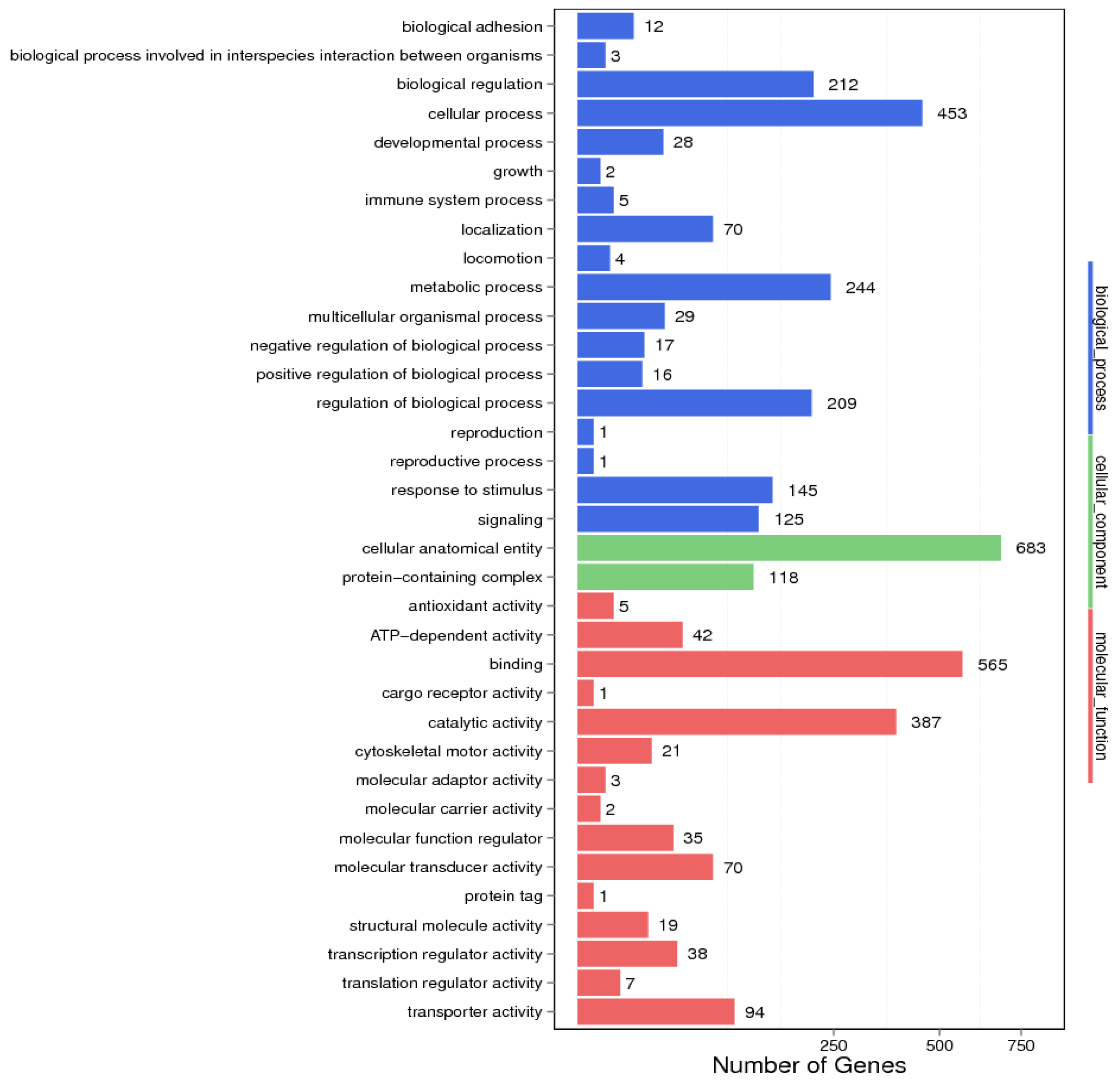
3.1.3. GO Annotation
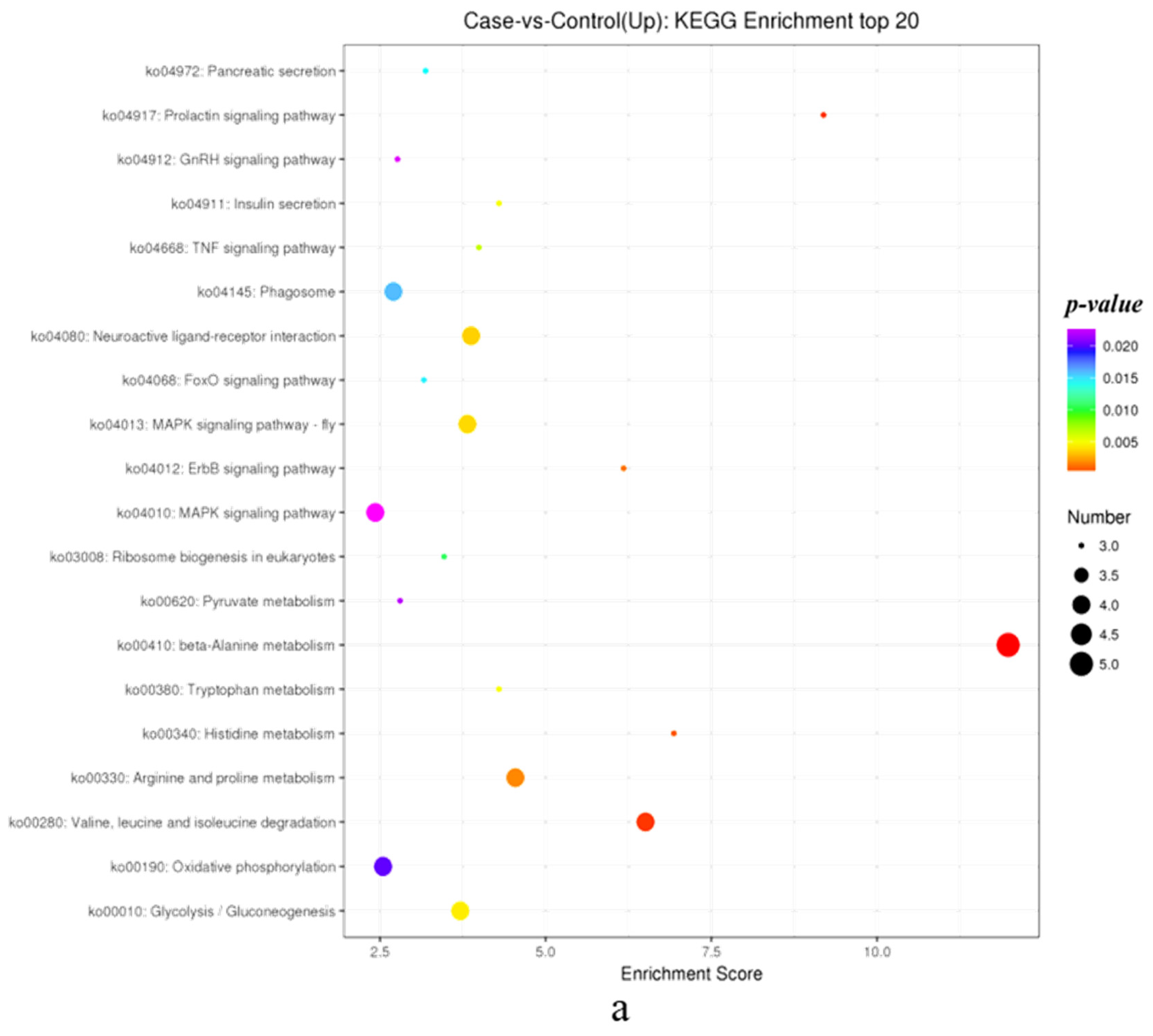
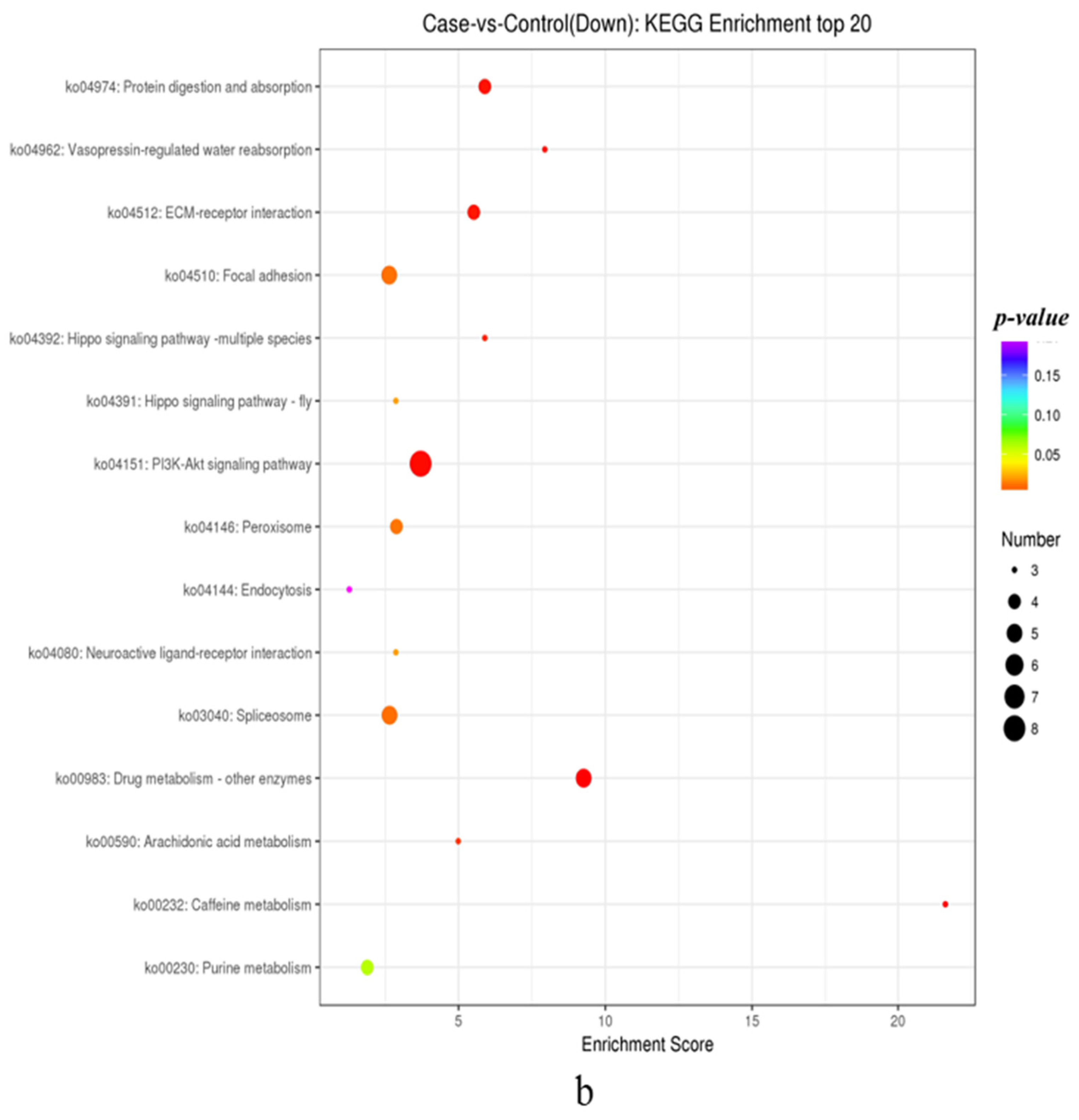
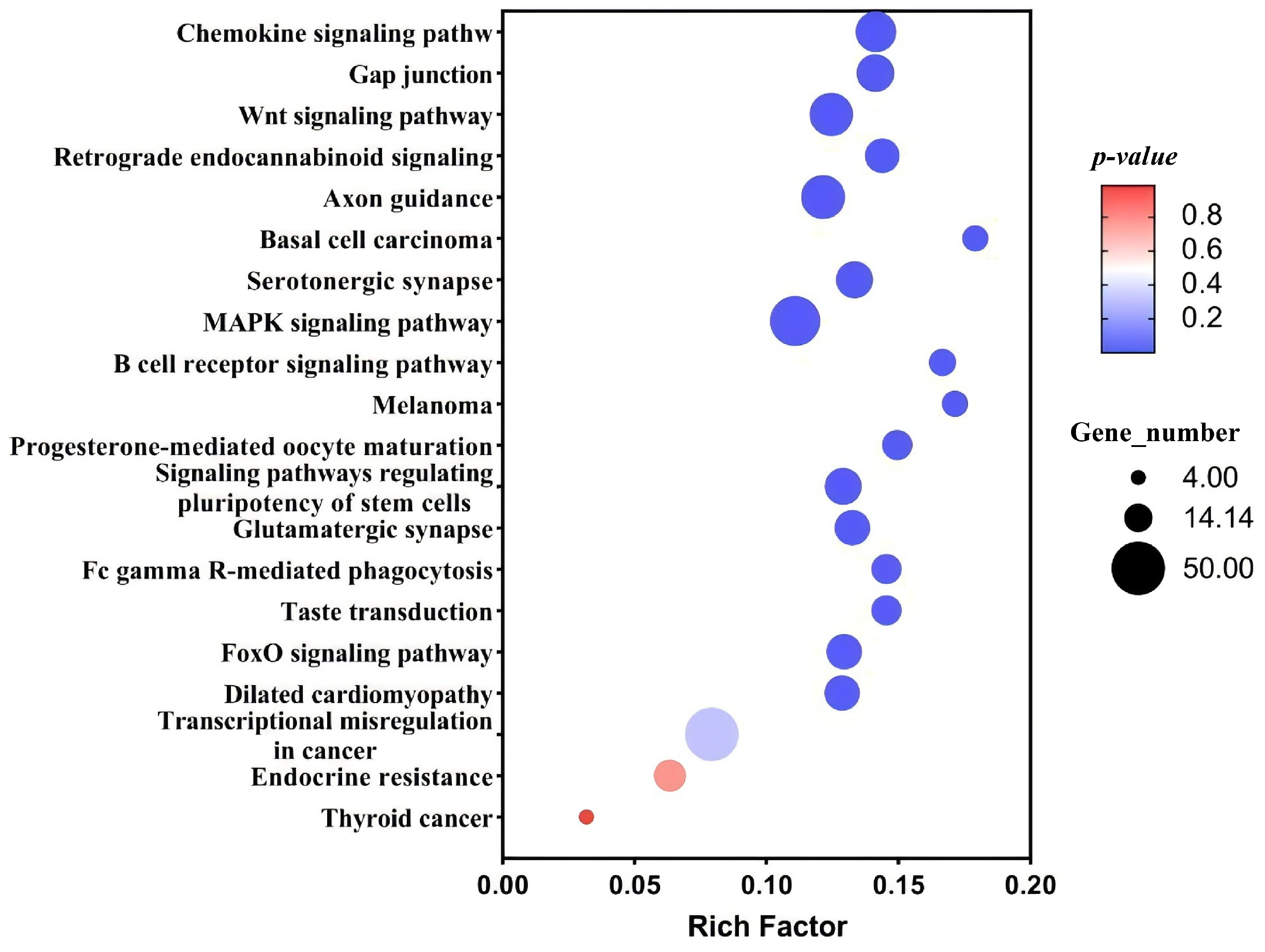
3.1.4. KEGG Annotation
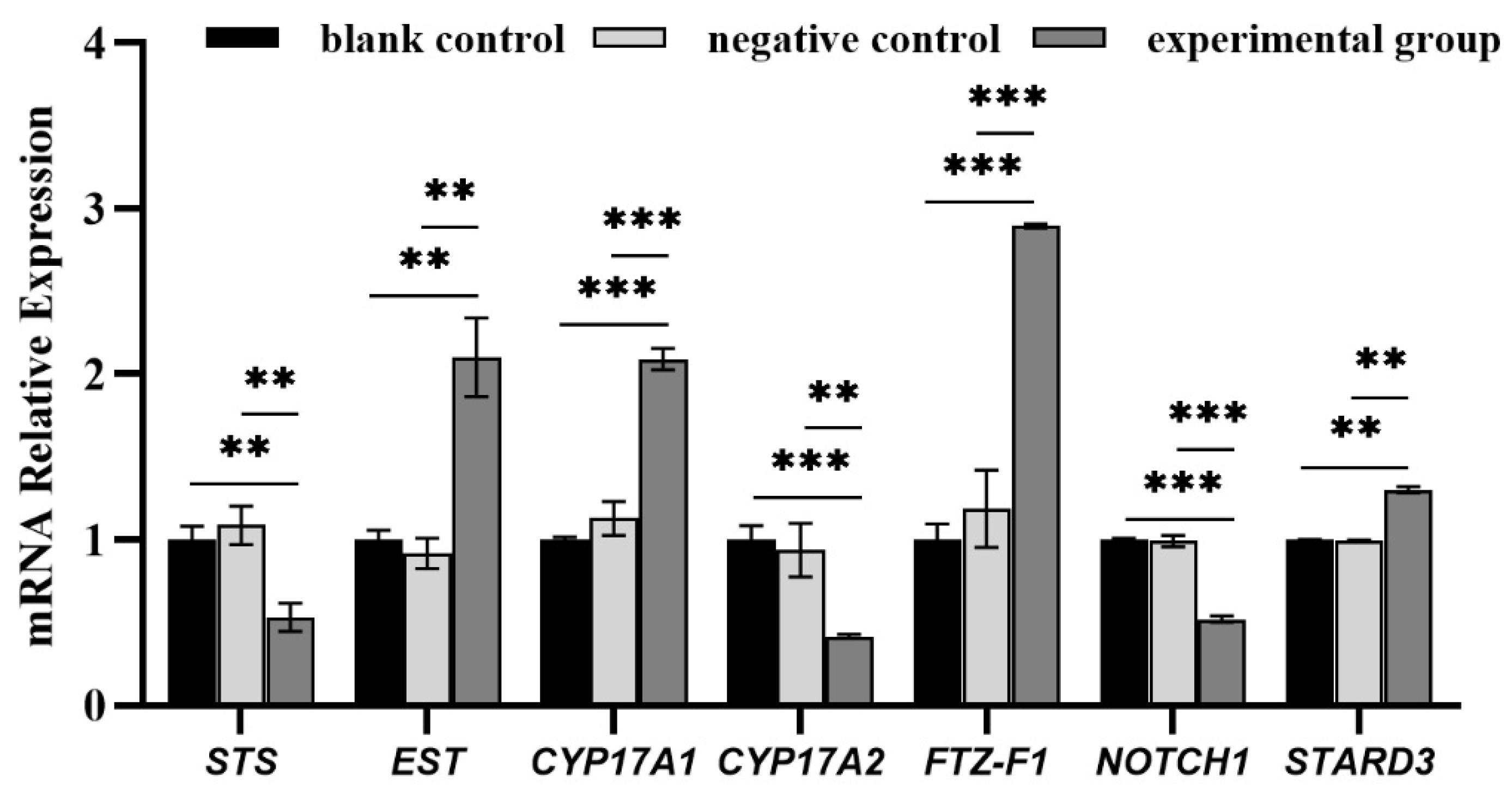
3.2. RT-qPCR Validation
3.3. ChIP-Seq Analysis
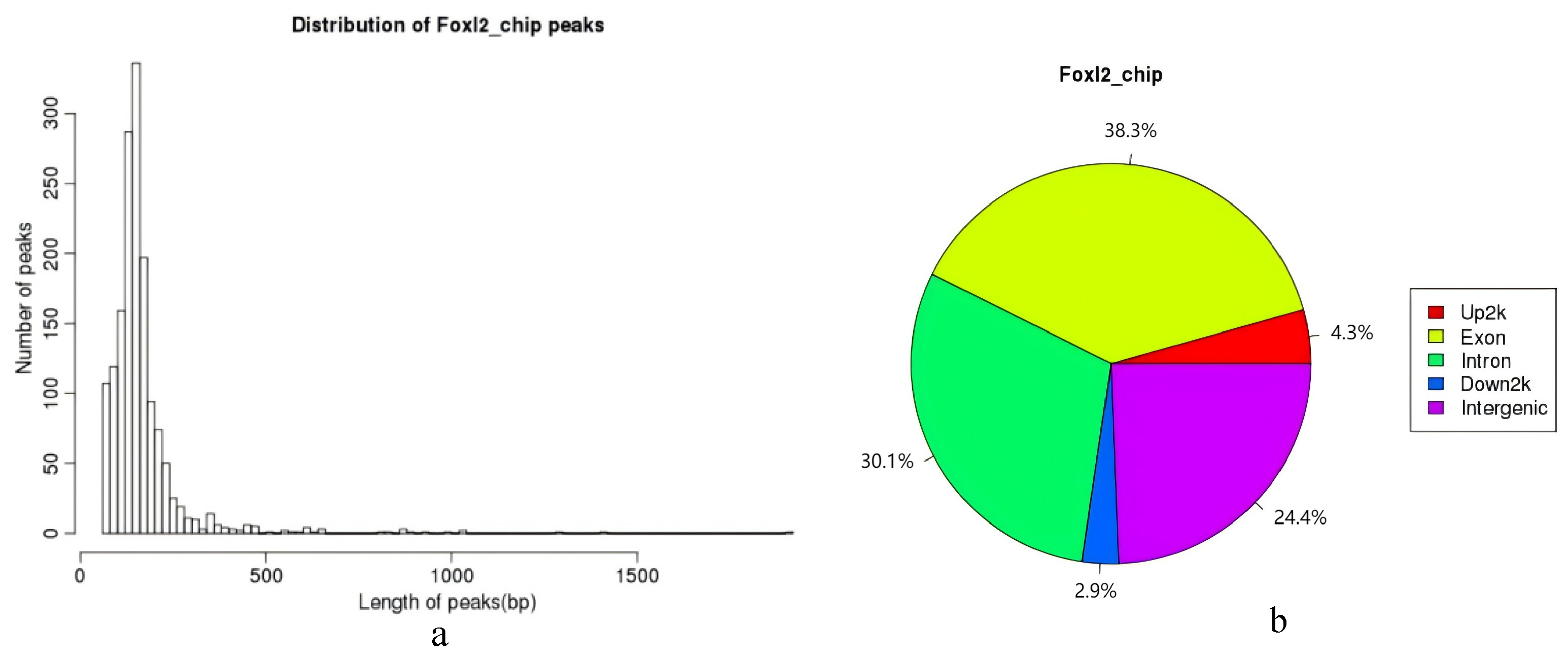
3.3.1. ChIP-Seq Peak Analysis
3.3.2. GO and KEGG Analysis of FOXL2 Target Sequences
3.3.3. Regulatory Candidate Genes of FOXL2
3.3.4. ChIP-qPCR
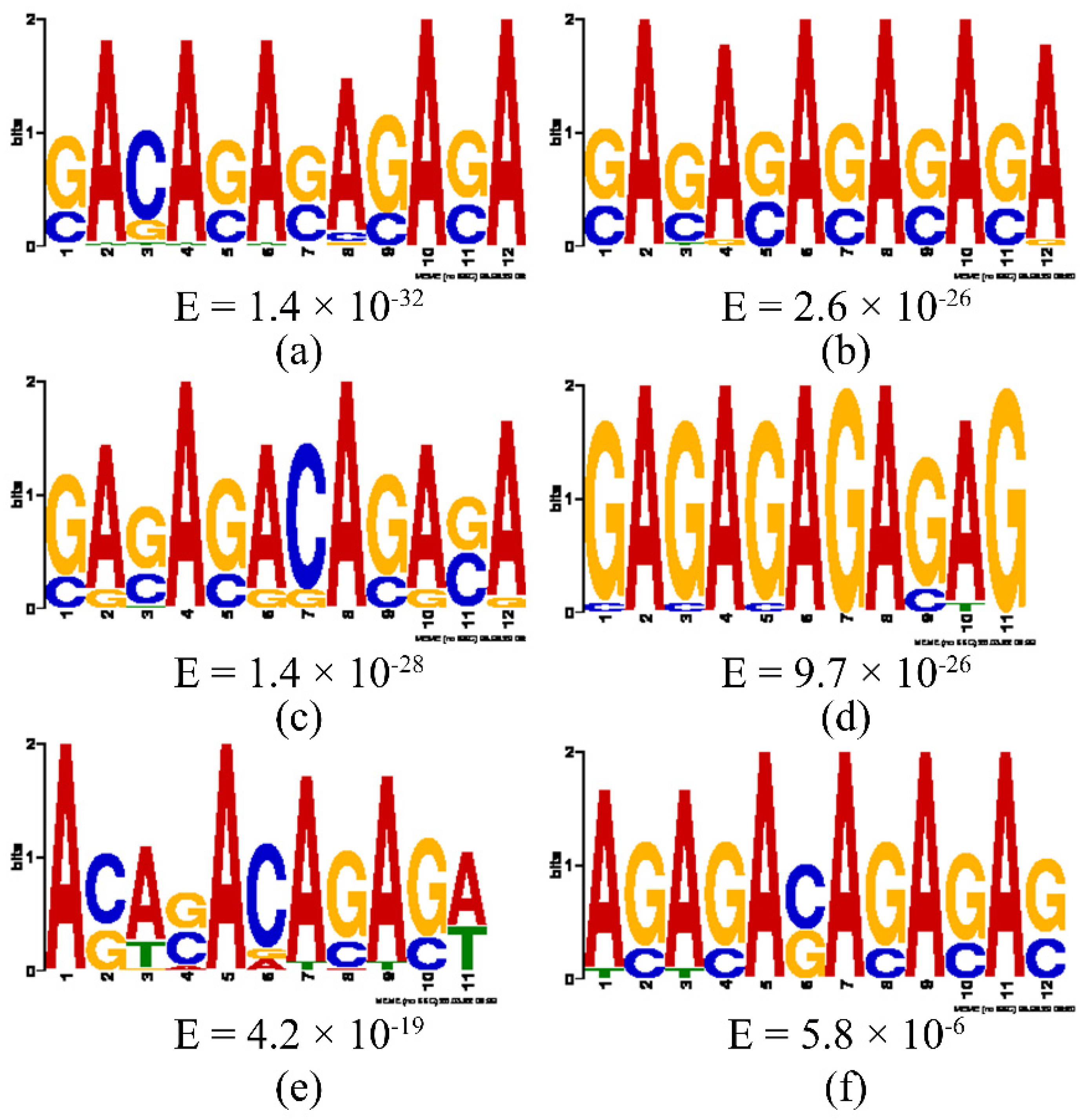
3.3.5. Motif Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pisaraka, M.D.; Bae, J.; Klein, C.; Hsueh, A.J.W. Forkhead L2 is expressed in the ovary and represses the promoter activity of the steroidogenic actute regulatory gene. Endocrinology 2004, 145, 3424–3433. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.S.; Kobayashi, T.; Zhou, L.Y.; Paul-Prasanth, B.; Ijiri, S.; Sakai, F.; Okubo, K.; Morohashi, K.I.; Nagahama, Y. Foxl2 up-regulates aromatase gene transcription in a female-specific manner by binding to the promoter as well as interacting with ad4bindingprotein/steroidogenic factor 1. Mol. Endocrinol. 2007, 21, 712–725. [Google Scholar] [CrossRef] [PubMed]
- Jakob, S. FOXL2 Represses the Testis-Specific Enhancer of Sox9 to Maintain Ovary Differentiation. Ph.D. Thesis, UCL (University College London), London, UK, 2010. [Google Scholar]
- Caburet, S.; Georges, A.; L’Hôte, D.; Veitia, R.A. The transcription factor FOXL2: At the crossroads of ovarian physiology and pathology. Mol. Cell. Endocrinol. 2011, 356, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Kashimada, K.; Pelosi, E.; Chen, H.; Schlessinger, D.; Wilhelm, D.; Koopman, P. FOXL2 and BMP2 Act Cooperatively to Regulate Follistatin Gene Expression during Ovarian Development. Endocrinology 2011, 152, 272–280. [Google Scholar] [CrossRef]
- Yang, X.W.; He, W.B.; Gong, F.; Li, W.; Li, X.R.; Zhong, C.G.; Lu, G.X.; Lin, G.; Du, J.; Tan, Y.Q. Novel FOXL2 mutations cause blepharophimosis-ptosis-epicanthus inversus syndrome with premature ovarian insufficiency. Mol. Genet. Genom. Med. 2017, 6, 261–267. [Google Scholar] [CrossRef]
- Hudson, Q.J.; Smith, C.A.; Sinclair, A.H. Aromatase inhibition reduces expression of FOXL2 in the embryonic chicken ovary. Dev. Dyn. 2005, 233, 1052–1055. [Google Scholar] [CrossRef]
- Kuo, F.T.; Fan, K.; Bentsi-Barnes, I.; Barlow, G.M.; Pisarska, M.D. Mouse forkhead L2 maintains repression of FSH-dependent genes in the granulosa cell. Reproduction 2012, 144, 485–494. [Google Scholar] [CrossRef]
- Pannetier, M. FOXL2 activates P450 aromatase gene transcription: Towards a better characterization of the early steps of mammalian ovarian development. J. Mol. Endocrinol. 2006, 36, 399–413. [Google Scholar] [CrossRef]
- Guiguen, Y.; Fostier, A.; Piferrer, F.; Chang, C.F. Ovarian aromatase and estrogens: A pivotal role for gonadal sex differentiation and sex change in fish. Gen. Comp. Endocrinol. 2010, 165, 352–366. [Google Scholar] [CrossRef]
- Liu, X.L.; Zhang, Z.F.; Shao, M.Y.; Liu, J.G.; Muhammad, F. Sexually dimorphic expression of foxl2 during gametogenesis in scallop Chlamys farreri, conserved with vertebrates. Dev. Genes. Evol. 2012, 222, 279–286. [Google Scholar] [CrossRef]
- Ye, R.H.; Ren, H.B. Molecular cloning and gene expression of foxl2 and sox14 gene from Hyriopsis cumingii. Oceanol. Limnol. Sin. 2018, 49, 160–167. [Google Scholar]
- Tang, L.; Hu, Q.; Zhao, R.Y.; Li, J.; Gulipari, A.; Zhang, J.J. Cloning and expression of foxl2 gene in Esox lucius. Guizhou Agric. Sci. 2020, 48, 97–101. [Google Scholar]
- He, J.Q.; Zhang, H.; She, Z.C.; Zhen, W.Q.; You, S.Y.; Wang, P.L.; Xu, Y.H.; Zhu, P. Cloning and expression pattern investigation of Crassostrea hongkongensis FOXL2 gene. Genom. Appl. Biol. 2020, 39, 1519–1528. [Google Scholar]
- Ren, X.L.; Liang, S.Q.; Zhong, Y.F.; Wang, Y.R.; Lao, J.T.; Li, G.L.; Huang, H.; Chen, H.P. Molecular cloning and tissue expression patterns of foxl2 gene in Bettas plendens. Genom. Appl. Biol. 2021, 40, 109–116. [Google Scholar]
- Zhao, D.; Zhou, L.Q.; Zheng, Y.X.; Sun, X.J.; Wu, B.; Liu, Z.H.; Wu, Z.; Wu, L. Expression pattern of the foxl2 gene in the scallop Patinopecten yessoensis during development. Prog. Fish. Sci. 2022, 43, 138–145. [Google Scholar]
- Liao, X.T. Study on the Gonadal Development and Expression of Sex Related Genes dmrt1 and foxl2 in clam Cyclina sinensis. Master’s Thesis, Jiangsu Ocean University, Lianyungang, China, 2022. [Google Scholar]
- Liu, X.L.; Liu, J.; Wang, Z.D.; Wang, Z.H.; Li, Y.S.; Cui, L.B. Necessity of foxl2 in the oogenesis of the scallop Chlamys farreri. Mar. Sci. 2018, 42, 127–131. [Google Scholar]
- Ning, J.H.; Cao, W.A.; Lu, X.; Chen, M.; Liu, B.; Wang, C.D. Identification and functional analysis of a sex-biased transcriptional factor Foxl2 in the bay scallop Argopecten irradians. Comp. Biochem. Physiol. B-Biochem. Mol. Biol. 2012, 256, 110638. [Google Scholar] [CrossRef]
- Barbara, N.; Grimm, S.A.; Artiom, G.; Scott, G.J.; Ray, M.K.; Yao, H.C. Genome-wide identification of FOXL2 binding and characterization of FOXL2 feminizing action in the fetal gonads. Hum. Mol. Genet. 2018, 27, 4273–4287. [Google Scholar] [CrossRef]
- Yang, D.D. Expression Characteristics and Functional Analysis of Krüppel-like Factor 4 in Chlamys farreri Spermatogenesis and Muscle Tissue. Master’s Thesis, Ocean University of China, Qingdao, China, 2018. [Google Scholar]
- Liu, X.L.; Wang, Z.D.; Sun, H.T.; Wang, Z.H.; Zhao, Z.J.; Li, J. Prokaryotic expression and purification of Foxl2 protein, preparation and detection of polyclonal antibody in Chlamys farreri. Mar. Fish. 2018, 40, 447–453. [Google Scholar]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Science Press: Beijing, China, 2002; pp. 518–531. [Google Scholar]
- Roberts, A.; Trapnell, C.; Donaghey, J.; Rinn, J.; Pachter, L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 2011, 12, R22. [Google Scholar] [CrossRef]
- Zhou, Q.R.; Shao, M.Y.; Qin, Z.K.; Kyoung, H.K.; Zhang, Z.F. Cloning, characterization, and expression analysis of the DEAD-box family genes, Fc-vasa and Fc-PL10a, in Chinese shrimp (Fenneropenaeus chinensis). Chin. J. Oceanol. Limnol. 2010, 28, 37–45. [Google Scholar] [CrossRef]
- Liu, F.Y.; Li, Y.L.; Yu, H.W.; Zhang, L.L.; Hu, J.J.; Bao, Z.M.; Wang, S. MolluscDB: An integrated functional and evolutionary genomics database for the hyper-diverse animal phylum Mollusca. Nucleic Acids Res. 2021, 49, 988–997. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, T.; Meyer, C.A.; Eeckhoute, J.; Johnson, D.S.; Bernstein, B.E.; Nusbau, C.; Myers, R.M.; Browm, M.; Li, W.; et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008, 9, R137. [Google Scholar] [CrossRef]
- Liu, X.L.; Liu, J.G.; Wang, D.; Zhang, Z.Y.; Zhang, Z.F. Expression pattern of the foxl2 gene in the scallop Chlamys farreri during development. JFSC 2013, 20, 205–210. [Google Scholar] [CrossRef]
- Liu, X.L.; Li, Y.; Liu, J.G.; Cui, L.B.; Zhang, Z.F. Gonadogenesis in scallop Chlamys farreri and Cf-foxl2 expression pattern during gonadal sex differentiation. Aquac. Res. 2016, 47, 1605–1611. [Google Scholar] [CrossRef]
- Wang, J. Study on the Molecular Mechanism of foxl2 in Chicken Geanulosa Cell Differentiation. Master’s Thesis, Huazhong Agricultural University, Wuhan, China, 2017. [Google Scholar]
- Thitiphuree, T.; Nagasawa, K.; Osada, M. Molecular identification of steroidogenesis-related genes in scallops and their potential roles in gametogenesis. J. Steroid Biochem. Mol. Biol. 2019, 186, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Mu, X.J. Sequence Characteristics and Expression Analysis of FTZ-F1, CYP17A1 and CYP17A2 Genes in Chlamys farreri. Master’s Thesis, Yantai University, Yantai, China, 2021. [Google Scholar]
- Guo, X.L.; Qiu, X.M.; Chang, Y.Q.; Zhang, W.J.; Cui, J.; Cong, Y.T.; Wu, L.Z.; Liu, Y.; Wang, X.L. Cloning and differential expression of CYP17 Gene in sea urchin Strongylocentrous intermedius. J. Dalian Ocean. Univ. 2013, 28, 1–6. [Google Scholar]
- Watari, H.; Arakane, F.; Moog-Lutz, C.; Kallen, C.B.; Tomasetto, C.; Gerton, G.L.; Rio, M.C.; Baker, M.E.; Strauss, J.F., III. MLN64contains a domain with homology to the steroidogenic acute regulatory protein (StAR) that stimulates steroidogenesis. Proc. Natl. Acad. Sci. USA 1997, 94, 8462–8467. [Google Scholar] [CrossRef]
- Tomasetto, C.; Régnier, C.; Moog-Lutz, C.; Mattei, M.G.; Chenard, M.P.; Lidereau, R.; Basset, P.; Rio, M.C. Identification of four novel human genes amplified and overexpressed in breast carcinoma and localized to the q11-q21.3 region of chromosome 17. Genomics 1995, 28, 367–376. [Google Scholar] [CrossRef]
- Akiyama, N.; Sasaki, H.; Ishizuka, T.; Kishi, T.; Sakamoto, H.; Onda, M.; Hirai, H.; Yazaki, Y.; Sugimura, T.; Terada, M. Isolation of a candidate gene, CAB1, for cholesterol transport to mitochondria from the c-ERBB-2 amplicon by a modified cDNA selection method. Cancer Res. 1997, 57, 3548–3553. [Google Scholar]
- Sugawara, T.; Holt, J.A.; Driscoll, D.; Strauss, J.F., 3rd; Lin, D.; Miller, W.L.; Patterson, D.; Clancy, K.P.; Hart, I.M.; Clark, B.J. Human steroidogenic acute regulatory protein: Functional activity in COS-1 cells, tissue-specific expression, and mapping of the structural gene to 8p11.2 and a pseudogene to chromosome13. Proc. Natl. Acad. Sci. USA 1995, 92, 4778–4782. [Google Scholar] [CrossRef]
- Luo, D.Y.; Zhai, X.T.; Liu, X.L.; Cui, L.B. Sequence analysis and expression pattern of STARD3 cDNA in Chlamys farreri. Mar. Fish. 2024, 46, 579–585. [Google Scholar]
- Takasawa, K.; Pelosi, E.; Takagi, M.; Morio, T.; Asahara, H.; Schlessinger, D.; Mizutani, S.; Koopman, P.; Kashimada, K. FOXL2 transcriptionally represses Sf1 expression by antagonizing WT1 during ovarian development in mice. FASEB J. 2014, 28, 2020–2028. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, L.L.; Li, R.J.; Xie, X.R.; Li, Y.P.; Li, W.R.; Bao, Z.M. Screening of interacting proteins of FOXL2 in scallop Patinopecten yessoensis by Yeast Two-Hybrid System. Period. Ocean. Univ. China 2019, 49, 54–59. [Google Scholar]
- Shao, C.; Yang, O.; Wang, H.; Gao, X.K.; Chen, B.Q.; Shao, G.X. Research on GABAR related proteins and ENOS responsive proteins in human spermatogenesis disorders. In Proceedings of the 5th National Male Science Academic Conference of the Chinese Medical Association, Nanjing, China, August 2004. Male Science Branch of the Chinese Medical Association, 21st Century Male Science. [Google Scholar]
- Wang, W.; Ruan, J.M.; Zhou, A.L.; Zhao, Y.N.; Cao, H.P.; Hu, K.; Yang, X.L. Cloning and Expression of Partial cDNA Encoding GABAB Receptor Subunit1 in Carassius auratus gibelio. Chin. J. Zool. 2013, 48, 905–911. [Google Scholar]
- Saito, M.; Kumamoto, K.; Robles, A.I.; Horikawa, I.; Furusato, B.; Okamura, S.; Goto, A.; Yamashita, T.; Nagashima, M.; Lee, T.L.; et al. Targeted disruption of Ing2 results in defective spermatogenesis and development of soft-tissue sarcomas. PLoS ONE 2010, 5, 0015541. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Ni, B.; Tian, Z.; Yang, F.; Gao, Y. Regulatory effects of autophagy on spermatogenesis. Biol. Reprod. 2017, 96, 525–530. [Google Scholar] [CrossRef] [PubMed]
- Tarique, I. Cellular Charactieristics of Autophagy and Secretions in the Make Reproductive System of the Chinese Soft-Shelled Turtle, Pelodiscus sinensis. Ph.D. Thesis, Nanjing Agricultural University, Nanjing, China, 2020. [Google Scholar]
- Tao, X.H.; Liu, M.; Qiu, H. Research progress on Sirt1 regulation of aging in mammals. Gansu Sci. Technol. 2016, 32, 77–80. [Google Scholar]
- Ni, J.B.; Zeng, Z.; Han, G.D.; Huang, H.Q.; Ke, C.H. Cloning and characterization of the follistatin gene from Crassostrea angulata and its expression during the reproductive cycle. Comp. Biochem. Physiol. B-Biochem. Mol. Biol. 2012, 163, 246–253. [Google Scholar] [CrossRef]
- Li, H.L. Cloning of Gene Wnt4, β-catenin, Dax1, and Their Correlation to the Gonadal Development in Scallop Chlamys farreri. Ph.D. Thesis, Ocean University of China, Qingdao, China, 2013. [Google Scholar]
- Su, Y.Q.; Sun, F.; Handel, M.A.; Schimenti, J.C.; Eppig, J.J. Meiosis arrest female 1 (MARF1) has nuage-like function in mammalian oocytes. Proc. Natl. Acad. Sci. USA 2012, 109, 18653–18660. [Google Scholar]
- Yao, Q.Q.; Cao, G.Y.; Li, M.Z.; Su, Y.Q. Ribonuclease activity of MARF1 controls oocyte RNA homeostasis and genome integrity in mice. Proc. Natl. Acad. Sci. USA 2018, 115, 11250–11255. [Google Scholar] [CrossRef]
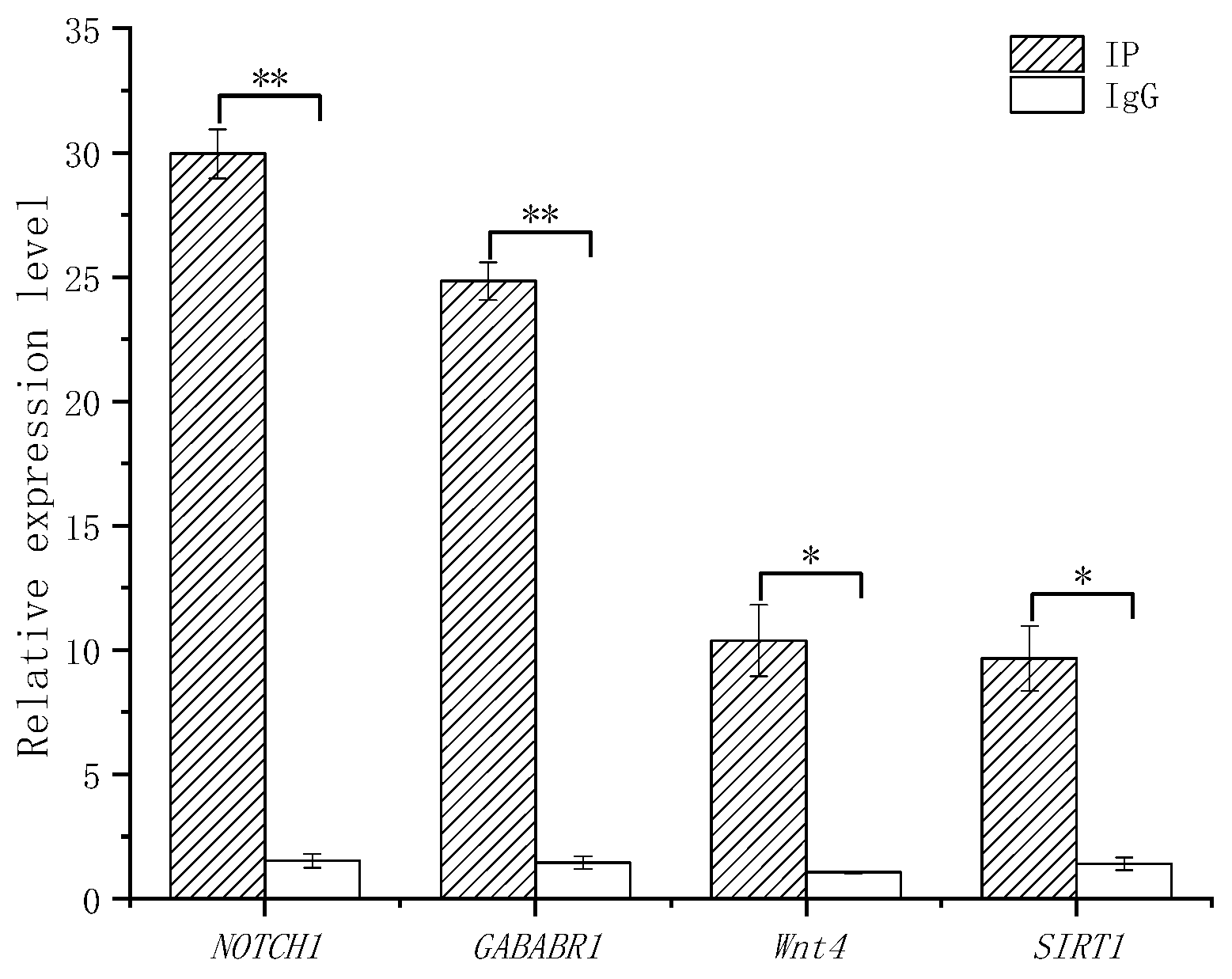
| Gene Name | Primer Sequence (5′→3′) | Product Length | Melting Temperature | Usage |
|---|---|---|---|---|
| Steroid Sulfatase (STS)-F | GGGTTCTTTTGTTCGTCTGGC | 146 bp | 58.97 | RT-qPCR |
| Steroid Sulfatase (STS)-R | TTTCCCGGGCTTCCAAGAATT | 59.35 | ||
| Estrogen Sulfotransferase (EST)-F | GGTGTGATGAAGGTCAAGGG | 105 bp | 60.67 | RT-qPCR |
| Estrogen Sulfotransferase (EST)-R | TACACATCGCCACAGAAGCA | 59.78 | ||
| Cytochrome P450c17A1 (CYP17A1)-F | CCAAGTAGCCGATTCAAAAAAGTGT | 144 bp | 59.81 | RT-qPCR |
| Cytochrome P450c17A1 (CYP17A1)-R | TCCAGCAAAGAAAATGTCAGCA | 60.87 | ||
| Cytochrome P450c17A2 (CYP17A2)-F | GATGTGGACGATGCTTTTCTC | 114 bp | 60.03 | RT-qPCR |
| Cytochrome P450c17A2 (CYP17A2)-R | TGTTTTGCCTGTTGCTGTTC | 59.73 | ||
| Fushi-Tarazu Factor-1 (FTZ-F1)-F | TAGAGGCAGTGAGACAGGATAGAA | 139 bp | 59.65 | RT-qPCR |
| Fushi-Tarazu Factor-1 (FTZ-F1)-R StAR-Related Lipid Transfer Protein 3 (STARD3)-F StAR-Related Lipid Transfer Protein 3 (STARD3)-R | GATTGCTGGGTCGGGTTTA GCGGGGTAGACAAACGGA ATGTCTCCTGTTCGCCGT | 128 bp | 60.78 60.54 60.02 | RT-qPCR |
| Neurogenic Locus Notch Homolog Protein 1 (NOTCH1)-F1 | GTTTACATCTGCTGAAGTGTGGAT | 155 bp | 59.59 | RT-qPCR |
| Neurogenic Locus Notch Homolog Protein 1 (NOTCH1)-R1 | CTCTGTGTCTTTTCTAGCCGTGTA | 60.15 | ||
| β-actin-F | TTCTTGGGAATGGAATCTGC | 119 bp | 58.89 | RT-qPCR |
| β-actin-R | GCCAGACTCGTCGTATTCCT | 58.12 | ||
| Neurogenic Locus Notch Homolog Protein 1 (NOTCH1)-F2 | GGAGAGGGACAACCAACACC | 117 bp | 58.7 | ChIP-qPCR |
| Neurogenic Locus Notch Homolog Protein 1 (NOTCH1)-R2 | TCACATTTGGATGGTTTCTGGA | 60.2 | ||
| γ-Aminobutyric Acid Type B Receptor Subunit 1 (GABABR1)-F | ATCAAGTGGTCCGCAACTCT | 49 bp | 56.8 | ChIP-qPCR |
| γ-Aminobutyric Acid Type B Receptor Subunit 1 (GABABR1)-R | TCATTGCATGACCTGTTGCC | 59.7 | ||
| Wingless-Type MMTV Integration Site Family, Member 4 (Wnt4)-F | AGGTTGGGAAACCCTTGC | 55 bp | 57 | ChIP-qPCR |
| Wingless-Type MMTV Integration Site Family, Member 4 (Wnt4)-R | CACAACTTGGCAGCACCA | 56.1 | ||
| Silent Mating Typeinformation Regulation 2 Homolog 1 (SIRT1)-F | GCCAAGCAGTTCAACATCAA | 74 bp | 56.8 | ChIP-qPCR |
| Silent Mating Typeinformation Regulation 2 Homolog 1 (SIRT1)-R | CTCCTGATGTTCCACAAATCC | 56.7 |
| Sample | Raw_Reads | Clean_Reads | Valid_Reads (%) | Q30 (%) | GC (%) |
|---|---|---|---|---|---|
| Con1 | 49,024,148 | 45,647,374 | 89.53 | 92.31 | 40.81 |
| KD1 | 44,400,934 | 42,162,166 | 91.97 | 93.62 | 41.92 |
| KD2 | 49,725,368 | 46,588,672 | 90.16 | 92.68 | 40.45 |
| Sample | Clean Reads | Clean Ratio | Mapped Reads | Mapped |
|---|---|---|---|---|
| FOXL2-IP | 23,700,286 | 99.80% | 10,424,229 | 43.98% |
| Input | 23,524,839 | 99.06% | 17,106,484 | 72.72% |
| Gene ID | Peak Start | Peak End | Peak Annotation | Gene Name |
|---|---|---|---|---|
| Steroid Hormone | ||||
| 110441780 | 1,660,594 | 1,660,681 | estradiol 17-beta-dehydrogenase 8-like | HSD17b8 |
| 110447098 | 2,005,851 | 2,005,913 | cytochrome P450 2J6-like | CYP2J6 |
| 110451342 | 431,036 | 431,098 | cytosolic phospholipase A2-like | cPLA2 |
| 110451344 | 520,879 | 520,970 | prostaglandin G/H synthase 2-like | PGE2 |
| 110461839 | 815,032 | 815,194 | adenylate cyclase type 9-like | ADCY9 |
| 110445220 | 505,918 | 506,027 | protein yippee-like 1 | YPEL1 |
| 110447714 | 109,981 | 110,076 | son of sevenless homolog 2-like | SOS2 |
| 110445266 | 925,477 | 925,554 | EF-hand calcium-binding domain-containing protein 11-like | EFCAB11 |
| 110459812 | 161,479 | 161,605 | RAC-beta serine/threonine-protein kinase B-like | AKT2 |
| 110460076 | 25,292 | 25,362 | SHC-transforming protein 4-like | SHC4 |
| 110460372 | 108,457 | 108,539 | inositol 1,4,5-trisphosphate receptor type 1-like | IP3R1 |
| 110461441 | 1,140,080 | 1,140,262 | 1-phosphatidylinositol 4,5-bisphosphate 2-phosphodiesterase beta-4-like | PLCB4 |
| 110445501 | 1,702,410 | 1,702,520 | GTPase HRas | Hras1 |
| 110463419 | 101,557 | 101,621 | hydroxysteroid 11-beta-dehydrogenase 1-like protein | HSD11B1 |
| 110464786 | 243,337 | 243,491 | lysosome membrane protein 2-like | LAMP2 |
| 110449851 | 223,863 | 224,009 | neurogenic locus notch homolog protein 1-like | NOTCH1 |
| Spermatogenesis | ||||
| 110459375 | 1,049,938 | 1,050,020 | gamma-aminobutyric acid type B receptor subunit 1-like | GABABR1 |
| 110456833 | 188,225 | 188,363 | golgin subfamily A member 3-like | GOLGA3 |
| Gonadal Development | ||||
| 110451002 | 72,929 | 73,016 | protein Wnt-4-like | Wnt4 |
| 110455012 | 1,822,037 | 1,822,131 | insulin-like peptide receptor | ILPR |
| Prevention of Premature Ovarian Failure | ||||
| 110460060 | 130,987 | 131,077 | NAD-dependent protein deacetylase sirtuin-1-like | SIRT1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Yun, H.; Xing, Y.; Wang, S.; Zhou, X.; Zhang, J. Study of FOXL2 Regulation on Ovarian Function in Chlamys farreri Through Comparative ChIP-Seq and Transcriptome Analysis Using RNA Interference. Biology 2025, 14, 1259. https://doi.org/10.3390/biology14091259
Liu X, Yun H, Xing Y, Wang S, Zhou X, Zhang J. Study of FOXL2 Regulation on Ovarian Function in Chlamys farreri Through Comparative ChIP-Seq and Transcriptome Analysis Using RNA Interference. Biology. 2025; 14(9):1259. https://doi.org/10.3390/biology14091259
Chicago/Turabian StyleLiu, Xiaoling, Han Yun, Yan Xing, Shuo Wang, Xueying Zhou, and Jianbai Zhang. 2025. "Study of FOXL2 Regulation on Ovarian Function in Chlamys farreri Through Comparative ChIP-Seq and Transcriptome Analysis Using RNA Interference" Biology 14, no. 9: 1259. https://doi.org/10.3390/biology14091259
APA StyleLiu, X., Yun, H., Xing, Y., Wang, S., Zhou, X., & Zhang, J. (2025). Study of FOXL2 Regulation on Ovarian Function in Chlamys farreri Through Comparative ChIP-Seq and Transcriptome Analysis Using RNA Interference. Biology, 14(9), 1259. https://doi.org/10.3390/biology14091259





