Whole-Genome Analysis of a Novel Multidrug-Resistant Escherichia coli Strain from Dairy Calves in Northeast China: Mechanisms of Antibiotic Resistance and Biofilm Formation
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Test Materials
2.2. Bacterial Isolation, Identification and Antibiotic Susceptibility Testing
2.3. Whole Genome Sequencing of Representative Strain
2.4. Gene Prediction and Functional Annotation
2.5. Bioinformatics Analysis
2.5.1. Analysis of Virulence Genes and Resistance Genes
2.5.2. Pathogenic Bacteria Secretion System and Protein Analysis
2.5.3. Transporter Protein Annotations
2.5.4. Analysis of Pathogen–Host Interactions
2.6. Metabolic System Analysis
2.7. Mobile Genetic Elements and Transferability Analysis
2.8. qRT-PCR Validation
2.9. Biofilm Formation Assay
2.10. Pathogenicity Test
2.11. Data Analysis
3. Results
3.1. Morphology and Identification of BA1
3.2. Whole Genome Sequencing and cgMLST Typing
3.3. Gene Prediction and Functional Annotation Results
3.4. Pathogenicity System Analysis
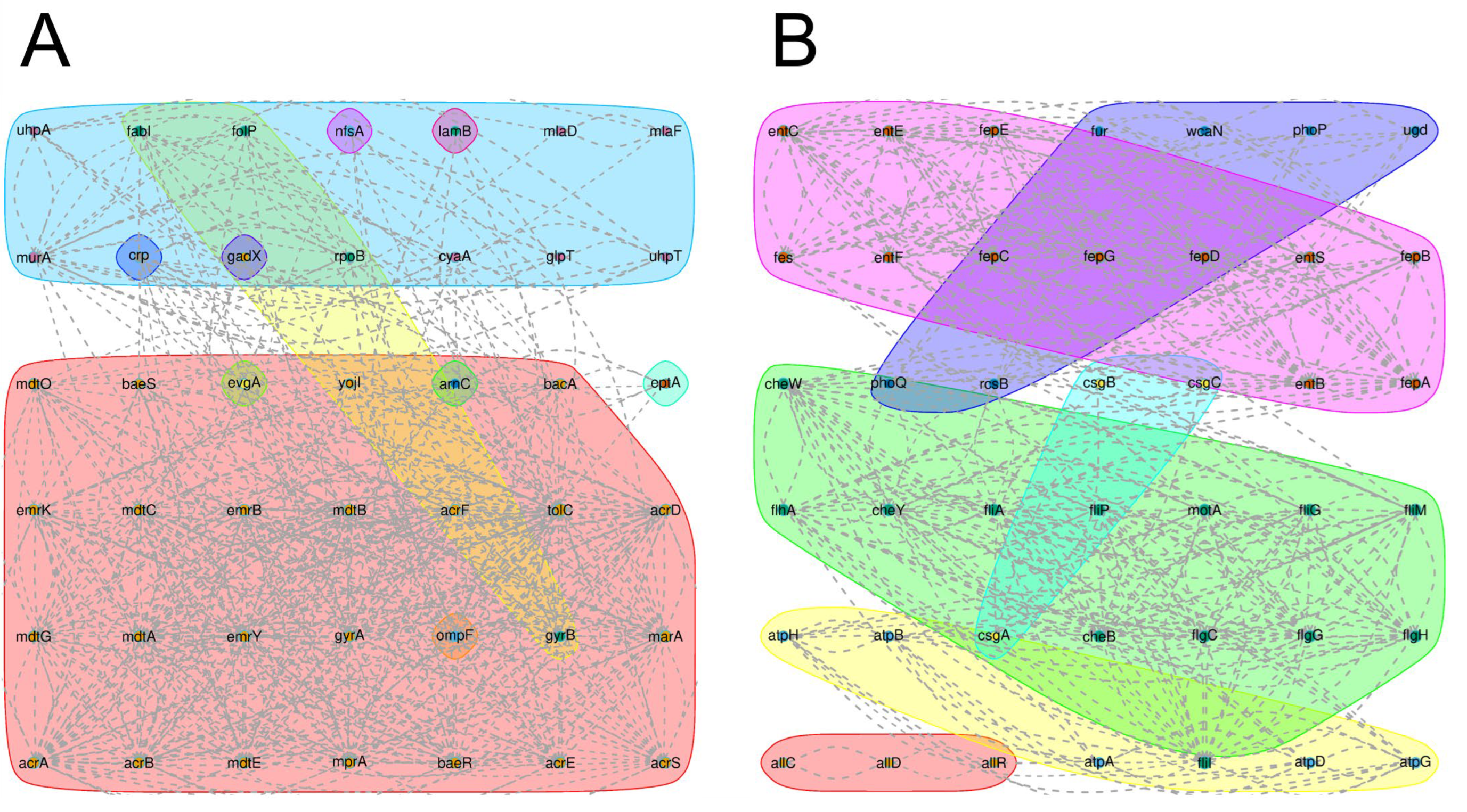
3.4.1. Resistance Gene and Virulence Factor Analysis
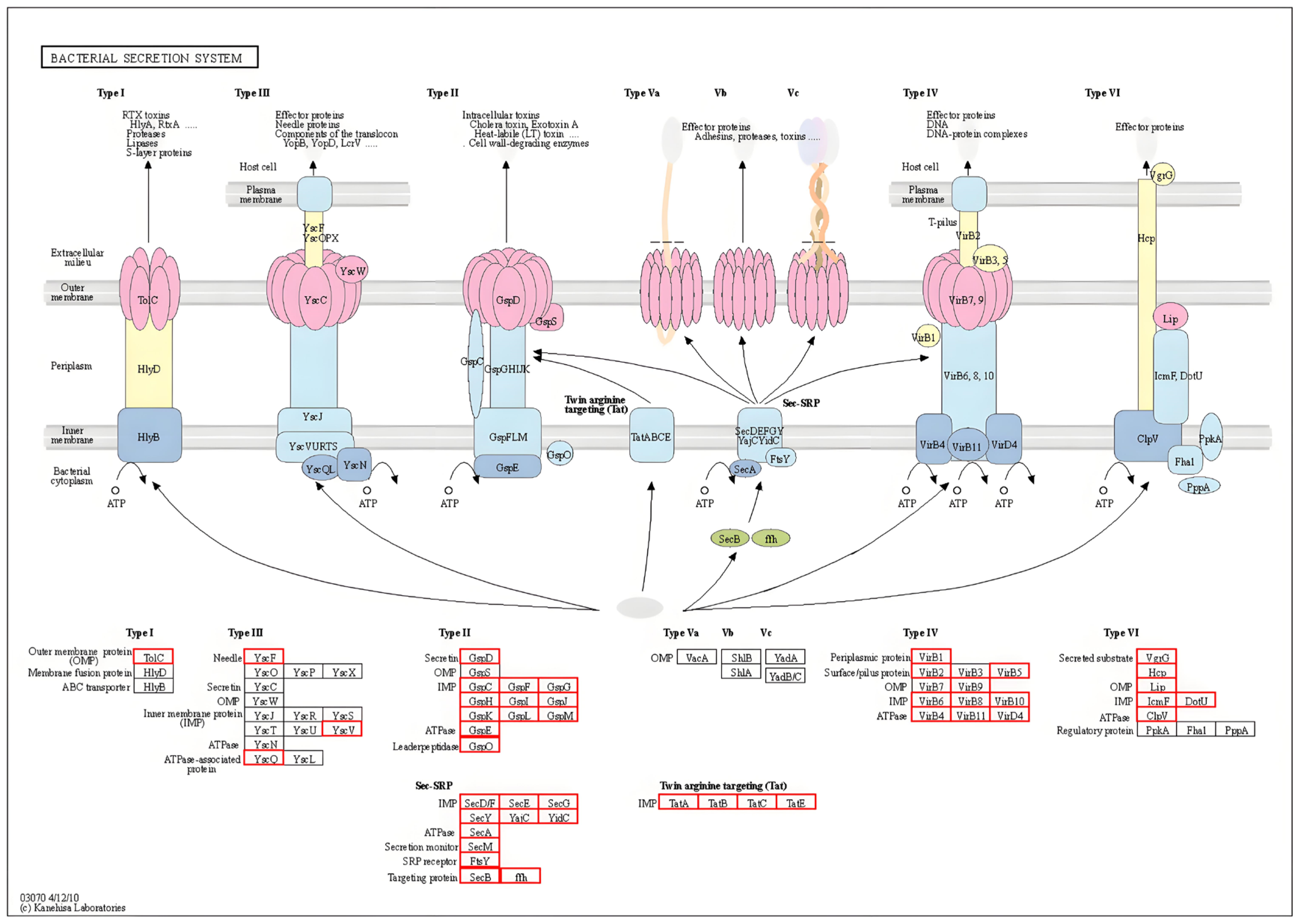
3.4.2. Secretory System and Protein Analysis
3.4.3. Transporter Protein Analysis
3.4.4. Pathogen-Host Interaction Analysis
3.5. Metabolic System Analysis Results
3.6. Mobile Genetic Elements and Zoonotic Transmission Risk
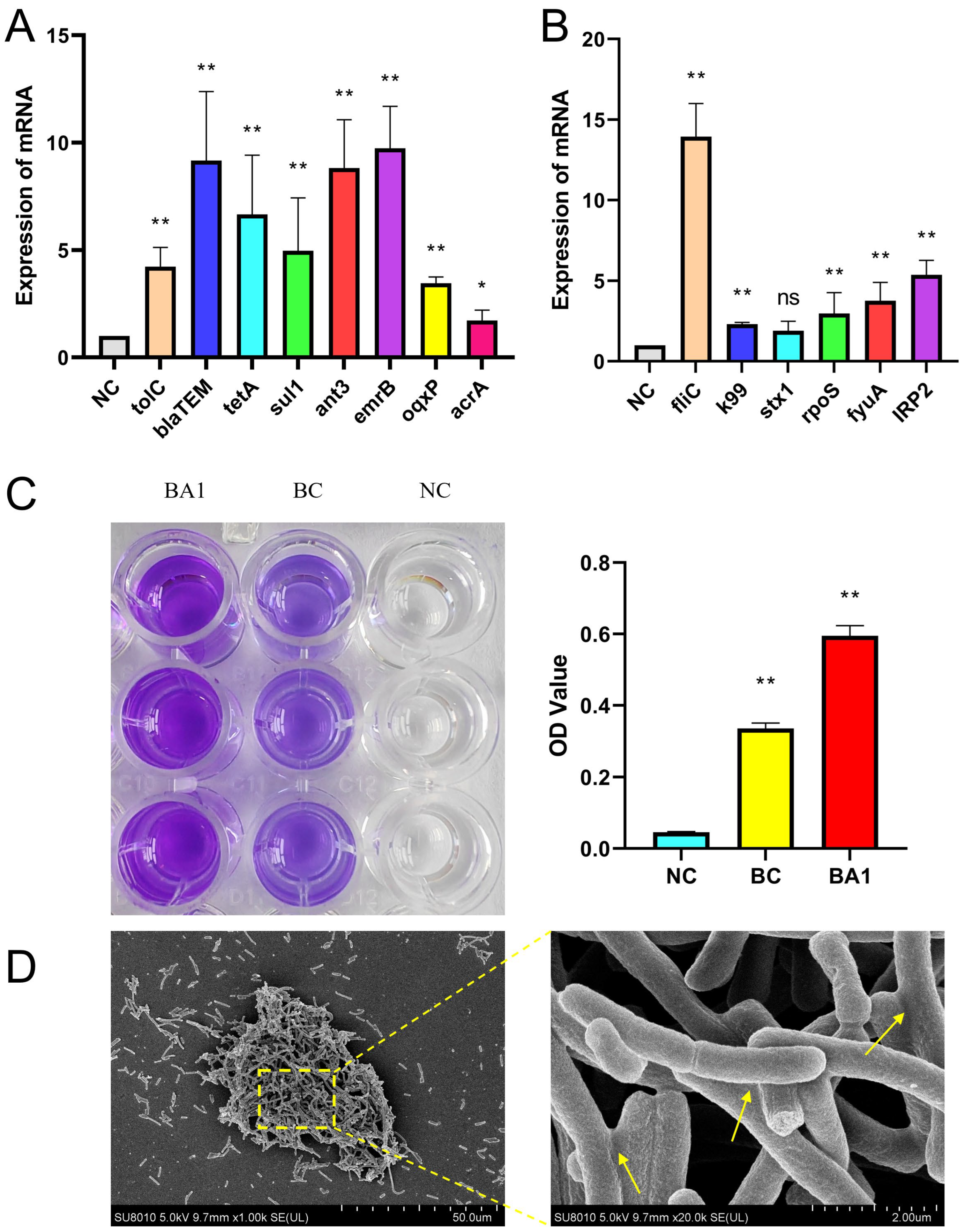
3.7. qRT-PCR Validation Results for Key Drug Resistance and Virulence Genes
3.8. Biofilm Test Results
3.9. Pathogenicity Test Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shoaib, M.; Ali, M.M.; Tang, M.; Weiwei, W.; Zhang, X.; He, Z.; Wu, Z.; Wang, S.; Hao, B.; Li, R.; et al. Genomic and phylogeographical analysis revealed CTX-M-55 producing Escherichia coli ST10 and ST2325 clones of One Health concern from dairy farm waste in Gansu, China. One Health 2025, 20, 101101. [Google Scholar] [CrossRef]
- Croxen, M.A.; Law, R.J.; Scholz, R.; Keeney, K.M.; Wlodarska, M.; Finlay, B.B. Recent Advances in Understanding Enteric Pathogenic Escherichia coli. Clin. Microbiol. Rev. 2013, 26, 822–880. [Google Scholar] [CrossRef]
- Braz, V.S.; Melchior, K.; Moreira, C.G. Escherichia coli as a Multifaceted Pathogenic and Versatile Bacterium. Front. Cell. Infect. Microbiol. 2020, 10, 548492. [Google Scholar] [CrossRef]
- Hao, R.; Shoaib, M.; Tang, M.; Cao, Z.; Liu, G.; Zhang, Y.; Wang, S.; Shang, R.; Zhang, H.; Pu, W. Genomic insights into resistome, virulome, and mobilome as organic contaminants of ESKAPE pathogens and E. coli recovered from milk, farm workers, and environmental settings in Hainan, China. Emerg. Contam. 2024, 10, 100385. [Google Scholar] [CrossRef]
- Fang, L.; Chen, C.; Li, S.; Ye, P.; Shi, Y.; Sharma, G.; Sarkar, B.; Shaheen, S.M.; Lee, S.S.; Xiao, R.; et al. A comprehensive and global evaluation of residual antibiotics in agricultural soils: Accumulation, potential ecological risks, and attenuation strategies. Ecotoxicol. Environ. Saf. 2023, 262, 115175. [Google Scholar] [CrossRef] [PubMed]
- Ng, P.C.; Kirkness, E.F. Whole genome sequencing. Genet. Var. Methods Protoc. 2010, 628, 215–226. [Google Scholar]
- Punina, N.V.; Makridakis, N.M.; Remnev, M.A.; Topunov, A.F. Whole-genome sequencing targets drug-resistant bacterial infections. Hum. Genom. 2015, 9, 1–20. [Google Scholar] [CrossRef]
- Shoaib, M.; He, Z.; Geng, X.; Tang, M.; Hao, R.; Wang, S.; Shang, R.; Wang, X.; Zhang, H.; Pu, W. The emergence of multi-drug resistant and virulence gene carrying Escherichia coli strains in the dairy environment: A rising threat to the environment, animal, and public health. Front. Microbiol. 2023, 14, 1197579. [Google Scholar] [CrossRef]
- Chen, L.; Yang, J.; Yu, J.; Yao, Z.; Sun, L.; Shen, Y.; Jin, Q. VFDB: A reference database for bacterial virulence factors. Nucleic Acids Res. 2005, 33, D325–D328. [Google Scholar] [CrossRef] [PubMed]
- McArthur, A.G.; Waglechner, N.; Nizam, F.; Yan, A.; Azad, M.A.; Baylay, A.J.; Bhullar, K.; Canova, M.J.; De Pascale, G.; Ejim, L.; et al. The Comprehensive Antibiotic Resistance Database. Antimicrob. Agents Chemother. 2013, 57, 3348–3357. [Google Scholar] [CrossRef]
- Humphries, R.; Bobenchik, A.M.; Hindler, J.A.; Schuetz, A.N. Overview of changes to the clinical and laboratory standards institute performance standards for antimicrobial susceptibility testing, M100. J. Clin. Microbiol. 2021, 59, e0021321. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zheng, D.; Zhou, S.; Chen, L.; Yang, J. VFDB 2022: A general classification scheme for bacterial virulence factors. Nucleic Acids Res. 2022, 50, D912–D917. [Google Scholar] [CrossRef] [PubMed]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef]
- Saier, M.H., Jr.; Reddy, V.S.; Tsu, B.V.; Ahmed, M.S.; Li, C.; Moreno-Hagelsieb, G. The Transporter Classification Database (TCDB): Recent advances. Nucleic Acids Res. 2016, 44, D372–D379. [Google Scholar] [CrossRef]
- Saier, M.H., Jr.; Tran, C.V.; Barabote, R.D. TCDB: The Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 2006, 34, D181–D186. [Google Scholar] [CrossRef]
- Urban, M.; Cuzick, A.; Rutherford, K.; Irvine, A.; Pedro, H.; Pant, R.; Sadanadan, V.; Khamari, L.; Billal, S.; Mohanty, S.; et al. PHI-base: A new interface and further additions for the multi-species pathogen–host interactions database. Nucleic Acids Res. 2017, 45, D604–D610. [Google Scholar] [CrossRef] [PubMed]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef]
- Johansson, M.H.K.; Bortolaia, V.; Tansirichaiya, S.; Aarestrup, F.M.; Roberts, A.P.; Petersen, T.N. Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: MobileElementFinder. J. Antimicrob. Chemother. 2021, 76, 101–109. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; Garcìa-Fernandez, A.; Larsen, M.; Lund, O.; Voldby Villa, L.; Møller Aarestrup, F.; Hasman, H. In Silico Detection and Typing of Plasmids. Antimicrob using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Ding, X.; Zhao, Y.; Li, J.; Zhang, S.; Yao, D.; Yang, H.; Zhang, Y.; Fan, H.; Basharat, Z. Mechanisms of Inhibitory and Anti-Inflammatory Effects of Aloe-Emodin Against Multidrug-Resistant Escherichia coli: A Network Pharmacology and Molecular Dynamics Approach. J. Food Biochem. 2025, 2025, 5709147. [Google Scholar] [CrossRef]
- Meroni, G.; Soares Filipe, J.F.; Drago, L.; Martino, P.A. Investigation on Antibiotic-Resistance, Biofilm Formation and Virulence Factors in Multi Drug Resistant and Non Multi Drug Resistant Staphylococcus pseudintermedius. Microorganisms 2019, 7, 702. [Google Scholar] [CrossRef] [PubMed]
- Azeredo, J.; Azevedo, N.F.; Briandet, R.; Cerca, N.; Coenye, T.; Costa, A.R.; Desvaux, M.; Di Bonaventura, G.; Hébraud, M.; Jaglic, Z.; et al. Critical review on biofilm methods. Crit. Rev. Microbiol. 2017, 43, 313–351. [Google Scholar] [CrossRef] [PubMed]
- Naves, P.; del Prado, G.; Huelves, L.; Gracia, M.; Ruiz, V.; Blanco, J.; Rodríguez-Cerrato, V.; Ponte, M.; Soriano, F. Measurement of biofilm formation by clinical isolates of Escherichia coli is method-dependent. J. Appl. Microbiol. 2008, 105, 585–590. [Google Scholar] [CrossRef]
- Chang, Q.; Wang, W.; Regev-Yochay, G.; Lipsitch, M.; Hanage, W.P. Antibiotics in agriculture and the risk to human health: How worried should we be? Evol. Appl. 2015, 8, 240–247. [Google Scholar] [CrossRef] [PubMed]
- Barathe, P.; Kaur, K.; Reddy, S.; Shriram, V.; Kumar, V. Antibiotic pollution and associated antimicrobial resistance in the environment. J. Hazard. Mater. Lett. 2024, 5, 100105. [Google Scholar] [CrossRef]
- Okaiyeto, S.A.; Sutar, P.P.; Chen, C.; Ni, J.-B.; Wang, J.; Mujumdar, A.S.; Zhang, J.-S.; Xu, M.-Q.; Fang, X.-M.; Zhang, C.; et al. Antibiotic resistant bacteria in food systems: Current status, resistance mechanisms, and mitigation strategies. Agric. Commun. 2024, 2, 100027. [Google Scholar] [CrossRef]
- Bleuel, C.; Große, C.; Taudte, N.; Scherer, J.; Wesenberg, D.; Krauß, G.J.; Nies, D.H.; Grass, G. TolC Is Involved in Enterobactin Efflux across the Outer Membrane of Escherichia coli. J. Bacteriol. 2005, 187, 6701–6707. [Google Scholar] [CrossRef]
- Kumawat, M.; Nabi, B.; Daswani, M.; Viquar, I.; Pal, N.; Sharma, P.; Tiwari, S.; Sarma, D.K.; Shubham, S.; Kumar, M. Role of bacterial efflux pump proteins in antibiotic resistance across microbial species. Microb. Pathog. 2023, 181, 106182. [Google Scholar] [CrossRef]
- Hussein, R.A.; Al-Kubaisy, S.H.; Al-Ouqaili, M.T. The influence of efflux pump, outer membrane permeability and β-lactamase production on the resistance profile of multi, extensively and pandrug resistant Klebsiella pneumoniae. J. Infect. Public Health 2024, 17, 102544. [Google Scholar] [CrossRef]
- Novelli, M.; Bolla, J.-M. RND Efflux Pump Induction: A Crucial Network Unveiling Adaptive Antibiotic Resistance Mechanisms of Gram-Negative Bacteria. Antibiotics 2024, 13, 501. [Google Scholar] [CrossRef]
- Haiko, J.; Westerlund-Wikström, B. The Role of the Bacterial Flagellum in Adhesion and Virulence. Biology 2013, 2, 1242–1267. [Google Scholar] [CrossRef] [PubMed]
- Lian, S.; Luo, Y.; Chen, Z.; Wei, X.; Liu, J.; Zhu, G.; Xia, P. Deficiency of the flagellin subunit FliC exacerbates the pathogenicity of extraintestinal pathogenic Escherichia coli in BALB/c mice by inducing a more intense inflammation. Int. J. Biol. Macromol. 2025, 289, 138761. [Google Scholar] [CrossRef] [PubMed]
- Mirghani, R.; Saba, T.; Khaliq, H.; Mitchell, J.; Do, L.; Chambi, L.; Diaz, K.; Kennedy, T.; Alkassab, K.; Huynh, T.; et al. Biofilms: Formation, drug resistance and alternatives to conventional approaches. AIMS Microbiol. 2022, 8, 239–277. [Google Scholar] [CrossRef]
- Grande, R.; Puca, V.; Muraro, R. Antibiotic resistance and bacterial biofilm. Expert Opin. Ther. Pat. 2020, 30, 897–900. [Google Scholar] [CrossRef]
- Fisher, R.A.; Gollan, B.; Helaine, S. Persistent bacterial infections and persister cells. Nat. Rev. Microbiol. 2017, 15, 453–464. [Google Scholar] [CrossRef]
- Oz, T.; Guvenek, A.; Yildiz, S.; Karaboga, E.; Tamer, Y.T.; Mumcuyan, N.; Ozan, V.B.; Senturk, G.H.; Cokol, M.; Yeh, P.; et al. Strength of Selection Pressure Is an Important Parameter Contributing to the Complexity of Antibiotic Resistance Evolution. Mol. Biol. Evol. 2014, 31, 2387–2401. [Google Scholar] [CrossRef]
- Green, E.R.; Mecsas, J. Bacterial Secretion Systems: An Overview. Microbiol. Spectr. 2016, 4, 1. [Google Scholar] [CrossRef]
- Emara, Y.; Jolliet, O.; Finkbeiner, M.; Heß, S.; Kosnik, M.; Siegert, M.-W.; Fantke, P. Comparative selective pressure potential of antibiotics in the environment. Environ. Pollut. 2023, 318, 120873. [Google Scholar] [CrossRef]
- Jin, M.; Osman, M.; Green, B.A.; Yang, Y.; Ahuja, A.; Lu, Z.; Cazer, C.L. Evidence for the transmission of antimicrobial resistant bacteria between humans and companion animals: A scoping review. One Health 2023, 17, 100593. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, N.; Joji, R.M.; Shahid, M. Evolution and implementation of One Health to control the dissemination of antibiotic-resistant bacteria and resistance genes: A review. Front. Cell. Infect. Microbiol. 2023, 12, 1065796. [Google Scholar] [CrossRef] [PubMed]
- Lessa, F.C.; Sievert, D.M. Antibiotic Resistance: A Global Problem and the Need to Do More. Clin. Infect. Dis. 2023, 77, S1–S3. [Google Scholar] [CrossRef] [PubMed]
- Årdal, C.; Outterson, K.; Hoffman, S.J.; Ghafur, A.; Sharland, M.; Ranganathan, N.; Smith, R.; Zorzet, A.; Cohn, J.; Pittet, D.; et al. International cooperation to improve access to and sustain effectiveness of antimicrobials. Lancet 2016, 387, 296–307. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, X.; An, Q.; Kang, H.; Li, S.; Zhang, S.; Yang, H.; Dou, X.; Ji, Y.; Zhao, Y.; Fan, H. Whole-Genome Analysis of a Novel Multidrug-Resistant Escherichia coli Strain from Dairy Calves in Northeast China: Mechanisms of Antibiotic Resistance and Biofilm Formation. Biology 2025, 14, 1257. https://doi.org/10.3390/biology14091257
Ding X, An Q, Kang H, Li S, Zhang S, Yang H, Dou X, Ji Y, Zhao Y, Fan H. Whole-Genome Analysis of a Novel Multidrug-Resistant Escherichia coli Strain from Dairy Calves in Northeast China: Mechanisms of Antibiotic Resistance and Biofilm Formation. Biology. 2025; 14(9):1257. https://doi.org/10.3390/biology14091257
Chicago/Turabian StyleDing, Xuanpan, Qiuyue An, Huijie Kang, Siyao Li, Shuai Zhang, Haotian Yang, Xinyi Dou, Yaxin Ji, Yuan Zhao, and Honggang Fan. 2025. "Whole-Genome Analysis of a Novel Multidrug-Resistant Escherichia coli Strain from Dairy Calves in Northeast China: Mechanisms of Antibiotic Resistance and Biofilm Formation" Biology 14, no. 9: 1257. https://doi.org/10.3390/biology14091257
APA StyleDing, X., An, Q., Kang, H., Li, S., Zhang, S., Yang, H., Dou, X., Ji, Y., Zhao, Y., & Fan, H. (2025). Whole-Genome Analysis of a Novel Multidrug-Resistant Escherichia coli Strain from Dairy Calves in Northeast China: Mechanisms of Antibiotic Resistance and Biofilm Formation. Biology, 14(9), 1257. https://doi.org/10.3390/biology14091257






