Gasdermin-Mediated Pyroptosis in Hidradenitis Suppurativa: Molecular Insights and Therapeutic Implications
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Type and Design
2.2. Study Population
2.2.1. Study Group―Patients with Hidradenitis Suppurativa (HS)
2.2.2. Control Groups
2.3. Biological Material
2.3.1. Blood
2.3.2. Skin Biopsies
2.4. Analytical Methods
2.4.1. Circulating GSDMD and GSDME
2.4.2. Skin Expression of GSDMD and GSDME
2.5. Statistical Analysis
3. Results
3.1. Characteristics of HS Patients
3.2. Gasdermins in HS Patients and Healthy Controls
3.3. Impact of Patients’ and the Disease Characteristics on Circulating Gasdermins
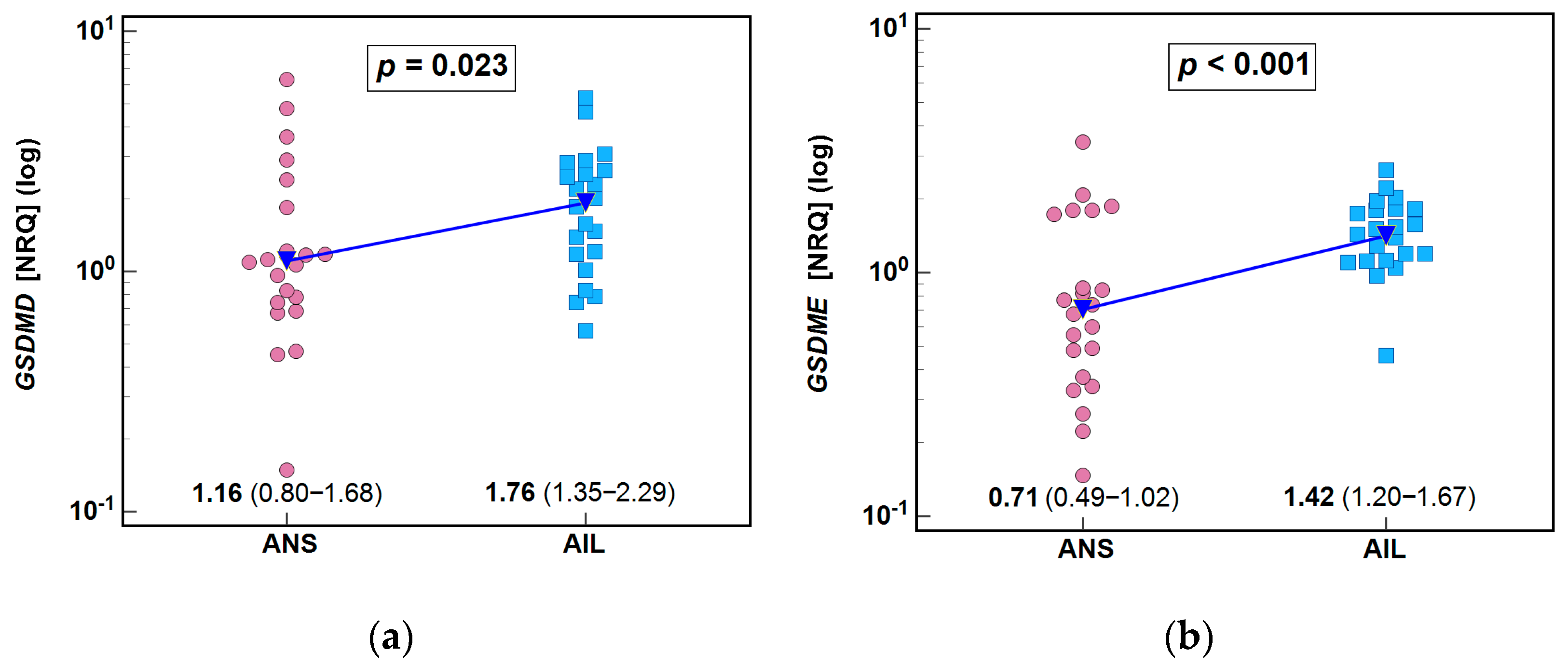
3.4. mRNA Expression of GSDMD and GSDME in the Skin
3.5. Association of Patients’ and Disease Characteristics with Skin Expression of GSDMD and GSDME
3.5.1. GSDMD
3.5.2. GSDME
3.6. Interplay Between Systemic and Local Gasdermins
4. Discussion
5. Limitations
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sabat, R.; Jemec, G.B.E.; Matusiak, Ł.; Kimball, A.B.; Prens, E.; Wolk, K. Hidradenitis suppurativa. Nat. Rev. Dis. Primers 2020, 6, 18. [Google Scholar] [CrossRef] [PubMed]
- Krajewski, P.K.; Matusiak, Ł.; von Stebut, E.; Schultheis, M.; Kirschner, U.; Nikolakis, G.; Szepietowski, J.C. Quality-of-Life Impairment among Patients with Hidradenitis Suppurativa: A Cross-Sectional Study of 1795 Patients. Life 2021, 11, 34. [Google Scholar] [CrossRef] [PubMed]
- Matusiak, Ł. Profound consequences of hidradenitis suppurativa: A review. Br. J. Dermatol. 2020, 183, e171–e177. [Google Scholar] [CrossRef] [PubMed]
- Diaz, M.J.; Aflatooni, S.; Abdi, P.; Li, R.; Anthony, M.R.; Neelam, S.; Farkouh, C.; Tran, J.T.; Svoboda, S.; Forouzandeh, M.; et al. Hidradenitis Suppurativa: Molecular Etiology, Pathophysiology, and Management—A Systematic Review. Curr. Issues Mol. Biol. 2023, 45, 4400–4415. [Google Scholar] [CrossRef]
- Sun, Q.; Broadaway, K.A.; Edmiston, S.N.; Fajgenbaum, K.; Miller-Fleming, T.; Westerkam, L.L.; Melendez-Gonzalez, M.; Bui, H.; Blum, F.R.; Levitt, B.; et al. Genetic Variants Associated With Hidradenitis Suppurativa. JAMA Dermatol. 2023, 159, 930–938. [Google Scholar] [CrossRef]
- Szukala, W.; Lichawska-Cieslar, A.; Krajewski, P.K.; Kulecka, M.; Rumienczyk, I.; Mikula, M.; Matusiak, Ł.; Jura, J.; Szepietowski, J.C. An Atlas of the Hidradenitis Suppurativa Transcriptome. Dermatol. Ther. 2024, 14, 409–420. [Google Scholar] [CrossRef]
- Ketelut-Carneiro, N.; Fitzgerald, K.A. Apoptosis, Pyroptosis, and Necroptosis-Oh My! The Many Ways a Cell Can Die. J. Mol. Biol. 2022, 434, 167378. [Google Scholar] [CrossRef]
- Aachoui, Y.; Sagulenko, V.; Miao, E.A.; Stacey, K.J. Inflammasome-mediated pyroptotic and apoptotic cell death, and defense against infection. Curr. Opin. Microbiol. 2013, 16, 319–326. [Google Scholar] [CrossRef]
- Shi, J.; Zhao, Y.; Wang, K.; Shi, X.; Wang, Y.; Huang, H.; Zhuang, Y.; Cai, T.; Wang, F.; Shao, F. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 2015, 526, 660–665. [Google Scholar] [CrossRef]
- Mulvihill, E.; Sborgi, L.; Mari, S.A.; Pfreundschuh, M.; Hiller, S.; Müller, D.J. Mechanism of membrane pore formation by human gasdermin-D. EMBO J. 2018, 37, e98321. [Google Scholar] [CrossRef]
- Sborgi, L.; Rühl, S.; Mulvihill, E.; Pipercevic, J.; Heilig, R.; Stahlberg, H.; Farady, C.J.; Müller, D.J.; Broz, P.; Hiller, S. GSDMD membrane pore formation constitutes the mechanism of pyroptotic cell death. EMBO J. 2016, 35, 1766–1778. [Google Scholar] [CrossRef]
- Krajewski, P.K.; Tsoukas, M.; Szepietowski, J.C. Pathological and Therapeutical Implications of Pyroptosis in Psoriasis and Hidradenitis Suppurativa: A Narrative Review. Curr. Issues Mol. Biol. 2024, 46, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Hurley, H. Axillary hyperhidrosis, apocrine bromhidrosis, hidradenitis suppurativa, and familial benign pemphigus: Surgical approach. In Dermatologic Surgery; Roenigk, R., Roenigk, H., Eds.; Marcel Dekker: New York, NY, USA, 1989; pp. 729–739. [Google Scholar]
- Zouboulis, C.C.; Tzellos, T.; Kyrgidis, A.; Jemec, G.B.E.; Bechara, F.G.; Giamarellos-Bourboulis, E.J.; Ingram, J.R.; Kanni, T.; Karagiannidis, I.; Martorell, A.; et al. Development and validation of the International Hidradenitis Suppurativa Severity Score System (IHS4), a novel dynamic scoring system to assess HS severity. Br. J. Dermatol. 2017, 177, 1401–1409. [Google Scholar] [CrossRef] [PubMed]
- Khazaal, M. Concise Review of Common Non-Traditional Dyslipidemic Indices in Clinical Practice. AlQalam J. Med. App. Sci. 2023, 6, 395–400. [Google Scholar]
- Hellemans, J.; Vandesompele, J. qPCR data analysis—Unlocking the secret to successful results. In PCR Troubleshooting and Optimization: The Essential Guide, 1st ed.; Kennedy, S., Oswald, N., Eds.; Caister Academic Press: Poole, UK, 2011. [Google Scholar]
- Kashyap, M.P.; Khan, J.; Sinha, R.; Jin, L.; Atigadda, V.; Deshane, J.S.; Ahmed, A.R.; Kilic, A.; Raman, C.; Mukhtar, M.S.; et al. Advances in molecular pathogenesis of hidradenitis suppurativa: Dysregulated keratins and ECM signaling. Semin. Cell Dev. Biol. 2022, 128, 120–129. [Google Scholar] [CrossRef]
- Chu, C.B.; Yang, C.C.; Hsueh, Y.Y.; Chen, P.C.; Hong, Y.K.; Kuo, Y.Y.; Tsai, S.J. Aberrant expression of interleukin-17A in mast cells contributes to the pathogenesis of hidradenitis suppurativa. Br. J. Dermatol. 2023, 189, 719–729. [Google Scholar] [CrossRef]
- Suh-Yun Joh, C.; Jeong, S.; Lee, C.; Lee, H.J.; Lee, J.H.; Choi, H.S.; Cho, S.; Kim, G.; Kim, J.; Krueger, J.G.; et al. Spatial transcriptomic profiling reveals the pathophysiology of early-stage hidradenitis suppurativa. Br. J. Dermatol. 2023, 189, 643–645. [Google Scholar] [CrossRef]
- Swanson, K.V.; Deng, M.; Ting, J.P. The NLRP3 inflammasome: Molecular activation and regulation to therapeutics. Nat. Rev. Immunol. 2019, 19, 477–489. [Google Scholar] [CrossRef]
- Witte, K.; Sabat, R.; Witte-Händel, E.; Ghoreschi, K.; Wolk, K. Phytotherapeuthics Affecting the IL-1/IL-17/G-CSF Axis: A Complementary Treatment Option for Hidradenitis Suppurativa? Int. J. Mol. Sci. 2022, 23, 9057. [Google Scholar] [CrossRef]
- Xue, Y.; Enosi Tuipulotu, D.; Tan, W.H.; Kay, C.; Man, S.M. Emerging Activators and Regulators of Inflammasomes and Pyroptosis. Trends Immunol. 2019, 40, 1035–1052. [Google Scholar] [CrossRef]
- Kanneganti, A.; Malireddi, R.K.S.; Saavedra, P.H.V.; Vande Walle, L.; Van Gorp, H.; Kambara, H.; Tillman, H.; Vogel, P.; Luo, H.R.; Xavier, R.J.; et al. GSDMD is critical for autoinflammatory pathology in a mouse model of Familial Mediterranean Fever. J. Exp. Med. 2018, 215, 1519–1529. [Google Scholar] [CrossRef]
- Xiao, J.; Wang, C.; Yao, J.C.; Alippe, Y.; Xu, C.; Kress, D.; Civitelli, R.; Abu-Amer, Y.; Kanneganti, T.D.; Link, D.C.; et al. Gasdermin D mediates the pathogenesis of neonatal-onset multisystem inflammatory disease in mice. PLoS Biol. 2018, 16, e3000047. [Google Scholar] [CrossRef]
- Piñero, D.J.; Hu, J.; Cook, B.M.; Scaduto, R.C., Jr.; Connor, J.R. Interleukin-1beta increases binding of the iron regulatory protein and the synthesis of ferritin by increasing the labile iron pool. Biochim. Biophys. Acta 2000, 1497, 279–288. [Google Scholar] [CrossRef]
- Li, Z.; Ji, S.; Jiang, M.L.; Xu, Y.; Zhang, C.J. The Regulation and Modification of GSDMD Signaling in Diseases. Front. Immunol. 2022, 13, 893912. [Google Scholar] [CrossRef] [PubMed]
- Badii, M.; Gaal, O.I.; Cleophas, M.C.; Klück, V.; Davar, R.; Habibi, E.; Keating, S.T.; Novakovic, B.; Helsen, M.M.; Dalbeth, N.; et al. Urate-induced epigenetic modifications in myeloid cells. Arthritis Res. Ther. 2021, 23, 202. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, J.M.; Moran, B.; Petrasca, A.; Smith, C.M. IL-17 in inflammatory skin diseases psoriasis and hidradenitis suppurativa. Clin. Exp. Immunol 2020, 201, 121–134. [Google Scholar] [CrossRef]
- Kim, J.; Lee, J.; Li, X.; Lee, H.S.; Kim, K.; Chaparala, V.; Murphy, W.; Zhou, W.; Cao, J.; Lowes, M.A.; et al. Single-cell transcriptomics suggest distinct upstream drivers of IL-17A/F in hidradenitis versus psoriasis. J. Allergy Clin. Immunol. 2023, 152, 656–666. [Google Scholar] [CrossRef]
- Nowowiejska, J.; Baran, A.; Hermanowicz, J.M.; Pryczynicz, A.; Sieklucka, B.; Pawlak, D.; Flisiak, I. Gasdermin D (GSDMD) Is Upregulated in Psoriatic Skin-A New Potential Link in the Pathogenesis of Psoriasis. Int. J. Mol. Sci. 2023, 24, 13047. [Google Scholar] [CrossRef]
- Fink, S.L.; Cookson, B.T. Caspase-1-dependent pore formation during pyroptosis leads to osmotic lysis of infected host macrophages. Cell. Microbiol. 2006, 8, 1812–1825. [Google Scholar] [CrossRef]
- Hu, J.J.; Liu, X.; Xia, S.; Zhang, Z.; Zhang, Y.; Zhao, J.; Ruan, J.; Luo, X.; Lou, X.; Bai, Y.; et al. FDA-approved disulfiram inhibits pyroptosis by blocking gasdermin D pore formation. Nat. Immunol. 2020, 21, 736–745. [Google Scholar] [CrossRef]
- Li, H.; Guan, Y.; Liang, B.; Ding, P.; Hou, X.; Wei, W.; Ma, Y. Therapeutic potential of MCC950, a specific inhibitor of NLRP3 inflammasome. Eur. J. Pharmacol. 2022, 928, 175091. [Google Scholar] [CrossRef]
- Jin, Y.; Liu, Y.; Xu, L.; Xu, J.; Xiong, Y.; Peng, Y.; Ding, K.; Zheng, S.; Yang, N.; Zhang, Z.; et al. Novel role for caspase 1 inhibitor VX765 in suppressing NLRP3 inflammasome assembly and atherosclerosis via promoting mitophagy and efferocytosis. Cell Death Dis. 2022, 13, 512. [Google Scholar] [CrossRef]
- Oymanns, M.; Kreuter, A.; Assaf, C. Anakinra therapy in a patient with multifocal pyoderma gangrenosum and hidradenitis suppurativa. Hautarzt 2021, 72, 435–438. [Google Scholar] [CrossRef] [PubMed]
- Russo, V.; Alikhan, A. Failure of Anakinra in a Case of Severe Hidradenitis Suppurativa. J. Drugs Dermatol. 2016, 15, 772–774. [Google Scholar]
- Houriet, C.; Seyed Jafari, S.M.; Thomi, R.; Schlapbach, C.; Borradori, L.; Yawalkar, N.; Hunger, R.E. Canakinumab for Severe Hidradenitis Suppurativa: Preliminary Experience in 2 Cases. JAMA Dermatol. 2017, 153, 1195–1197. [Google Scholar] [CrossRef] [PubMed]
- Tekin, B.; Salman, A.; Ergun, T. Hidradenitis suppurativa unresponsive to canakinumab treatment: A case report. Indian J. Dermatol. Venereol. Leprol. 2017, 83, 615–617. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.X.; Dai, Y.Z.; Zhao, Y.Z.; Nie, K. Gasdermin E: A Prospective Target for Therapy of Diseases. Front. Pharmacol. 2022, 13, 855828. [Google Scholar] [CrossRef]
- Zheng, Z.; Yang, S.; Dai, W.; Xue, P.; Sun, Y.; Wang, J.; Zhang, X.; Lin, J.; Kong, J. The role of pyroptosis in metabolism and metabolic disease. Biomed. Pharmacother. 2024, 176, 116863. [Google Scholar] [CrossRef]
- Wei, Y.; Lan, B.; Zheng, T.; Yang, L.; Zhang, X.; Cheng, L.; Tuerhongjiang, G.; Yuan, Z.; Wu, Y. GSDME-mediated pyroptosis promotes the progression and associated inflammation of atherosclerosis. Nat. Commun. 2023, 14, 929. [Google Scholar] [CrossRef]
- Nowowiejska, J.; Baran, A.; Pryczynicz, A.; Hermanowicz, J.M.; Sieklucka, B.; Pawlak, D.; Flisiak, I. Gasdermin E (GSDME)—A New Potential Marker of Psoriasis and Its Metabolic Complications: The First Combined Study on Human Serum, Urine and Tissue. Cells 2023, 12, 2149. [Google Scholar] [CrossRef]
- Wei, Y.; Wu, Y.; Yuan, Z.; Wang, J.; Yin, D.L. GSDME Deficiency Sensitizes Mice to Diet-Induced Obesity by Suppressing Lipolysis. In Endocrine Abstracts, Proceedings of the 26th European Congress of Endocrinology, Stockholm, Sweden, 11–14 May 2024; Bioscientifica: Bristol, UK, 2024. [Google Scholar] [CrossRef]
- Gold, D.A.; Reeder, V.J.; Mahan, M.G.; Hamzavi, I.H. The prevalence of metabolic syndrome in patients with hidradenitis suppurativa. J. Am. Acad. Dermatol. 2014, 70, 699–703. [Google Scholar] [CrossRef]
- Łapa, W.; Kutaj, A.; Kowalczyk, E.; Głuszak, P.; Welc, N.; Jałowska, M. Glucose metabolism disorders in hidradenitis suppurativa: Current evidence and future directions. Forum Dermatologicum 2025, 104845. [Google Scholar] [CrossRef]
- Krajewski, P.K.; Matusiak, Ł.; Ständer, S.; Thaçi, D.; Szepietowski, J.C.; Zirpel, H. Risk of cardiovascular disorders in hidradenitis suppurativa patients: A large-scale, propensity-matched global retrospective cohort study. Int. J. Dermatol. 2024, 63, 799–805. [Google Scholar] [CrossRef]
- Liu, J.; Jiang, Y.; Diao, Z.; Chen, D.; Xia, R.; Wang, B.; Yang, S.; Yin, Z. Gasdermin D-mediated neutrophil pyroptosis drives inflammation in psoriasis. Elife 2024, 13, RP101248. [Google Scholar] [CrossRef]
- Lu, Y.; Sun, Y.; Peng, Y.; Zhao, X.; Wang, D.; Zhang, T.; Qian, F.; Wang, J. Inhibition of gasdermin D (GSDMD) as a promising therapeutic approach for atopic dermatitis. Int. Immunopharmacol. 2023, 124, 110958. [Google Scholar] [CrossRef]



| Factor | Protein Analysis | Gene Expression Analysis | ||||
|---|---|---|---|---|---|---|
| HC | HS | P | Controls | HS | P | |
| Sex [F/M], n Females | 14/12 54% | 26/36 42% | 0.353 1 | 4/8 58% | 8/14 36% | 1.0 1 |
| Age [yrs.] median (95% CI) | 42 (31–49) | 37 (32–44) | 0.253 2 | 57 (20–73) | 43 (27–49) | 0.341 2 |
| Gene | Encoded Protein | Primer Sequence (5′→3′) | Amplicon Size [bp] |
|---|---|---|---|
| GSDMD | Gasdermin D | F: atgaggtgcctccacaacttcc R: ccagttccttggagatggtctc | 109 |
| GSDME | Gasdermin E | F: gatctctgagcacatgcaggtc R: gttggagtccttggtgacattcc | 112 |
| YWHAZ | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein ζ | F: accgttacttggctgaggttgc R: cccagtctgataggatgtgttgg | 130 |
| GUSB | β-Glucuronidase | F: ctgtcaccaagagccagttcct R: ggttgaagtccttcaccagcag | 126 |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | F: gtctcctctgacttcaacagcg R: accaccctgttgctgtagccaa | 131 |
| Factor | Patients with HS | p Value 1 | ||
|---|---|---|---|---|
| All | Females | Males | ||
| N | 62 | 26 (42%) | 36 (58%) | - |
| Age [yrs.] | 37 (32–44) | 35 (31–45) | 39 (29.5–46.5) | 0.849 2 |
| BMI [kg/m2] | 29 (28–34) | 30.5 (24–35) | 29 (26–35) | 0.678 2 |
| BMI [%] overweight/obese | 27.9/48.8 | 15/55 | 39.1/43.5 | 0.197 3 |
| BMI abnormal [%] | 76.7 | 70 | 82.6 | 0.473 4 |
| Severe HS by Hurley [%] | 33.3 | 36.4 | 30.8 | 0.764 4 |
| IHS4 | 16 (14–22.5) | 15 (10–24) | 18.5 (15–24) | 0.474 2 |
| Severe HS by IHS4 [%] | 70.8 | 59.1 | 80.8 | 0.122 4 |
| Disease duration [yrs.] | 6 (5–9) | 7 (5–11.5) | 6 (3–10) | 0.496 2 |
| Smoking habit [%] yes/in the past | 52.5/10 | 44.4/11.1 | 59.1/9.1 | 0.649 3 |
| Anemia [%] | 13.6 | 10 | 16.7 | 0.673 4 |
| Glucose [mg/dL] | 88.5 (81–94) | 92 (81.5–108.5) | 86 (77–93) | 0.273 2 |
| IFG [%] | 25 | 40 | 11.8 | 0.106 4 |
| Insulin [mU/mL] | 14.1 (11.9–18.1) | 16 (12.1–52.7) | 13.5 (9.65–47) | 0.341 2 |
| HI: >10 mU/mL [%] | 75 | 90/40 | 64.3 | 0.341 3 |
| HOMA-IR | 3.25 (2.2–4.62) | 3.35 (2.94–31.69) | 2.55 (2.08–11.1) | 0.260 2 |
| IR by HOMA [%] | 68.2 | 87.5 | 57.1 | 0.193 4 |
| TyG | 4.59 (4.48–4.7) | 4.53 (4.37–4.69) | 4.65 (4.49–4.82) | 0.255 5 |
| IR by TyG ≥ 4.5 | 51.6 | 40 | 62.5 | 0.289 4 |
| tCHOL [mg/dL] | 184 (172–196) | 180 (165–196) | 188 (167–208) | 0.557 5 |
| HyperCHOL [%] | 35.3 | 33.3 | 37.5 | 1.0 4 |
| HDL-CHOL [mg/dL] | 48.5 (43.6–53.3) | 54.9 (48–61.9) | 41.6 (36.1–47) | 0.003 5 |
| ↓ HDL-CHOL [%] | 42.4 | 29.4 | 56.2 | 0.166 4 |
| LDL-CHOL [mg/dL] | 110 (98–122) | 105 (92–118) | 117 (93–141) | 0.348 5 |
| ↑ LDL-CHOL [%] | 43.3 | 35.3 | 53.8 | 0.460 4 |
| Non-HDL-CHOL [mg/dL] | 137 (124–150) | 129 (112–145) | 146 (125–167) | 0.176 5 |
| ↑ Non-HDL-CHOL [%] | 55.9 | 44.4 | 68.7 | 0.185 4 |
| TG [mg/dL] | 101 (86–120) | 92.5 (69–115) | 116 (85–157) | 0.143 2 |
| HyperTG [%] | 52.9 | 44.4 | 62.5 | 0.327 4 |
| ↑ CV risk by AIP [%] | 30.3 | 11.8 | 50 | 0.025 4 |
| ↑ CV risk by AC [%] | 48.5 | 23.5 | 75 | 0.005 4 |
| ↑ CV risk by CRI-I [%] | 30.3 | 17.6 | 43.7 | 0.141 4 |
| ↑ CV risk by CRI-II [%] | 21.2 | 11.8 | 31.2 | 0.225 4 |
| ↑ CV risk by LCI [%] | 39.4 | 23.5 | 56.2 | 0.080 4 |
| CREA [mg/dL] | 0.76 (0.73–0.79) | 0.73 (0.68–0.77) | 0.78 (0.75–0.82) | 0.041 5 |
| Urea [mg/dL] | 23.6 (21.3–25.9) | 24.5 (21.3–27.7) | 22.7 (19.2–26.2) | 0.428 5 |
| UA [mg/dL] | 5.46 (4.98–5.94) | 4.87 (4.21–5.54) | 6.05 (5.42–6.68) | 0.010 5 |
| HyperUA [%] | 40 | 33.3 | 46.7 | 0.710 5 |
| WBC [×103/μL] | 8.16 (7.34–9.07) | 8.19 (6.81–9.85) | 8.13 (7.13–9.27) | 0.731 5 |
| Leukocytosis [%] | 25 | 20 | 29.2 | 0.728 4 |
| NEU [×103/μL] | 5.19 (4.83–5.8) | 5.31 (4.38–5.8) | 5.0 (4.69–6.12) | 0.906 2 |
| Neutrophilia [%] | 13.6 | 10 | 16.7 | 0.673 4 |
| LYM [×103/μL] | 2.09 (1.84–2.36) | 2.26 (1.74–2.79) | 2.08 (1.8–2.32) | 0.582 2 |
| PLT [×103/μL] | 282 (251–303) | 277 (239–345) | 287 (246–306) | 0.841 2 |
| NLR | 2.3 (2.0–2.8) | 2.1 (1.7–2.9 | 2.3 (1.7–3.0) | 0.723 2 |
| ↑ NLR [%] | 34.9 | 31.6 | 37.5 | 0.755 4 |
| PLR | 133 (119–149) | 131 (111–155) | 134 (114–158) | 0.837 5 |
| SII | 681 (567–819) | 685 (503–933) | 678 (531–866) | 0.960 5 |
| CRP [mg/L] | 6.2 (4.5–9.2) | 6.6 (2.7–10.5) | 5.6 (4.4–10.5) | 0.764 2 |
| Inflammation by CRP [%] | 38.9 | 50 | 27.8 | 0.196 4 |
| tBIL [mg/dL] | 0.46 (0.35–0.5) | 0.4 (0.3–0.5) | 0.5 (0.37–0.6) | 0.177 2 |
| ALT [U/L] | 20 (18–25.5) | 17 (10.8–20.2) | 25 (19–31.2) | 0.014 2 |
| AST [U/L] | 18 (15.5–19) | 17 (13.8–19.2) | 19 (16.6–23.2) | 0.364 2 |
| GGT [U/L] | 24 (22–28.9) | 22 (16.8–28.2) | 27.5 (23–52.4) | 0.018 2 |
| ALP [U/L] | 69.1 (61.8–77.2) | 61.7 (53.2–71.6) | 78.7 (67.3–92) | 0.024 5 |
| Factor | Categories | GSDMD | GSDME | ||
|---|---|---|---|---|---|
| Median | p Value | Median | p Value | ||
| Sex | females | 0.34 (0.25–0.54) | 0.364 | 0.62 (0.5–0.73) | 0.221 |
| males | 0.31 (0.20–0.45) | 0.72 (0.63–0.77) | |||
| Hurley score | 1/2 | 0.42 (0.32–0.67) | 0.793 | 0.71 (0.61–0.77) | 0.205 |
| 3 | 0.34 (0.25–1.32) | 0.62 (0.5–0.72) | |||
| HS severity by IHS4 | moderate | 0.58 (0.32–1.56) | 0.063 | 0.56 (0.46–0.79) | 0.352 |
| severe | 0.33 (0.26–0.54) | 0.69 (0.62–0.76) | |||
| Smoking habit | no | 0.54 (0.31–1.49) | 0.404 | 0.72 (0.47–0.76) | 0.716 |
| yes | 0.34 (0.27–0.59) | 0.70 (0.59–0.77) | |||
| in the past | 0.38 (0.19–0.80) | 0.73 (0.57–0.84) | |||
| BMI | normal | 0.47 (0.35–1.13) | 0.438 | 0.76 (0.64–0.82) | 0.093 |
| elevated | 0.34 (0.29–0.74) | 0.65 (0.54–0.74) | |||
| Anemia | no | 0.39 (0.32–0.62) | 0.811 | 0.72 (0.65–0.76) | 0.005 |
| yes | 0.38 (0.17–6.19) | 0.48 (0.37–0.61) | |||
| IR (by TyG) F | no | 0.36 (0.19–1.31) | 0.594 | 0.76 (0.61–0.9) | 0.007 |
| yes | 0.98 (0.09–1.89) | 0.48 (0.43–0.63) | |||
| HyperCHOL F | no | 0.43 (0.2–1.08) | 0.281 | 0.71 (0.6–0.79) | 0.031 |
| yes | 0.99 (0.3–1.92) | 0.48 (0.43–0.85) | |||
| ↑ LDL-CHOL F | no | 0.36 (0.17–1.34) | 0.546 | 0.72 (0.58–0.8) | 0.063 |
| yes | 0.74 (0.3–1.88) | 0.52 (0.43–0.85) | |||
| ↑ non-HDL-CHOL F | no | 0.34 (0.16–1.26) | 0.230 | 0.73 (0.62–0.81) | 0.029 |
| yes | 0.76 (0.31–1.47) | 0.54 (0.46–0.71) | |||
| HyperTG F | no | 0.41 (0.23–1.26) | 0.450 | 0.73 (0.59–0.81) | 0.050 |
| yes | 0.76 (0.27–1.47) | 0.54 (0.46–0.72) | |||
| ↑ CV risk by CRI-1 F | no | 0.34 (0.28–1.13) | 0.068 | 0.71 (0.48–0.79) | 0.283 |
| yes | 1.34 (0.8–1.89) | 0.58 (0.52–0.59) | |||
| ↑ CV risk by AC F | no | 0.32 (0.24–1.0) | 0.047 | 0.72 (0.54–0.81) | 0.089 |
| yes | 1.23 (0.87–1.7) | 0.54 (0.48–0.59) | |||
| ↑ CV risk by AIP F | no | 0.36 (0.29–1.05) | 0.044 | 0.7 (0.51–0.78) | 0.296 |
| yes | 1.7 (1.34–2.06) | 0.54 (0.5–0.58) | |||
| ↑ CV risk by LCI F | no | 0.32 (0.23–0.84) | 0.036 | 0.72 (0.50–0.81) | 0.023 |
| yes | 1.23 (0.98–1.66) | 0.48 (0.46–0.54) | |||
| Inflammation by CRP M | no | 0.32 (0.13–0.76) | 0.218 | 0.76 (0.71–0.82) | 0.043 |
| yes | 0.48 (0.33–2.31) | 0.49 (0.44–0.73) | |||
| Factor | Category | N | GSDMD | GSDME | ||
|---|---|---|---|---|---|---|
| ρ | p Value | ρ | p Value | |||
| Age | all | 51 | −0.09 | 0.522 | 0.0 | 0.992 |
| IHS4 | all | 48 | −0.12 | 0.427 | 0.02 | 0.883 |
| HS duration | all | 41 | 0.02 | 0.913 | 0.03 | 0.830 |
| RBC | all | 44 | −0.35 | 0.020 | 0.11 | 0.521 |
| Hb | all | 44 | −0.16 | 0.295 | 0.30 | 0.051 |
| F | 20 | 0.24 | 0.302 | 0.14 | 0.543 | |
| M | 24 | −0.21 | 0.316 | 0.47 | 0.019 | |
| BMI | all | 43 | −0.33 | 0.032 | −0.04 | 0.819 |
| ≥30 | 21 | −0.43 | 0.051 | 0.24 | 0.296 | |
| GLU | F | 15 | −0.14 | 0.629 | −0.59 | 0.020 |
| M | 17 | −0.20 | 0.450 | 0.47 | 0.058 | |
| TyG | F | 15 | 0.36 | 0.186 | −0.62 | 0.013 |
| M | 16 | −0.19 | 0.471 | 0.37 | 0.161 | |
| tCHOL | F | 18 | 0.31 | 0.216 | −0.50 | 0.034 |
| M | 13 | 0.12 | 0.687 | 0.20 | 0.463 | |
| LDL-CHOL | F | 17 | 0.04 | 0.885 | −0.51 | 0.037 |
| M | 13 | 0.10 | 0.734 | −0.03 | 0.915 | |
| Non-HDL-CHOL | F | 18 | 0.19 | 0.464 | −0.45 | 0.061 |
| M | 13 | 0.04 | 0.892 | 0.17 | 0.535 | |
| TG | F | 18 | 0.46 | 0.058 | −0.50 | 0.036 |
| M | 16 | −0.08 | 0.782 | 0.21 | 0.434 | |
| LCI | F | 17 | 0.26 | 0.313 | −0.55 | 0.032 |
| M | 16 | −0.02 | 0.936 | 0.21 | 0.441 | |
| AIP | F | 17 | 0.36 | 0.155 | −0.43 | 0.084 |
| M | 16 | −0.38 | 0.148 | 0.05 | 0.867 | |
| CREA | all | 37 | −0.03 | 0.872 | 0.24 | 0.015 |
| AST | F | 19 | 0.09 | 0.702 | −0.45 | 0.056 |
| ALP | F | 16 | −0.49 | 0.055 | −0.29 | 0.283 |
| Ferritin | F | 14 | 0.58 | 0.029 | −0.13 | 0.657 |
| Fe | F | 7 | 0.76 | 0.047 | −0.13 | 0.782 |
| M | 13 | −0.07 | 0.825 | 0.52 | 0.071 | |
| Factor | Cat. | Skin Expression of GSDMD | |||||
|---|---|---|---|---|---|---|---|
| AIL | p | ANS | p | Fold Change | p | ||
| Sex | F | 1.4 (0.9–2.3) | 0.293 | 1.4 (0.6–3.2) | 0.555 | 1.1 (0.5–2.2) | 0.156 |
| M | 1.9 (1.4–2.7) | 1.1 (0.7–1.7) | 1.8 (1.2–2.8) | ||||
| Hurley | 2 | 1.7 (1.1–2.4) | 0.374 | 1.1 (0.6–1.8) | 0.858 | 1.6 (1–2.4) | 0.407 |
| 3 | 1.3 (0.9–2) | 1.2 (0.4–3.3) | 1.1 (0.5–2.7) | ||||
| IHS4 | moderate | 2.2 (0.8–6.2) | 0.215 | 1.3 (0.2–8.2) | 0.783 | 1.7 (0.4–7) | 0.599 |
| severe | 1.5 (1.1–1.9) | 1.1 (0.7–1.9) | 1.3 (0.9–2) | ||||
| Duration | <10 yrs. | 1.6 (1.2–2) | 0.322 | 1.1 (0.6–1.9) | 0.683 | 1.5 (0.9–2.5) | 0.890 |
| ≥10 yrs. | 2.1 (0.7–5.8) | 1.3 (0.3–6.1) | 1.6 (0.5–4.9) | ||||
| Smoking | no | 2.0 (1.0–3.8) | 0.407 | 1.0 (0.6–1.5) | 0.801 | 2.1 (1.3–3.2) | 0.342 |
| yes | 1.6 (1.1–2.2) | 1.1 (0.5–2.2) | 1.5 (0.8–2.7) | ||||
| HyperUA | no | 2.3 (1.6–3.3) | 0.038 | 2.2 (0.9–5.2) | 0.228 | 1.1 (0.5–2.5) | 0.877 |
| yes | 1.4 (0.9–2.2) | 1.2 (0.5–2.8) | 1.1 (0.5–2.7) | ||||
| Factor | AIL | ANS | Fold Change | |||
|---|---|---|---|---|---|---|
| Corr. Coeff. | p | Corr. Coeff. | p | Corr. Coeff. | p | |
| Age | r = 0.20 | 0.368 | r = 0.08 | 0.716 | r = 0.06 | 0.778 |
| IHS4 | τ = −0.24 | 0.141 | τ = −0.26 | 0.106 | τ = 0.14 | 0.408 |
| HS duration | τ = 0.14 | 0.484 | τ = 0.09 | 0.689 | τ = 0.07 | 0.764 |
| UA | r = −0.58 | 0.062 | r = −0.41 | 0.214 | r = 0.10 | 0.778 |
| tBIL | τ = 0 | 0.955 | τ = 0.37 | 0.077 | τ = −0.37 | 0.060 |
| GGT | r = 0.46 | 0.085 | r = 0.20 | 0.478 | r = 0.08 | 0.777 |
| Factor | Cat. | Skin Expression of GSDME | |||||
|---|---|---|---|---|---|---|---|
| AIL | p | ANS | p | Fold Change | p | ||
| Sex | F | 1.6 (1.2–2.1) | 0.284 | 1.0 (0.5–1.8) | 0.244 | 1.7 (0.9–3.1) | 0.478 |
| M | 1.3 (1.1–1.7) | 0.6 (0.4–1) | 2.2 (1.4–3.4) | ||||
| Hurley | 2 | 1.4 (1–1.9) | 0.820 | 0.8 (0.5–1.4) | 0.515 | 1.7 (1.1–2.8) | 0.414 |
| 3 | 1.5 (1.2–1.9) | 0.6 (0.3–1.5) | 2.4 (1.1–5.3) | ||||
| IHS4 | moderate | 1.5 (1–2.2) | 0.944 | 0.6 (0–8.6) | 0.708 | 2.4 (1.1–39) | 0.660 |
| severe | 1.4 (1.2–1.8) | 0.8 (0.5–1.2) | 1.9 (1.3–2.8) | ||||
| Duration | <10 yrs. | 1.5 (1.3–1.8) | 0.449 | 0.6 (0.3–1.2) | 0.324 | 2.4 (1.3–4.4) | 0.430 |
| ≥10 yrs. | 1.7 (0.9–3.3) | 1.1 (0.4–3) | 1.6 (0.4–5.5) | ||||
| Smoking | no | 1.5 (1.1–2.1) | 0.936 | 0.8 (0.3–2.2) | 0.971 | 2 (0.8–5.4) | 0.948 |
| yes | 1.5 (1.3–1.8) | 0.8 (0.5–1.2) | 2 (1.2–3.1) | ||||
| IR (TyG) | no | 1.8 (1.3–2.3) | 0.255 | 0.6 (0.3–1.3) | 0.163 | 3 (1.5–6) | 0.055 |
| yes | 1.5 (1.1–1.9) | 1.2 (0.5–3.1) | 1.2 (0.5–2.8) | ||||
| ↓ HDL-C | no | 1.8 (1.3–2.5) | 0.099 | 0.8 (0.4–1.8) | 0.822 | 2.3 (1–5.1) | 0.736 |
| yes | 1.4 (1.2–1.7) | 0.7 (0.3–1.6) | 1.9 (1–3.8) | ||||
| HyperTG | no | 1.7 (1.3–2.1) | 0.410 | 0.6 (0.3–1.1) | 0.112 | 2.8 (1.7–4.7) | 0.039 |
| yes | 1.5 (1.1–1.9) | 1.2 (0.5–3.1) | 1.2 (0.5–2.8) | ||||
| ↑AIP | no | 1.5 (1.2–2) | 0.432 | 0.5 (0.3–0.9) | 0.027 | 2.8 (1.7–4.7) | 0.049 |
| yes | 1.7 (1.2–2.5) | 1.4 (0.6–3.6) | 1.2 (0.5–2.9) | ||||
| ↑CRI2 | no | 1.6 (1.2–2) | 0.641 | 0.7 (0.4–1.1) | 0.056 | 2.3 (1.5–3.6) | 0.078 |
| yes | 1.7 (1.4–2.1) | 1.7 (0.3–12) | 1 (0.1–7.3) | ||||
| Factor | AIL | ANS | Fold Change | |||
|---|---|---|---|---|---|---|
| Corr. Coeff. | p | Corr. Coeff. | p | Corr. Coeff. | p | |
| Age | r = 0.28 | 0.208 | r = 0.62 | 0.002 | r = −0.52 | 0.014 |
| IHS4 | τ = −0.06 | 0.693 | τ = −0.28 | 0.091 | τ = 0.26 | 0.122 |
| HS duration | τ = 0.20 | 0.318 | τ = 0.34 | 0.089 | τ = −0.30 | 0.110 |
| HOMA–IR | τ = 0.07 | 0.814 | τ = 0.29 | 0.239 | τ = −0.40 | 0.071 |
| CRI-2 | τ = −0.27 | 0.169 | τ = 0.18 | 0.409 | τ = −0.35 | 0.069 |
| LCI | τ = −0.36 | 0.073 | τ = 0.10 | 0.665 | τ = −0.23 | 0.240 |
| Ferritin | r = 0.05 | 0.885 | r = 0.69 | 0.012 | r = −0.71 | 0.010 |
| IgA | r = −0.32 | 0.317 | r = 0.45 | 0.147 | r = −0.57 | 0.045 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tyczyńska, K.; Krajewski, P.K.; Sójka, A.; Nowicka-Suszko, D.; Bednarz-Misa, I.; Fleszar, M.; Krzystek-Korpacka, M.; Szepietowski, J.C. Gasdermin-Mediated Pyroptosis in Hidradenitis Suppurativa: Molecular Insights and Therapeutic Implications. Biology 2025, 14, 1258. https://doi.org/10.3390/biology14091258
Tyczyńska K, Krajewski PK, Sójka A, Nowicka-Suszko D, Bednarz-Misa I, Fleszar M, Krzystek-Korpacka M, Szepietowski JC. Gasdermin-Mediated Pyroptosis in Hidradenitis Suppurativa: Molecular Insights and Therapeutic Implications. Biology. 2025; 14(9):1258. https://doi.org/10.3390/biology14091258
Chicago/Turabian StyleTyczyńska, Kinga, Piotr K. Krajewski, Aleksandra Sójka, Danuta Nowicka-Suszko, Iwona Bednarz-Misa, Mariusz Fleszar, Małgorzata Krzystek-Korpacka, and Jacek C. Szepietowski. 2025. "Gasdermin-Mediated Pyroptosis in Hidradenitis Suppurativa: Molecular Insights and Therapeutic Implications" Biology 14, no. 9: 1258. https://doi.org/10.3390/biology14091258
APA StyleTyczyńska, K., Krajewski, P. K., Sójka, A., Nowicka-Suszko, D., Bednarz-Misa, I., Fleszar, M., Krzystek-Korpacka, M., & Szepietowski, J. C. (2025). Gasdermin-Mediated Pyroptosis in Hidradenitis Suppurativa: Molecular Insights and Therapeutic Implications. Biology, 14(9), 1258. https://doi.org/10.3390/biology14091258








