Whole Transcriptome Sequencing and Differential Analysis of Testes in Pre- and Post-Sexual Maturity Bactrian Camels (Camelus bactrianus)
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Sample Collection
2.2. Micromorphological Examination
2.3. RNA Extraction, Quality, and Integrity Assessment
2.4. Quality Assessment, Alignment Optimization, and Transcriptome Reconstruction of RNA-Seq Data
2.5. Identification of Differentially Expressed Genes
2.6. Functional Enrichment Analysis of Differentially Expressed Genes
2.7. Constructed ceRNA Network
2.8. Validation of Differentially Expressed Genes
3. Results and Analysis
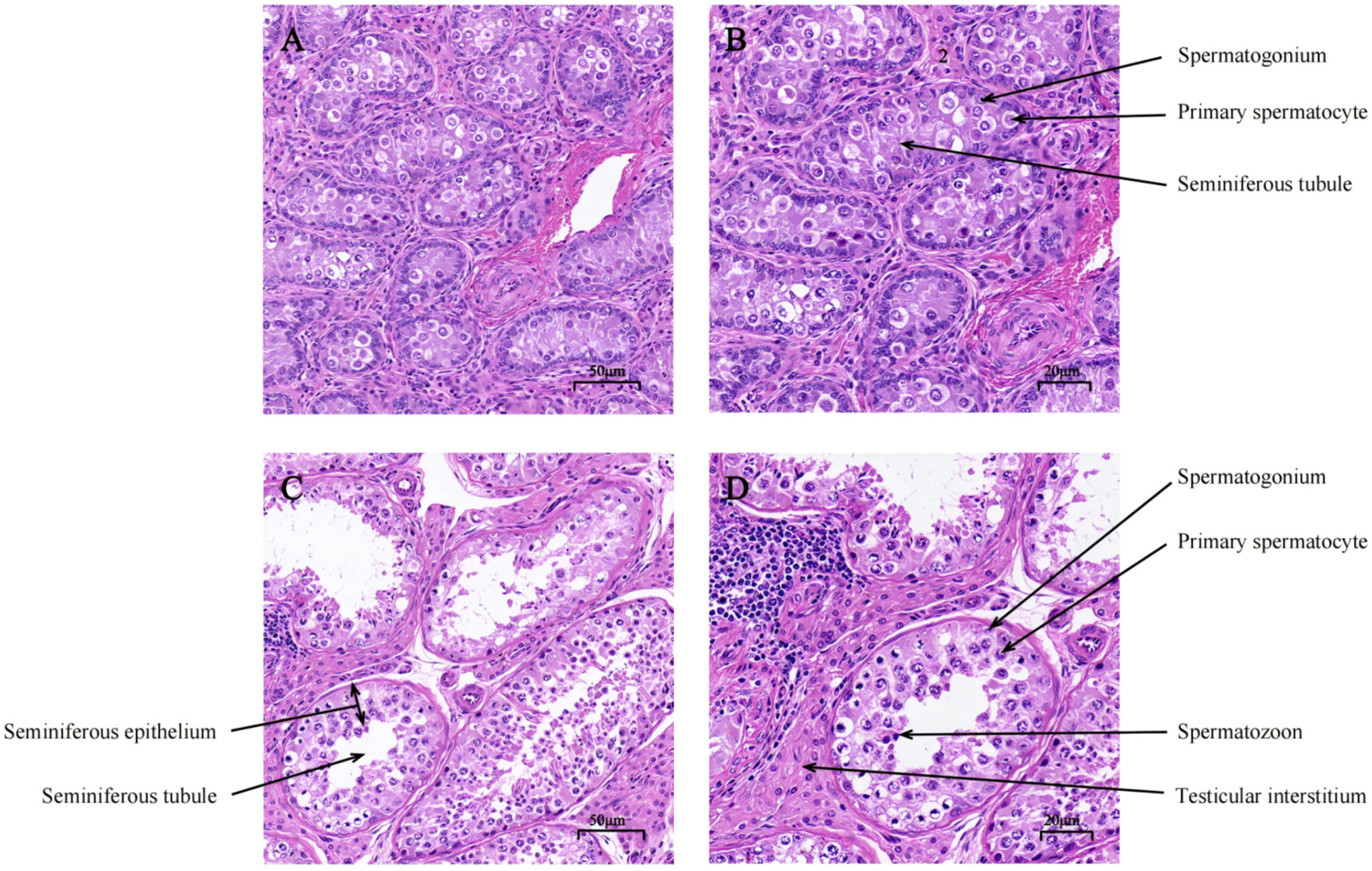
3.1. Histological Analysis of Testicular Tissues
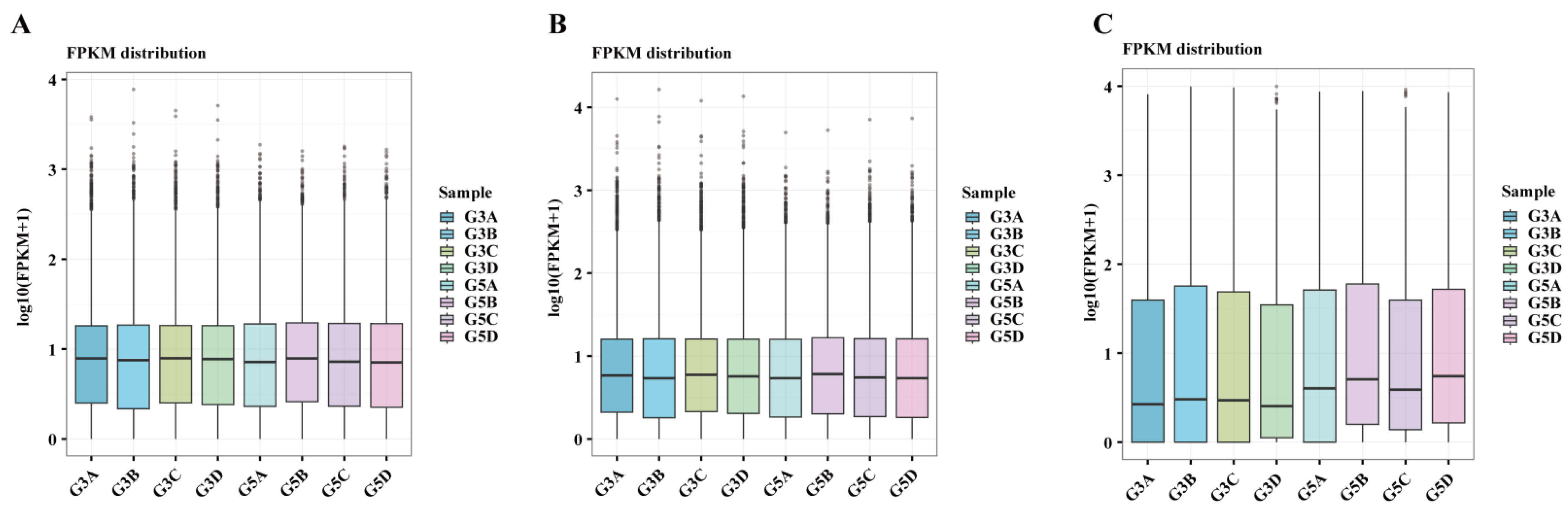
3.2. RNA-Seq Data Analysis
3.3. Gene Expression Quantification
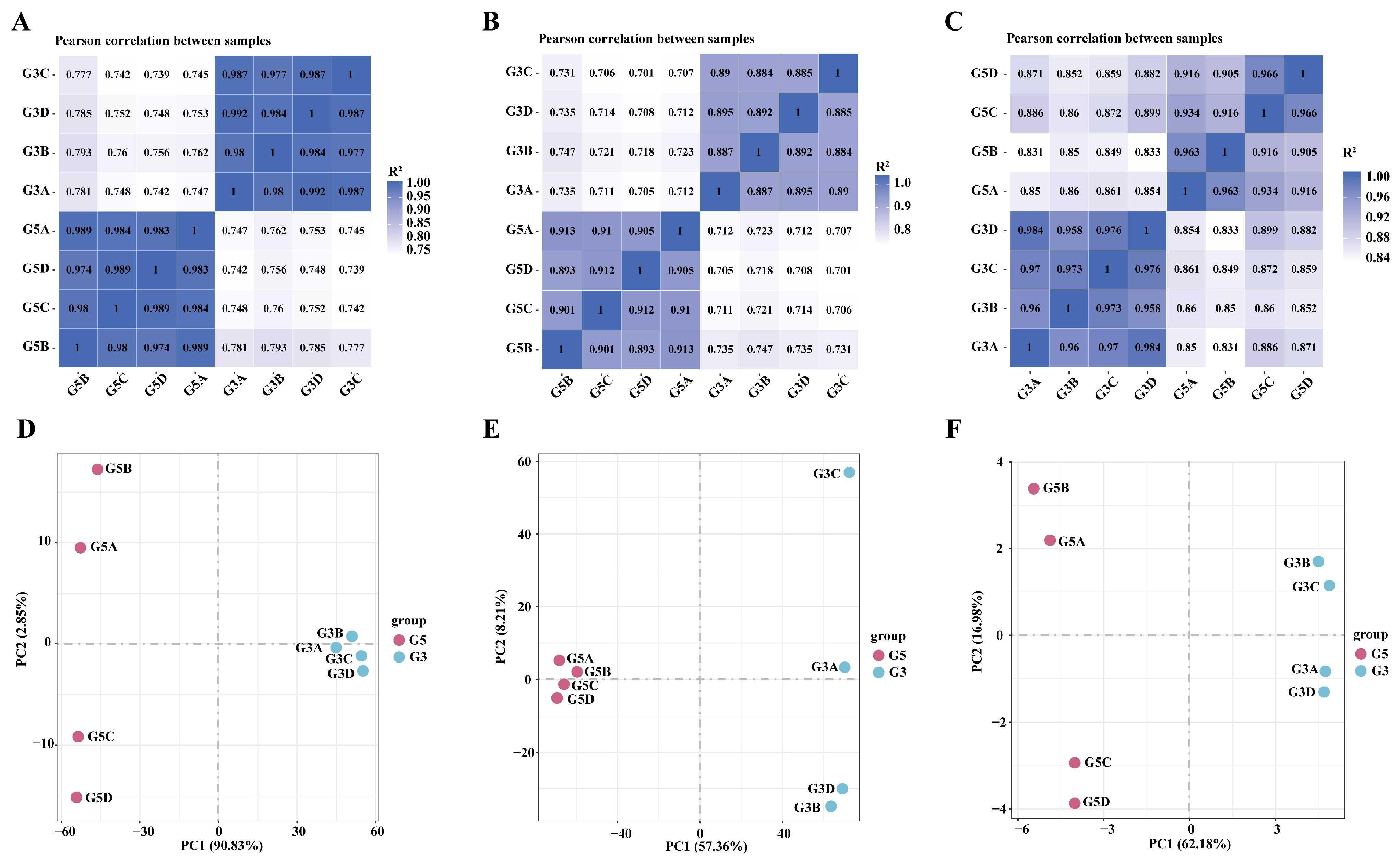
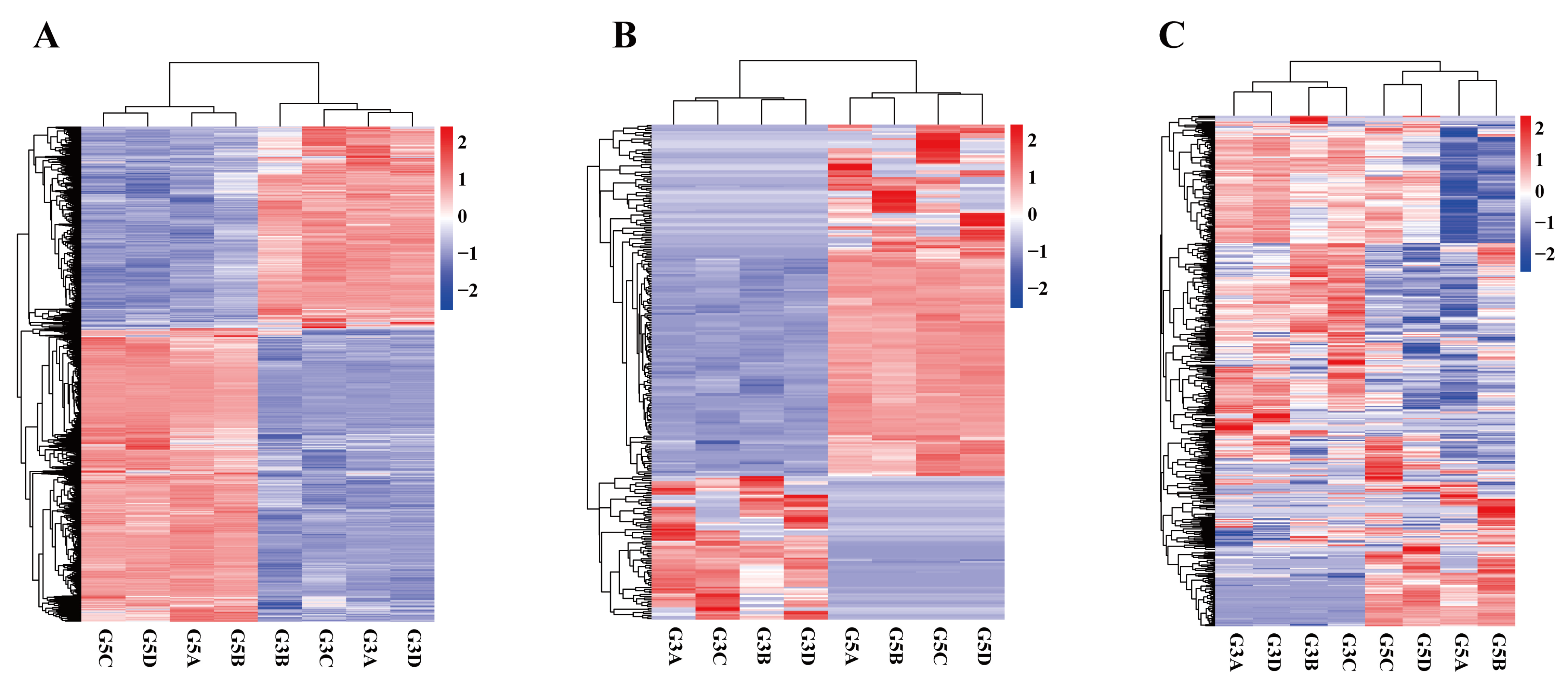
3.4. Correlation Analysis of Differentially Expressed Genes in Testicular Tissues at Different Developmental Stages
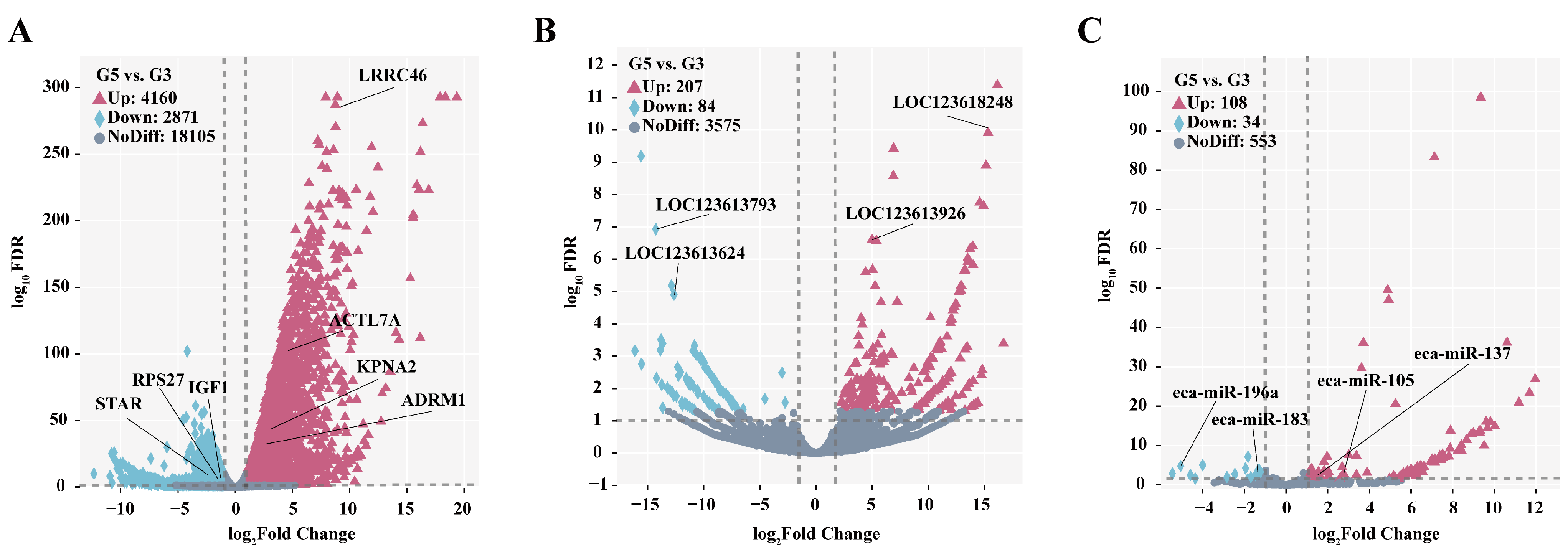
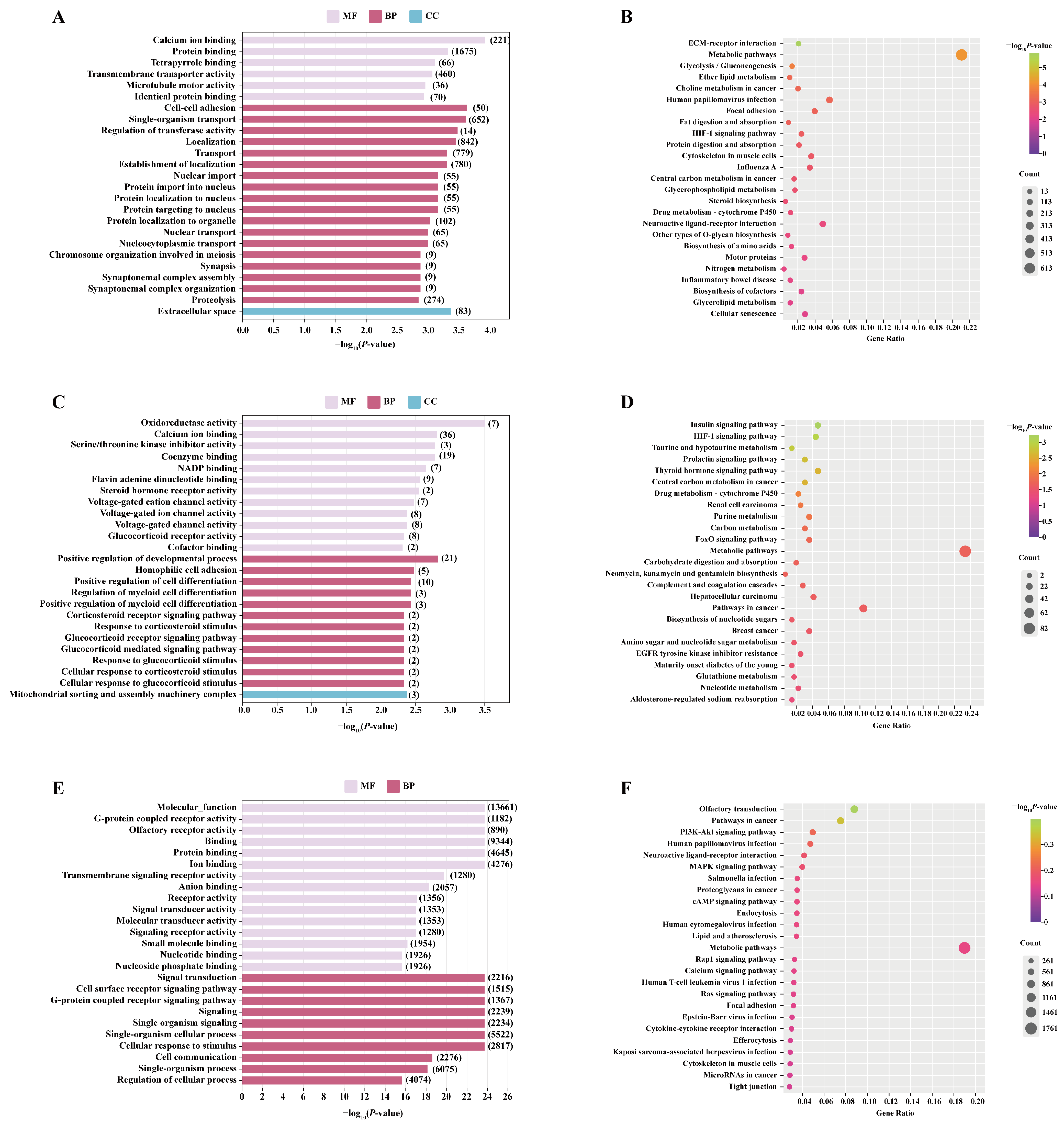
3.5. Identification and Bioinformatics Analysis of Differentially Expressed mRNA
3.6. Characteristics of Differentially Expressed LncRNA
3.7. Screening of Differentially Expressed miRNA and Target Gene Enrichment Analysis
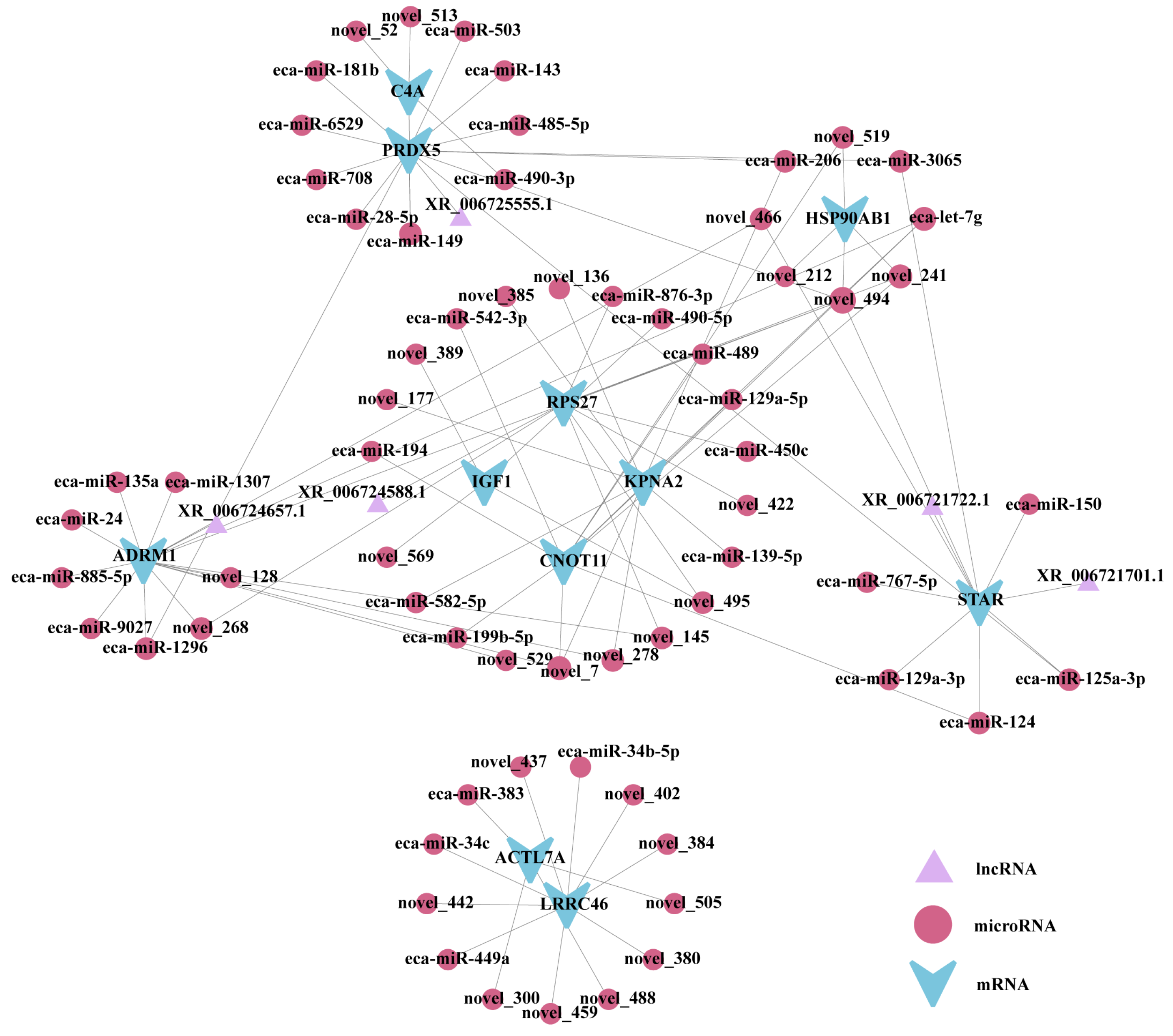
3.8. LncRNA-miRNA-mRNA in the ceRNA Network
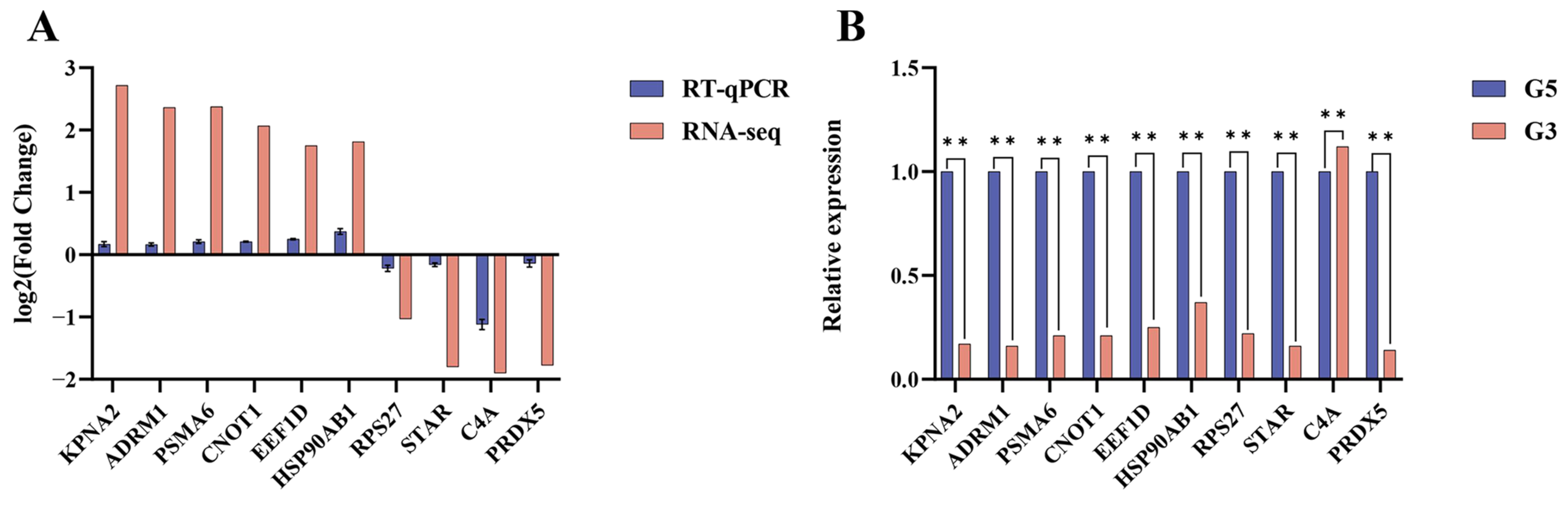
3.9. Validation of RNA-Seq Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Si, R.; Ming, L.; Yun, X.; He, J.; Yi, L.; Na, Q.; Ji, R.; Dong, T. Proteomics integrated with metabolomics: Analysis of the internal mechanism underlying changes in meat quality in different muscles from Bactrian camels. Food Chem. X 2025, 26, 102230. [Google Scholar] [CrossRef]
- Burger, P.A.; Ciani, E.; Faye, B. Old World camels in a modern world-a balancing act between conservation and genetic improvement. Anim. Genet. 2019, 50, 598–612. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Yuan, L.; Ma, X.; Qi, Y.; Cheng, S.; Zhang, Y. Histological study of Bactrian camel cryptorchidism and expression of immunoglobulin λ light chain in the testicular and epididymis of cryptorchid Bactrian camel. Reprod. Domest. Anim. 2024, 59, e14512. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.; Liang, X.; Dou, Z.; Zhao, Z.; Ma, W.; Hao, Z.; Yang, J. Transcriptome analysis to identify candidate genes related to mammary gland development of Bactrian camel (Camelus bactrianus). Front. Vet. Sci. 2023, 10, 1196950. [Google Scholar] [CrossRef] [PubMed]
- Hasi, G.; Wu, L.; Sodnompil, T.; Yi, R.; Wu, R.; Zhang, R.; Nasenochir, N. Differential expression and localisation of heat shock protein 70 (HSP70) and glutathione peroxidase 5 (GPX5) in the testis and epididymis of Sonid Bactrian camels. Reprod. Fertil. Dev. 2023, 35, 552–562. [Google Scholar] [CrossRef]
- Li, X.L.; Wu, Q.; Wang, T.; Zhang, L.; Wu, X.; Zhang, Y.; Liu, B. Integrated testis transcriptome and whole genome analysis of sexual maturity in Large White and Tongcheng pigs. Reprod. Domest. Anim. 2023, 58, 955–964. [Google Scholar] [CrossRef]
- Rawlings, N.; Evans, A.C.O.; Chandolia, R.K.; Bagu, E.T. Sexual maturation in the bull. Reprod. Domest. Anim. 2008, 43, 295–301. [Google Scholar] [CrossRef]
- Ebel, F.; Ulloa, O.; Strobel, P.; Ramírez-Reveco, A. Semen quality and freezability analyses in the ejaculates of two Poitou donkeys in the southern hemisphere. Front. Vet. Sci. 2021, 8, 662887. [Google Scholar] [CrossRef]
- Zhang, F.L.; Zhang, X.Y.; Zhao, J.X.; Zhu, K.X.; Liu, S.Q.; Zhang, T.; Shen, W. Multispecies comparative analysis reveals transcriptional specificity during Mongolian horse testicular development. Reprod. Domest. Anim. 2022, 57, 1295–1306. [Google Scholar] [CrossRef]
- Zhao, W.; Adjei, M.; Zhang, Z.; Yuan, Z.; Cisang, Z.; Song, T. The role of GnRH in Tibetan male sheep and goat reproduction. Reprod. Domest. Anim. 2023, 58, 1179–1187. [Google Scholar]
- Su, J.; Yang, Y.; Wang, D.; Su, H.; Zhao, F.; Zhang, C.; Zhang, M.; Li, X.; He, T.; Li, X. A dynamic transcriptional cell atlas of testes development after birth in Hu sheep. BMC Biol. 2025, 23, 78. [Google Scholar] [CrossRef] [PubMed]
- Słowińska, M.; Paukszto, Ł.; Jastrzębski, J.P.; Bukowska, J.; Kozłowski, K.; Jankowski, J.; Ciereszko, A. Transcriptome analysis of turkey (Meleagris gallopavo) reproductive tract revealed key pathways regulating spermatogenesis and post-testicular sperm maturation. Poult. Sci. 2020, 99, 6094–6118. [Google Scholar] [CrossRef] [PubMed]
- La, Y.; Ma, X.; Bao, P.; Chu, M.; Yan, P.; Liang, C.; Guo, X. Genome-wide Landscape of mRNAs, LncRNAs, and circRNAs during Testicular Development of Yak. Int. J. Mol. Sci. 2023, 24, 4420. [Google Scholar] [CrossRef]
- Xi, B.; Lu, Z.; Zhang, R.; Zhao, S.; Li, J.; An, X.; Yue, Y. Comprehensive analysis of the transcriptome-wide m6A Methylome in sheep testicular development. Genomics 2025, 117, 111005. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Yuan, L.; Zeng, J.; Qi, Y.; Ma, L.; Li, H.; Lv, J.; Chen, Y. Comparative proteomic and transcriptomic analysis of testicular tissue of yaks with or without cryptorchidism. Theriogenology 2025, 239, 117376. [Google Scholar] [CrossRef]
- Yao, H.; Liu, M.; Ma, W.; Yue, H.; Su, Z.; Song, R.; Ma, Q.; Li, L.; Wu, Z.; Ma, Y. Prevalence and pathology of Cephalopina titillator infestation in Camelus bactrianus from Xinjiang, China. BMC Vet. Res. 2022, 18, 360. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Kong, L.; Zhang, Y.; Ye, Z.-Q.; Liu, X.-Q.; Zhao, S.-Q.; Wei, L.; Gao, G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 35 (Suppl. S2), W345–W349. [Google Scholar] [CrossRef]
- Lara, N.d.L.e.M.; Sakib, S.; Dobrinski, I. Regulation of cell types within testicular organoids. Endocrinology 2021, 162, bqab033. [Google Scholar] [CrossRef] [PubMed]
- Oliver, E.; Stukenborg, J.B. Rebuilding the human testis in vitro. Andrology 2020, 8, 825–834. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Sun, L.; Li, S.; Liu, H.; Chen, Z.; Huang, S.; Mo, Y.; Li, G. miR-124-3p regulates the involvement of Ptpn1 in testicular development and spermatogenesis in mouse. Gene 2024, 893, 147967. [Google Scholar] [CrossRef] [PubMed]
- Koskenniemi, J.J.; Virtanen, H.E.; Toppari, J. Testicular growth and development in puberty. Curr. Opin. Endocrinol. Diabetes Obes. 2017, 24, 215–224. [Google Scholar] [CrossRef]
- Contreras-Ortiz, A.J.; Vigueras, R.M.; Mendoza-Elvira, S.E.; Martínez-Castañeda, F.E.; Gutiérrez-Pérez, O.; Trujillo-Ortega, M.E. Postnatal testicular development in Vietnamese pot-bellied pigs. Acta Histochem. 2021, 123, 151741. [Google Scholar] [CrossRef]
- Hasi, G.; Sodnompil, T.; Na, H.; Liu, H.; Ji, M.; Xie, W.; Nasenochir, N. Whole transcriptome sequencing reveals core genes related to spermatogenesis in Bactrian camels. J. Anim. Sci. 2023, 101, skad115. [Google Scholar] [CrossRef]
- Navarrete-López, P.; Maroto, M.; Pericuesta, E.; Fernández-González, R.; Lombó, M.; Ramos-Ibeas, P.; Gutiérrez-Adán, A. Loss of the importin Kpna2 causes infertility in male mice by disrupting the translocation of testis-specific transcription factors. Iscience 2023, 26, 107134. [Google Scholar] [CrossRef]
- Li, H.; Yang, B.-Y.; Liu, M.-M.; Zhao, S.-W.; Xie, S.-Z.; Wang, H.; Zhang, S.; Xuan, X.-N.; Jia, L.-J. Reproductive injury in male BALB/c mice infected with Neospora caninum. Parasites Vectors 2021, 14, 158. [Google Scholar] [CrossRef]
- Wu, Y.; Li, Q.; Qi, X.; Liu, Z.; Wang, C.; Zhao, X.; Ma, Y. Molecular characteristics and regulatory role of insulin-like growth factor 1 gene in testicular Leydig cells of Tibetan sheep. Sci. Rep. 2024, 14, 24799. [Google Scholar] [CrossRef]
- Huang, Y.H.; Chin, C.C.; Ho, H.N.; Chou, C.K.; Shen, C.N.; Kuo, H.C.; Ling, T.Y. Pluripotency of mouse spermatogonial stem cells maintained by IGF-1-dependent pathway. FASEB J. 2009, 23, 2076–2087. [Google Scholar] [CrossRef]
- Wang, J.; Ren, Q.; Hua, L.; Chen, J.; Zhang, J.; Bai, H.; Li, H.; Xu, B.; Shi, Z.; Cao, H.; et al. Comprehensive analysis of differentially expressed mRNA, LncRNA and circRNA and their ceRNA networks in the Longissimus Dorsi muscle of two different pig breeds. Int. J. Mol. Sci. 2019, 20, 1107. [Google Scholar] [CrossRef]
- Yan, Q.; Wang, Q. Exploring the characters of non-coding RNAs in spermatogenesis and male infertility. Int. J. Mol. Sci. 2025, 26, 1128. [Google Scholar] [CrossRef] [PubMed]
- Han, G.; Hong, S.H.; Lee, S.J. Transcriptome analysis of testicular aging in mice. Cells 2021, 10, 2895. [Google Scholar] [CrossRef]
- Ran, M.; Chen, B.; Li, Z.; Wu, M.; Liu, X.; He, C.; Li, Z. Systematic identification of long noncoding RNAs in immature and mature porcine testes. Biol. Reprod. 2016, 94, 77. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Li, S.; Lai, Z.; Zhou, Z.; Wu, F.; Huang, Y.; Dang, R. Analysis of long non-coding RNA and mRNA expression profiling in immature and mature bovine (Bos taurus) testes. Front. Genet. 2019, 10, 646. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Du, M.; Zhang, L.; Wang, N.; He, Q.; Cao, J.; Dugarjaviin, M. Comparative analysis of mRNA and LncRNA expression profiles in testicular tissue of sexually immature and sexually mature mongolian horses. Animals 2024, 14, 1717. [Google Scholar] [CrossRef] [PubMed]
- Sakaguchi, M. The role of insulin signaling with FOXO and FOXK transcription factors. Endocr. J. 2024, 71, 939–944. [Google Scholar] [CrossRef]
- Liu, W.; Meng, P.; Li, Z.; Shen, Y.; Meng, X.; Hu, M. The multifaceted impact of physical exercise on FoxO signaling pathways. Front. Cell Dev. Biol. 2025, 13, 1614732. [Google Scholar] [CrossRef]
- Yildirim, O.G.; Guney, C.; Alcigir, M.E.; Akar, F. High-fructose consumption suppresses insulin signaling pathway accompanied by activation of macrophage and apoptotic markers in rat testis. Reprod. Biol. 2023, 23, 100815. [Google Scholar] [CrossRef]
- Ling, Y.H.; Ren, C.H.; Guo, X.F.; Xu, L.N.; Huang, Y.F.; Luo, J.C.; Zhang, Y.H.; Zhang, X.R.; Zhang, Z.J. Identification and characterization of microRNAs in the ovaries of multiple and uniparous goats (Capra hircus) during follicular phase. BMC Genom. 2014, 15, 339. [Google Scholar] [CrossRef]
- Chen, W.; Cui, Y.; Ning, M.; Zhang, H.; Yin, C.; He, Z. The mechanisms and functions of microRNAs in mediating the fate determinations of human spermatogonial stem cells and Sertoli cells. Semin. Cell Dev. Biol. 2022, 121, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Yan, Z.; Gao, Y.; Li, J.; Wang, Z.; Wang, P.; Gun, S. Integrated analysis of miRNA and mRNA expression profiles in testes of Landrace and Hezuo boars. Front. Vet. Sci. 2022, 9, 942669. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Blondin, P.; Vigneault, C.; Labrecque, R.; Sirard, M.A. Sperm miRNAs-Potential mediators of bull age and early embryo development. BMC Genom. 2020, 21, 798. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Su, X.; Li, B.; Chu, C.; Sun, H.; Zhang, N.; Han, B.; Li, C.; Zou, B.; Niu, Y.; et al. PM2.5 exposure impairs sperm quality through testicular damage dependent on NALP3 inflammasome and miR-183/96/182 cluster targeting FOXO1 in mouse. Ecotoxicol. Environ. Saf. 2019, 169, 551–563. [Google Scholar] [CrossRef]
- Zhang, S.; Guo, J.; Liang, M.; Qi, J.; Wang, Z.; Jian, X.; Zhang, Z.; Sun, B.; Li, Z. miR-196a Promotes Proliferation and Inhibits Apoptosis of Immature Porcine Sertoli Cells. DNA Cell Biol. 2019, 38, 41–48. [Google Scholar] [CrossRef]
- Tang, Y.; Sun, L.; Li, S.; Luo, L.; Liu, H.; Chen, Z.; Li, G. miR-9-5p regulates Sirt1 involved in testicular development and spermatogenesis in mouse. Theriogenology 2024, 230, 61–71. [Google Scholar] [CrossRef]
- La, Y.; Ma, X.; Bao, P.; Chu, M.; Guo, X.; Liang, C.; Yan, P. Identification and profiling of microRNAs during yak’s testicular development. BMC Vet. Res. 2023, 19, 53. [Google Scholar] [CrossRef]
- Tian, H.; Cao, Y.X.; Zhang, X.S.; Liao, W.P.; Yi, Y.H.; Lian, J.; Sun, F. The targeting and functions of miRNA-383 are mediated by FMRP during spermatogenesis. Cell Death Dis. 2013, 4, e617. [Google Scholar] [CrossRef]
- Zhou, X.; Liu, Z.; Jia, W.; Hou, M.; Zhang, X. Actl7a deficiency in mice leads to male infertility and fertilization failure. Biochem. Biophys. Res. Commun. 2022, 623, 154–161. [Google Scholar] [CrossRef]
- Yin, Y.; Mu, W.; Yu, X.; Wang, Z.; Xu, K.; Wu, X.; Cai, Y.; Zhang, M.; Lu, G.; Chan, W.Y. LRRC46 accumulates at the midpiece of sperm flagella and is essential for spermiogenesis and male fertility in mouse. Int. J. Mol. Sci. 2022, 23, 8525. [Google Scholar] [CrossRef]
| Gene | Accession Number | Primer Sequence (5′ → 3′) | Product Length (bp) |
|---|---|---|---|
| β-actin | XM_010965866.2 | F: TCACTCACACTGTGCCCATC | 276 |
| R: CGATGGTGATGACCTGACCG | |||
| KPNA2 | XM_010948667.2 | F: TTGAAGAATGTGGAGGTTTGGA | 231 |
| R: AACAAACAACTTTACGCCTCAGCT | |||
| ADRM1 | XM_045516234.1 | F: CGACGACTCCCTCATTCACTTT | 216 |
| R: ACCTTCCGGCAGTGTTCCTC | |||
| PSMA6 | XM_010967859.2 | F: TTGACCGCCACATTACCATT | 274 |
| R: TTAGCTGCCTCATAGCGTGCC | |||
| CNOT1 | XM_010967251.2 | F: TGAAAGCACCACCAGAGGATGA | 145 |
| R: TCTGGACAGTGTTTGATAGGGAAG | |||
| HSP90AB1 | XM_045509278.1 | F: GATGCCTTGGACAAGATTCGTT | 250 |
| R: CAACACCAAACTGCCCAATCA | |||
| EEF1D | XM_010961152.2 | F: CAAGTACGACGATGCAGAAAGGA | 139 |
| R: AGCGAGGGACTTCTGGATGTT | |||
| RPS27 | XM_010954019.2 | F: GGAGGATTTCCGCTTTCGCT | 248 |
| R: CTGGCAGAGGACAGTAGAGC | |||
| STAR | XM_045509996.1 | F: AGACGTGGGCAAGGTGTTCC | 116 |
| R: CCTTGACATTCGGATTCCACTC | |||
| C4A | XM_010961471.2 | F: GCGGAAGAAGGAGGTGTATGCT | 134 |
| R: ACTGGGTGCTGCTGTTGGAAT | |||
| PRDX5 | XM_045518137.1 | F: ACGGAAGGCAAGGTTCAGCT | 163 |
| R: GCTCCACATTCAAGGACTTCACTA |
| Sample | Raw Reads | Clean Reads | Q20 | Q30 | GC Content | Mapped Reads |
|---|---|---|---|---|---|---|
| G3A | 107,793,310 | 107,593,776 | 97.98% | 94.26% | 51.53% | 84,447,363 (78.49%) |
| G3B | 106,556,608 | 106,350,854 | 97.91% | 93.96% | 52.56% | 80,037,945 (75.26%) |
| G3C | 104,963,176 | 104,776,860 | 98.01% | 94.37% | 50.86% | 82,017,719 (78.28%) |
| G3D | 105,845,590 | 105,667,472 | 97.99% | 94.26% | 50.99% | 83,238,145 (78.77%) |
| G5A | 106,950,304 | 106,752,926 | 98.00% | 94.53% | 52.11% | 83,811,285 (78.51%) |
| G5B | 105,116,432 | 104,942,932 | 98.22% | 94.83% | 51.76% | 83,279,099 (79.36%) |
| G5C | 104,563,528 | 104,384,630 | 98.01% | 94.45% | 51.09% | 83,145,960 (79.65%) |
| G5D | 107,799,328 | 107,629,916 | 98.21% | 94.72% | 50.32% | 87,684,364 (81.47%) |
| Sample | Raw Reads | Clean Reads | Q20 | Q30 | GC Content | Mapped Reads |
|---|---|---|---|---|---|---|
| G3A | 16,926,078 | 15,008,051 | 97.29% | 93.32% | 77.75% | 12,583,375 (83.84%) |
| G3B | 15,945,398 | 12,588,361 | 92.78% | 85.45% | 77.91% | 11,070,688 (87.94%) |
| G3C | 16,923,305 | 15,112,777 | 95.52% | 90.17% | 77.26% | 13,308,521 (88.06%) |
| G3D | 16,930,364 | 14,851,069 | 96.82% | 92.19% | 77.96% | 13,521,818 (91.05%) |
| G5A | 12,995,234 | 10,307,133 | 93.79% | 86.86% | 78.54% | 86,165,400 (83.60%) |
| G5B | 15,946,548 | 13,550,212 | 95.43% | 88.71% | 78.06% | 11,290,231 (83.32%) |
| G5C | 16,924,183 | 15,521,055 | 91.96% | 83.07% | 76.42% | 13,735,688 (88.50%) |
| G5D | 13,005,404 | 10,033,266 | 97.66% | 93.71% | 78.34% | 8,979,809 (89.50%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, X.; Yao, X.; Meng, J.; Wang, J.; Zeng, Y.; Li, L.; Ren, W. Whole Transcriptome Sequencing and Differential Analysis of Testes in Pre- and Post-Sexual Maturity Bactrian Camels (Camelus bactrianus). Biology 2025, 14, 1254. https://doi.org/10.3390/biology14091254
Chang X, Yao X, Meng J, Wang J, Zeng Y, Li L, Ren W. Whole Transcriptome Sequencing and Differential Analysis of Testes in Pre- and Post-Sexual Maturity Bactrian Camels (Camelus bactrianus). Biology. 2025; 14(9):1254. https://doi.org/10.3390/biology14091254
Chicago/Turabian StyleChang, Xiaokang, Xinkui Yao, Jun Meng, Jianwen Wang, Yaqi Zeng, Linling Li, and Wanlu Ren. 2025. "Whole Transcriptome Sequencing and Differential Analysis of Testes in Pre- and Post-Sexual Maturity Bactrian Camels (Camelus bactrianus)" Biology 14, no. 9: 1254. https://doi.org/10.3390/biology14091254
APA StyleChang, X., Yao, X., Meng, J., Wang, J., Zeng, Y., Li, L., & Ren, W. (2025). Whole Transcriptome Sequencing and Differential Analysis of Testes in Pre- and Post-Sexual Maturity Bactrian Camels (Camelus bactrianus). Biology, 14(9), 1254. https://doi.org/10.3390/biology14091254






