Proteomic Analysis of Antheraea pernyi Larvae After Injection of Vitamin C and Preliminary Exploration of the Function of Antheraea pernyi Aldonolactonase
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Materials
2.2. Proteomic Analysis of Midgut Proteins in A. pernyi Larvae After VC Injection
2.2.1. Protein Extraction
2.2.2. Proteolysis After Quality Control of Protein Extraction
2.2.3. High-Performance Liquid Chromatography
2.2.4. Mass Spectrometry Detection
2.2.5. Protein Identification
2.2.6. MaxQuant Parameter Configuration
2.2.7. Protein Quantification and Differential Analysis
2.3. Cloning and Bioinformatic Analysis of ApALase Gene
2.4. Effect of VC Injection on ApALase Gene Expression in Fourth A. pernyi Larvae
2.5. Data Analysis
3. Results
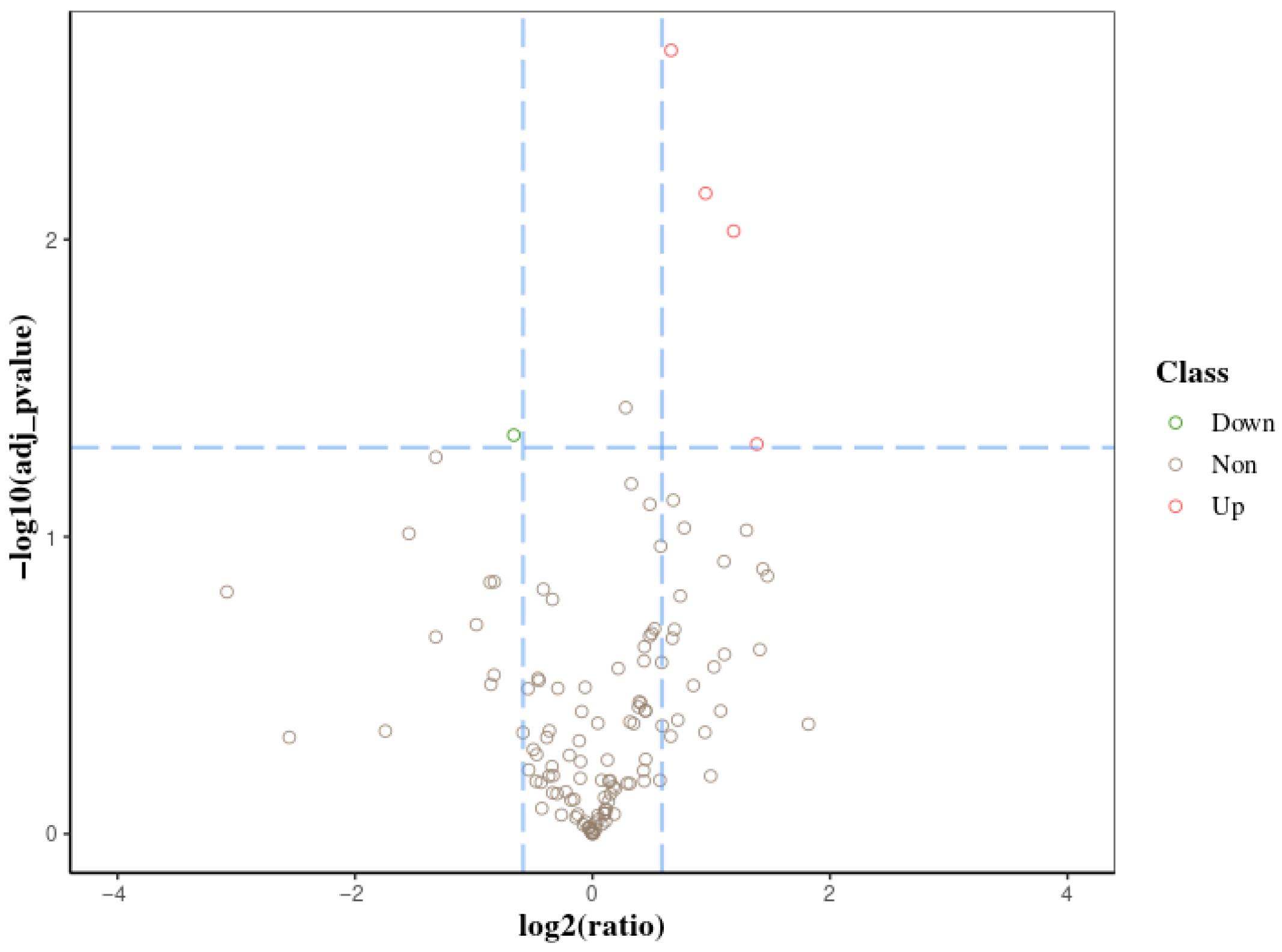
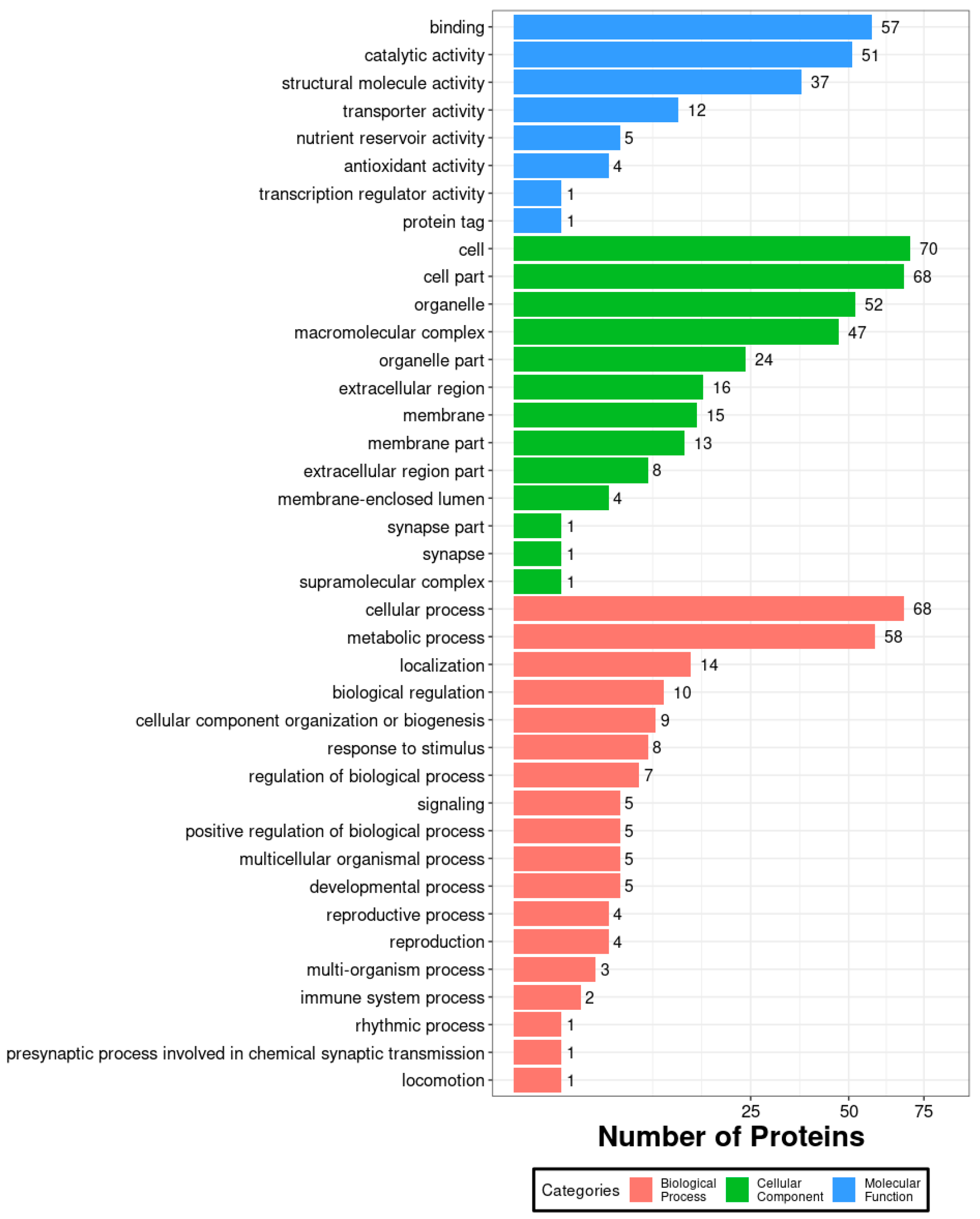
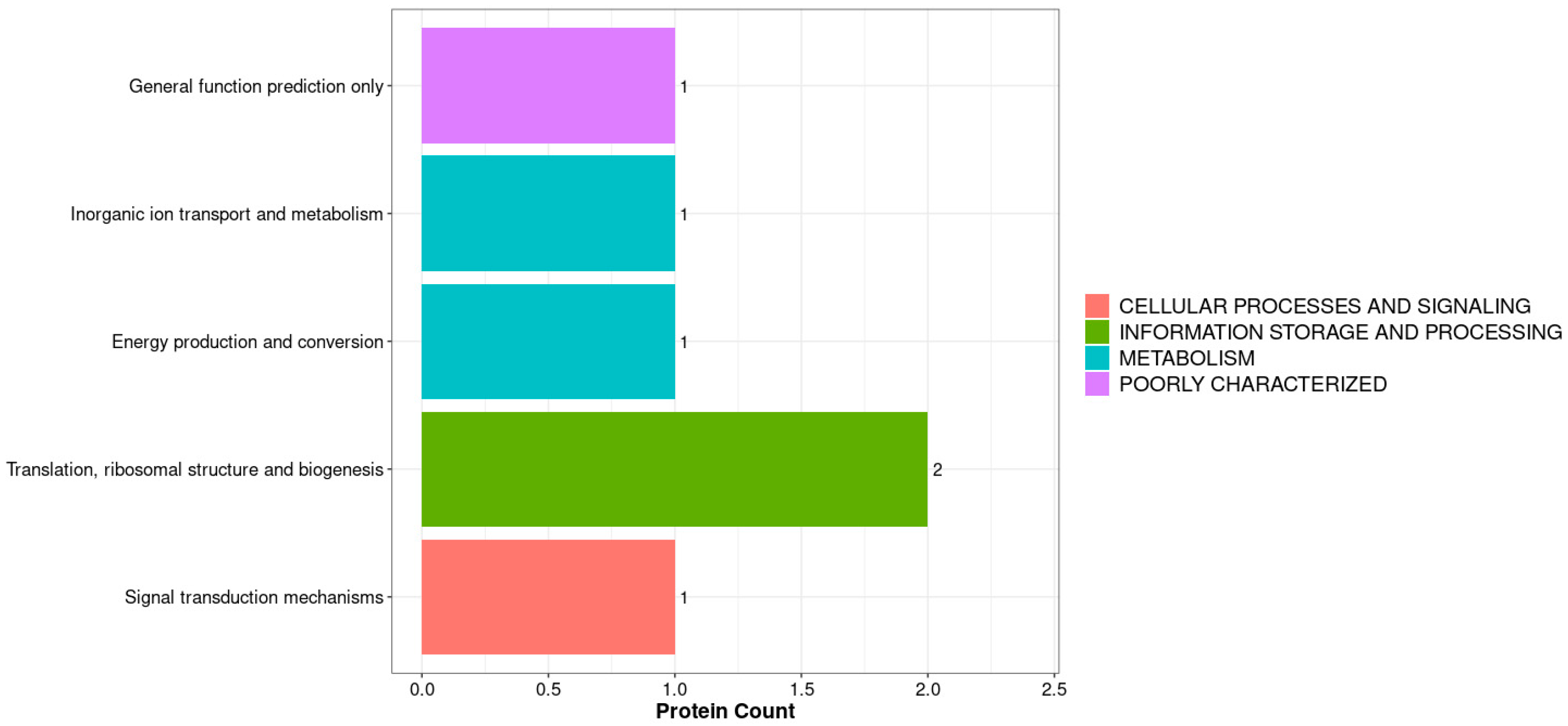
3.1. Proteomic Data of Midgut Proteins in A. pernyi Larvae After VC Injection and GO Functional Annotation
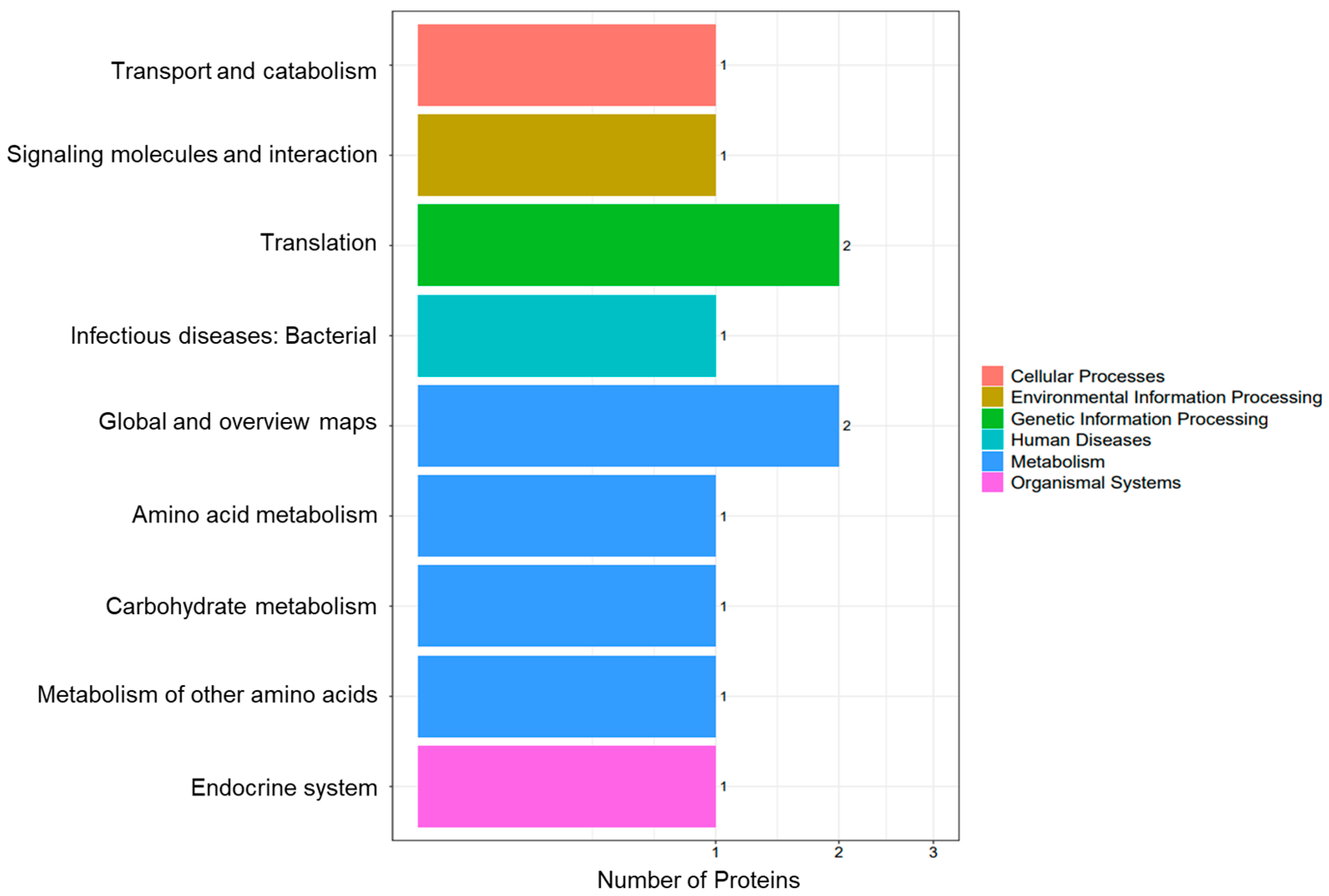
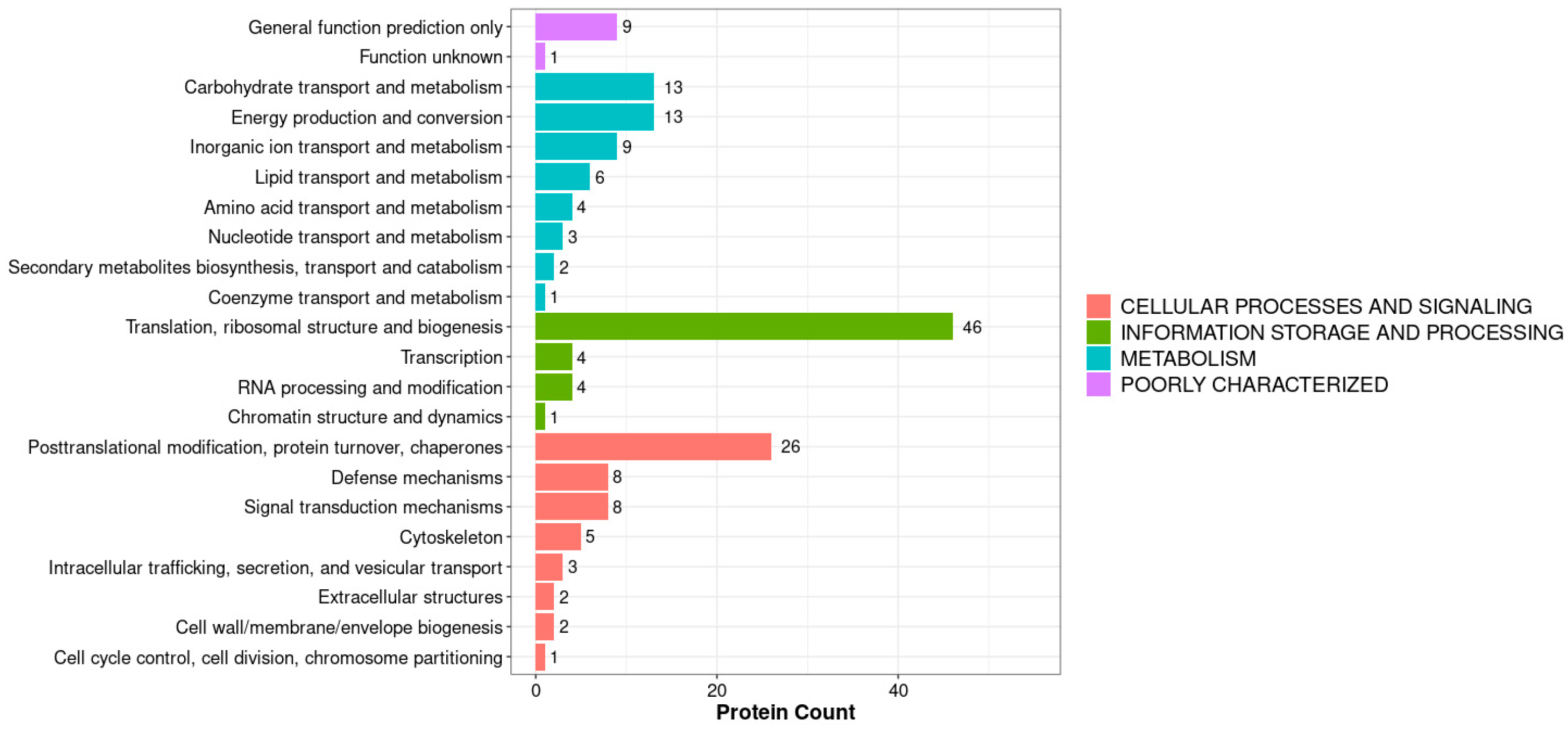
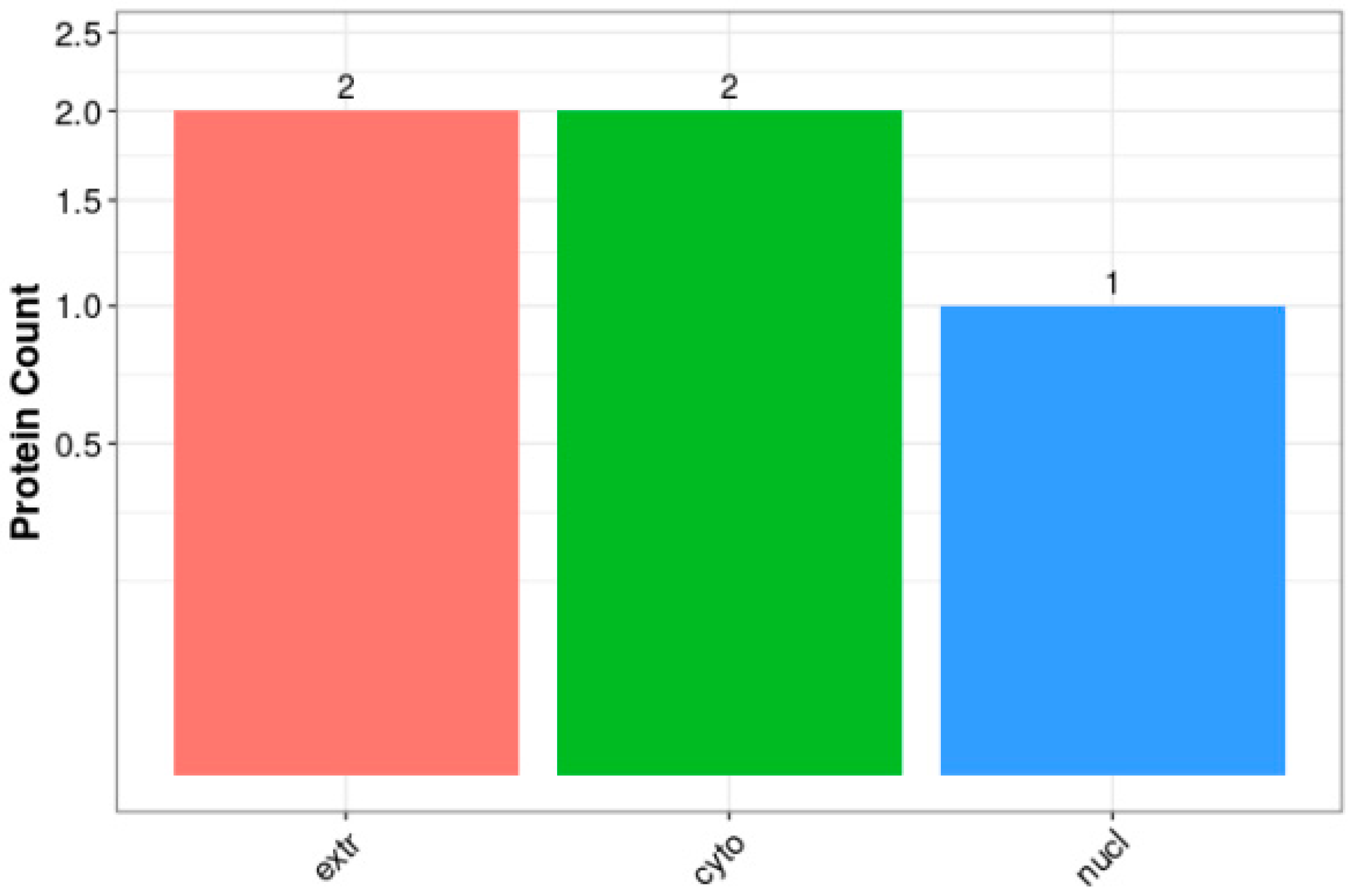
3.2. Functional Annotation and Subcellular Localization Analysis of the Midgut Proteome in A. pernyi Larvae Following VC Injection
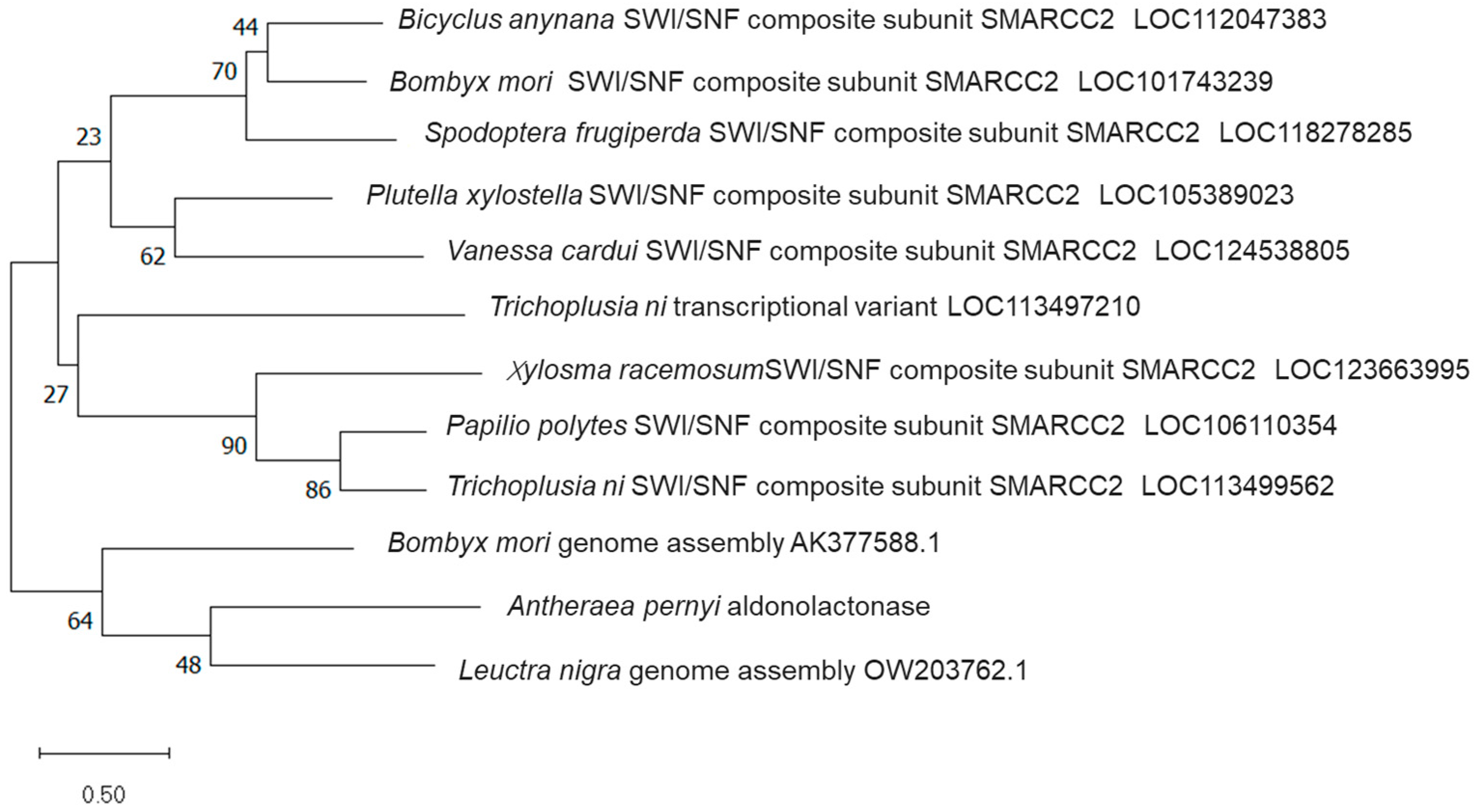
3.3. Cloning Sequence and Biological Analysis of ApALase
3.4. The Effect of 24-h VC Injection on the Relative Expression Level of ApALase Gene in Fourth Instar A. pernyi
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, G.F.; Wang, Y.J. Physiological functions of Vitamin C in metabolism. Chin. J. Food Nutr. 2014, 20, 71–74. [Google Scholar]
- Griffiths, H.R.; Lunec, J. Ascorbic acid in the 21st century more than a simple antioxidant. Environ. Toxicol. Pharmacol. 2001, 10, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Feng, Y.; He, Z.; Ma, T.; Zhang, X. Studies on alkaline solution extraction of polysaccharide from silkworm pupa and its immumomodulating activities. For. Res. 2007, 20, 782–786. [Google Scholar]
- Dong, G.; Mu, L.X.; Liao, S.T.; Zou, Y.X.; Liu, F.; Shen, W.Z.; Liu, J.P. Research on the main nutrients and active substances of some edible insects at different development stages. Seric. Sci. 2014, 40, 737–742. [Google Scholar]
- Wei, X.; Bin, C.; Haibin, Y.; Shusen, S. Analysis of Vitamin content and antioxidant enzyme activity in different developmental stages of yellow brown oil gourd. J. Northeast For. Univ. 2005, 33, 86–88. [Google Scholar]
- Pu Sheng, H.X. The relationship between the development of silkworms and Vitamin C after the emergence of. changes in Vitamin C content in mulberry leaves. Jpn. Silkworm Var. 1941, 12, 65–78. [Google Scholar]
- Ito, T. Research on the nutrition of silkworms IV: The Effect of Vitamin C. Trial Rep. 1961, 17, 119–136. [Google Scholar]
- Ito, T. Effect of dietary ascorbic acid on the silkworm, Bombyx mori. Nature 1961, 192, 951–952. [Google Scholar] [CrossRef]
- Cui, W.Z.; Wang, Y.W.; Mou, Z.M.; Juan, H.Z.; Liang, X.J. The taste electrophysiological response of silkworms to Vitamin C. Seric. Sci. 2001, 21, 92–95. [Google Scholar]
- Banhegyi, G.; Braun, L.; Csala, M.; Puskas, F.; Mandl, J. Ascorbate metabolism and its regulation in animals. Free. Radic. Biol. Med. 1997, 23, 793–803. [Google Scholar] [CrossRef] [PubMed]
- Scott, S.H.; Bahnson, B.J. Senescence marker protein 30: Functional and structural insights to its unknown physiological function. Biomol. Concepts 2011, 2, 469–480. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.R. Detection of Microsporidia in Tussah silkworms and Transcriptome and Proteomic Study of Tussah Intestine Infection; Shenyang Agricultural University: Shenyang, China, 2012. [Google Scholar]
- Su, G.M.; Ding, J. Observation on the infection and tissue lesions of tussah tissue cells by tussah microsporidia. North. Seric. 2003, 34–35. [Google Scholar]
- Yong, W.; Wei, L.; Binhe, W.; Yiren, J.; Ying, S.; Ruisheng, Y.; Li, Q. QRT PCR detection of the response of ApHSC70, a heat shock protein gene in the midgut of oak silkworms, to the invasion of microsporidia. Seric. Sci. 2015, 41, 300–307. [Google Scholar]
- Gong, X.; Niu, C.J. The synthesis ability of animal Vitamin C and its influencing factors. Biol. Bull. 2013, 48, 1–3. [Google Scholar]
- Ito, T. Vitamin nutrition of silkworms. J. Seric. 1985, 5, 12–19. [Google Scholar]
- Ishikawa, T.; Nishikawa, H.; Gao, Y.; Sawa, Y.; Shibata, H.; Yabuta, Y.; Maruta, T.; Shigeoka, S. The pathway via D-galacturonate/L-galactonate is significant for ascorbate biosynthesis in euglena gracilis-identification and functional characterization of aldonolactonase. J. Biol. Chem. 2008, 283, 31133–31141. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wu, Y.; Lan, F.; Chen, Y.; Cui, H.; Liu, Q.; Mou, Z.; Cui, W.; Meng, F. Changes in ascorbic acid content in silkworms at different developmental stages. Seric. Sci. 2016, 42, 15–24. [Google Scholar]




| Primers | Primer Sequences | Use |
|---|---|---|
| ApALase-F | TGTTTATGGACATGAAGCCG | Gene cloning |
| ApALase-R | CAGCATTATCGAATCGAAAGG | |
| ApA-F | CCAAAGGCCAACAGAGAGAAGA | |
| ApA-R | CAAGAATGAGGGCTGGAAGAGA | |
| q-ApALase-F | CCCATTGGAGAAGTGAAGT | qRT-PCR |
| q-ApALase-R | ACAAGAAGTGGAAGCGATA | |
| q-ApA-F | CGCTCCATCTACGATGAAGATC | |
| q-ApA-R | ACTCGTCGTACTCCTGTTTCG |
| Sample | Total Spectra | Identified Spectra | Identified Peptides | Identified Proteins |
|---|---|---|---|---|
| LCK 1 | 28,367 | 3830 | 1284 | 149 |
| LCK 2 | 28,147 | 3421 | 1255 | 146 |
| LCK 3 | 27,179 | 3482 | 1279 | 146 |
| LW 1 | 29,429 | 3402 | 1292 | 153 |
| LW 2 | 28,071 | 3691 | 1324 | 152 |
| LW 3 | 26,221 | 3475 | 1307 | 150 |
| Gene Ontology Term | Cluster Frequency | Protein Frequency of Use | p-Value |
|---|---|---|---|
| ribonucleoprotein complex | 12 out of 36 genes, 33.3% | 90 out of 766 genes, 11.7% | 0.000389 |
| intracellular | 30 out of 36 genes, 83.3% | 511 out of 766 genes, 66.7% | 0.019471 |
| chromatin | 2 out of 36 genes, 5.6% | 6 out of 766 genes, 0.8% | 0.028608 |
| intracellular part | 29 out of 36 genes, 80.6% | 503 out of 766 genes, 65.7% | 0.036246 |
| macromolecular complex | 14 out of 36 genes, 38.9% | 197 out of 766 genes, 25.7% | 0.052778 |
| cell | 32 out of 36 genes, 88.9% | 592 out of 766 genes, 77.3% | 0.059895 |
| non-membrane-bounded organelle | 5 out of 36 genes, 13.9% | 47 out of 766 genes, 6.1% | 0.062586 |
| endoplasmic reticulum | 2 out of 36 genes, 5.6% | 9 out of 766 genes, 1.2% | 0.062811 |
| chromosome | 2 out of 36 genes, 5.6% | 11 out of 766 genes, 1.4% | 0.090476 |
| cytoplasmic part | 5 out of 36 genes, 13.9% | 80 out of 766 genes, 10.4% | 0.319755 |
| cytoplasm | 5 out of 36 genes, 13.9% | 81 out of 766 genes, 10.6% | 0.32958 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, X.; Liu, Z.; Zhang, Y.; Zhang, Y.; Lian, Y.; Zhang, Z.; Zhu, X. Proteomic Analysis of Antheraea pernyi Larvae After Injection of Vitamin C and Preliminary Exploration of the Function of Antheraea pernyi Aldonolactonase. Biology 2025, 14, 1236. https://doi.org/10.3390/biology14091236
Xu X, Liu Z, Zhang Y, Zhang Y, Lian Y, Zhang Z, Zhu X. Proteomic Analysis of Antheraea pernyi Larvae After Injection of Vitamin C and Preliminary Exploration of the Function of Antheraea pernyi Aldonolactonase. Biology. 2025; 14(9):1236. https://doi.org/10.3390/biology14091236
Chicago/Turabian StyleXu, Xin, Zhongwen Liu, Yongjun Zhang, Yaoting Zhang, Yanxian Lian, Zhen Zhang, and Xuwei Zhu. 2025. "Proteomic Analysis of Antheraea pernyi Larvae After Injection of Vitamin C and Preliminary Exploration of the Function of Antheraea pernyi Aldonolactonase" Biology 14, no. 9: 1236. https://doi.org/10.3390/biology14091236
APA StyleXu, X., Liu, Z., Zhang, Y., Zhang, Y., Lian, Y., Zhang, Z., & Zhu, X. (2025). Proteomic Analysis of Antheraea pernyi Larvae After Injection of Vitamin C and Preliminary Exploration of the Function of Antheraea pernyi Aldonolactonase. Biology, 14(9), 1236. https://doi.org/10.3390/biology14091236





