1. Introduction
Kazakh horses, one of the most representative traditional livestock breeds, possess skeletal muscle characteristics closely linked to their athletic performance, such as endurance for long-distance running and load-bearing capacity. These traits constitute critical targets for breed selection and functional optimization [
1,
2]. Skeletal muscle is a principal component of the mammalian locomotor system. Its development involves mechanisms such as muscle fiber differentiation, metabolic regulation, and regeneration, all of which directly influence athletic performance [
3,
4,
5]. Beyond locomotion, skeletal muscle also plays a pivotal role in overall physiological function and metabolic health. Strength training can improve maximum voluntary contraction (MVC) activity, while low-intensity resistance exercises can increase the oxygen consumption of skeletal muscles [
6,
7]. In equines, athletic capacity is fundamentally determined by the structure and function of skeletal muscles, which are regulated by muscle fiber types, metabolic mechanisms, genetic background, postnatal training, and nutritional management [
8]. As the driving force of locomotion, the structure and function of skeletal muscles ultimately determine performance metrics such as speed, endurance, and explosive power during physical exertion [
9].
Energy supply in skeletal muscle operates through two primary metabolic pathways: aerobic and anaerobic metabolism. Aerobic metabolism relies on mitochondrial oxidation of fatty acids and glycogen to generate abundant ATP with minimal lactate accumulation, thereby supporting prolonged exercise. In contrast, anaerobic metabolism rapidly generates energy through glycolysis, which results in lactate accumulation and leads to muscle acidification and fatigue [
10].
This study focused on three functionally distinct muscles: the longissimus dorsi, the rectus abdominis, and the diaphragm. The latissimus dorsi comprises a greater proportion of slow-twitch muscle fibers, which are well suited for oxidative metabolism and maintain muscle endurance [
11,
12]. The rectus abdominis contributes to core stability and affects muscle explosive power through fast-twitch muscle fibers [
4,
13]. The diaphragm, as the primary respiratory muscle, directly regulates oxygen intake and influences aerobic performance [
14].
Sexual dimorphism affects athletic performance through mechanisms such as hormone regulation, metabolic pathways, and muscle structure remodeling [
15]. Androgens promote muscle protein synthesis by activating the mTOR signaling pathway, thereby influencing muscle fiber composition and enhancing athletic performance in males [
16,
17]. Androgen receptors (ARs) in mesenchymal stem cells regulate the expression of IGF1, which promotes skeletal muscle protein synthesis and improves male athletic performance [
18]. In contrast, estrogens in females enhance mitochondrial biogenesis and fatty acid oxidation efficiency through the AMPK signaling pathway, thereby improving aerobic endurance [
19]. During post-exercise recovery, estrogen enhances antioxidant and anti-inflammatory responses by regulating
BDNF expression, thus accelerating muscle repair [
20].
Existing studies primarily focused on age-related changes and region-specific differences in the skeletal muscles of Kazakh horses of the same sex [
1]. However, few involved gender differences at the transcriptional level in muscle sites. In this study, although direct performance traits such as oxygen consumption and lactate threshold were not measured, RNA sequencing (RNA-seq) was employed to compare sex-specific gene expression profiles across three functionally distinct muscle groups: longissimus dorsi, rectus abdominis, and diaphragm muscles. By integrating molecular data with quantitative histology and metabolite composition analyses, these findings provide molecular insights into the mechanisms underlying sex differences in equine skeletal muscle growth.
2. Materials and Methods
2.1. Experimental Animals and Sample Collection
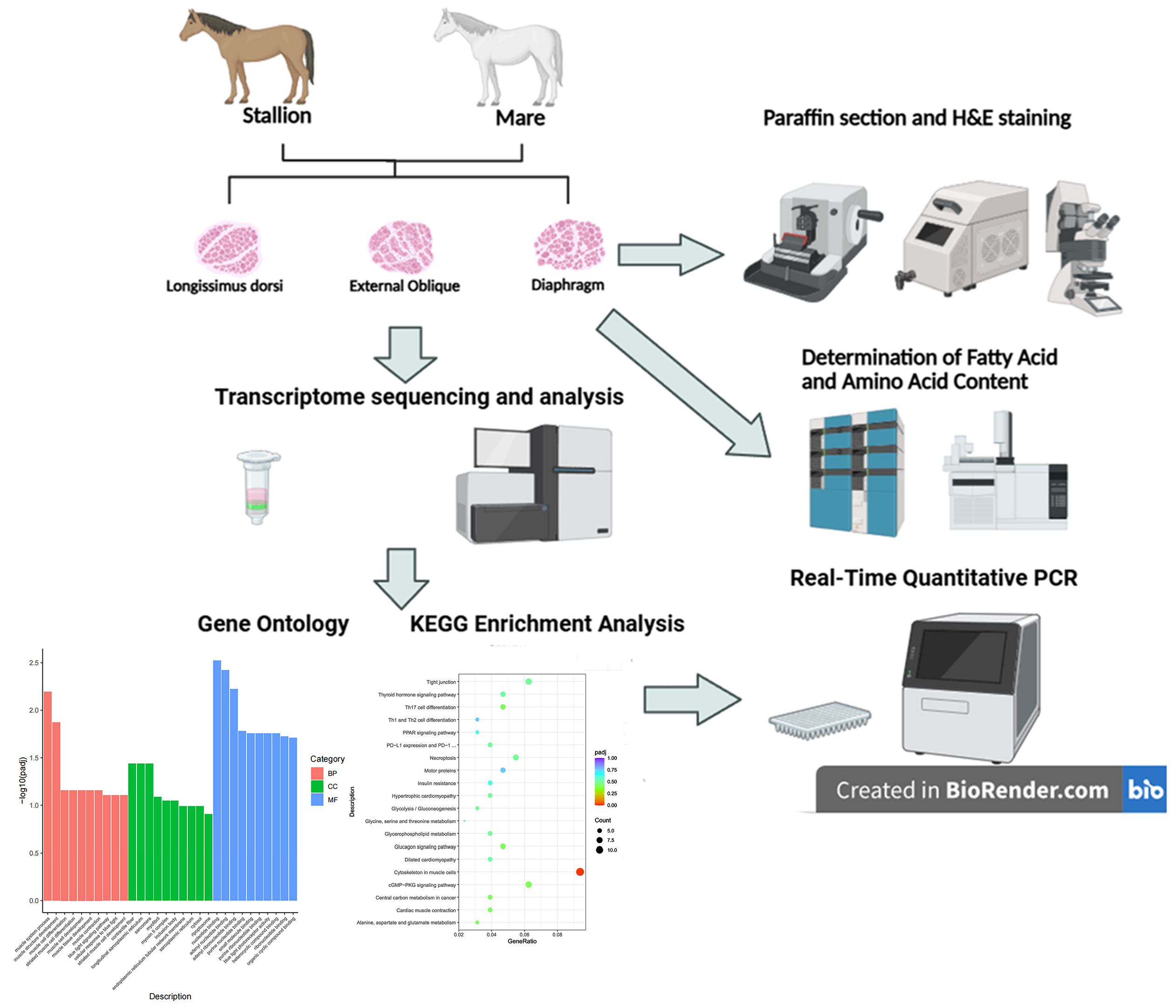
Eight clinically healthy Kazakh horses, consisting of four three-year-old stallions and four three-year-old mares, were selected from Tacheng, Xinjiang, China. All horses were raised under the same feeding conditions, provided with high-quality alfalfa hay and corn kernels, and had free access to water. Immediately after slaughter, muscle specimens were collected from each stallion and mare as follows: longissimus dorsi, 3 cm lateral to the transverse process of the 13th thoracic vertebra; rectus abdominis, 3 cm lateral to the external sheath of the linea alba; and diaphragm, at the rib attachment site near the costal angle. Full-thickness muscle samples with a depth of 3 cm were synchronously obtained. Each group comprised four biological replicates, with individual sample weights standardized at 30 g.
Figure 1 illustrates the experimental procedure: a portion of the samples was immediately flash-frozen in liquid nitrogen (−196 °C) for subsequent RNA sequencing and biochemical analyses (fatty acid and amino acid content determination), while another portion was quickly fixed in 4% paraformaldehyde (PAF) for paraffin section preparation.
2.2. Histochemical Analysis
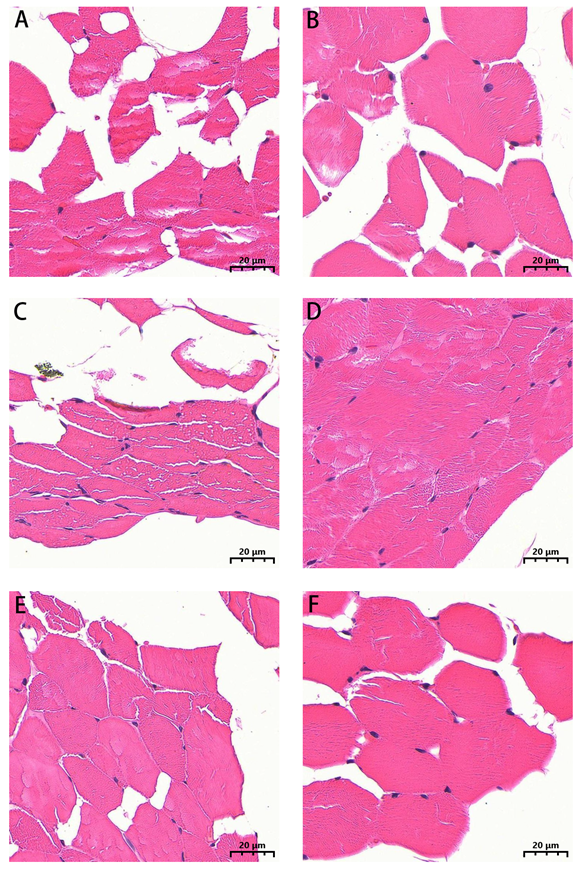
Muscle samples fixed in paraformaldehyde were subjected to dehydration using a gradient of ethanol (75%, 85%, 90%, 95%, and 100%). Samples were then cleared with xylene, embedded in paraffin, and sectioned into 5 μm cross-sections using a rotary microtome. After being stained with hematoxylin and eosin (H&E), the tissue slices were photographed using a light microscope (Eclipse E100 Nikon, Nikon Corporation, Tokyo, Japan) connected to a camera system.
2.3. Determination of Fatty Acid and Amino Acid Content
For fatty acid analysis, approximately 50 mg of the sample was homogenized. Then, 3 mL of n-hexane was added, and the mixture was shaken at 50 °C for 30 min. Subsequently, 3 mL of methanolic KOH solution (0.4 mol/L) was added and shaken at 50 °C for another 30 min for derivatization. Upon standing and cooling to room temperature, 1 mL of water was added and mixed thoroughly. The supernatant was collected after standing and diluted fivefold. From the diluted solution, 90 μL was taken, to which 10 μL of internal standard (methyl nonadecanoate, 125 μg/mL) was added, followed by analysis using gas chromatography-mass spectrometry (GC-MS, Agilent 7890B-5977B, Agilent Technologies, Santa Clara, CA, USA).
For amino acid analysis, 2 mL of 6 mol/L hydrochloric acid was added to the sample under nitrogen protection. Acid hydrolysis was performed at 110 °C for 24 h. Then, 100 μL of the hydrolysate was taken and evaporated to dryness at 40 °C under nitrogen using a nitrogen blower. The residue was reconstituted with 1 mL of water. For both mixed standards and test samples, 50 μL of solution was mixed with 50 μL of protein precipitant (10% sulfosalicylic acid containing NVL), vortexed, and centrifuged at 13,200 rpm under cooling conditions for 4 min. Next, 8 μL of the supernatant was mixed with 42 μL of borate buffer (pH 8.5), vortexed, and briefly centrifuged. Then, 20 μL of AQC derivatization reagent was added, vortexed, briefly centrifuged, and incubated at 55 °C for 15 min for derivatization. The resulting sample was cooled in a refrigerator, mixed thoroughly, and centrifuged again. A 50 µL of the supernatant was analyzed using an ultra-high-performance liquid chromatography-quadrupole ion trap tandem mass spectrometer (UHPLC-MS/MS, Waters ACQUITY UPLC I-Class/Xevo TQ-S, Waters Corp., Milford, MA, USA).
Quantitative data for fatty acids and amino acids were initially processed using Microsoft Excel. Independent-sample t-tests were then performed in SPSS 19.0. Data are presented as mean ± standard deviation (mean ± SD). Differences were considered highly significant at p < 0.01 and significant at p < 0.05.
2.4. Transcriptomic Sequencing
RNAs were extracted from the longissimus dorsi, rectus abdominis, and diaphragm tissues. After quality control, high-quality RNAs were purified, fragmented, and reverse-transcribed into complementary DNAs (cDNAs) to construct transcriptomic libraries. Sequencing was performed by Hangzhou Repugene Technology Co., Ltd. on the Illumina NovaSeq6000 platform (Repugene Technology, Hangzhou, China) [
21].
2.5. Bioinformatics Analysis
FastQC (fastqc_v0.11.8) was employed to assess the quality metrics of raw Illumina sequencing data. Adapter sequences and low-quality reads were filtered using fastp (fastp 0.23.1), yielding high-quality clean reads. Bismark (version 0.24.0) was subsequently applied to map the clean reads to the Equus Caballus reference genome (EquCab3.0). Reads successfully aligned to the reference genome were designated as target sequences for subsequent standardized and customized analyses [
22].
Filtering parameters included the elimination of reads containing adapter contamination, those with terminal base quality scores below 3, or ambiguous bases (N). A sliding window approach, with a four-base window and a quality threshold of 15, was applied to truncate reads once the average quality within the window dropped below the threshold. Reads shorter than 36 nt post-trimming or lacking valid pairing were excluded from further analysis.
2.6. GO and KEGG Enrichment Analyses
In order to annotate the differentially expressed mRNAs, GO and KEGG enrichment analyses were performed. KOBAS software (version 3.0) was used to assess the statistical enrichment of DEGs in KEGG pathways, while GOseq software (version 1.56.0) was adopted for GO functional analysis. Statistical significance for enrichment was set at p < 0.05.
2.7. Protein–Protein Interaction (PPI) Network Analysis
Based on the intersection of DEGs and known protein interaction pairs retrieved from the STRING database (
https://cn.string-db.org, accessed on 25 July 2025), a PPI network was constructed. Homologous protein interaction relationships were integrated into the network. Cytoscape (version 3.10.0) was employed for network visualization and analysis. Specifically, the “Betweenness-unDir” plugin within the CytoNCA module was utilized to identify key targets and visually analyze the results.
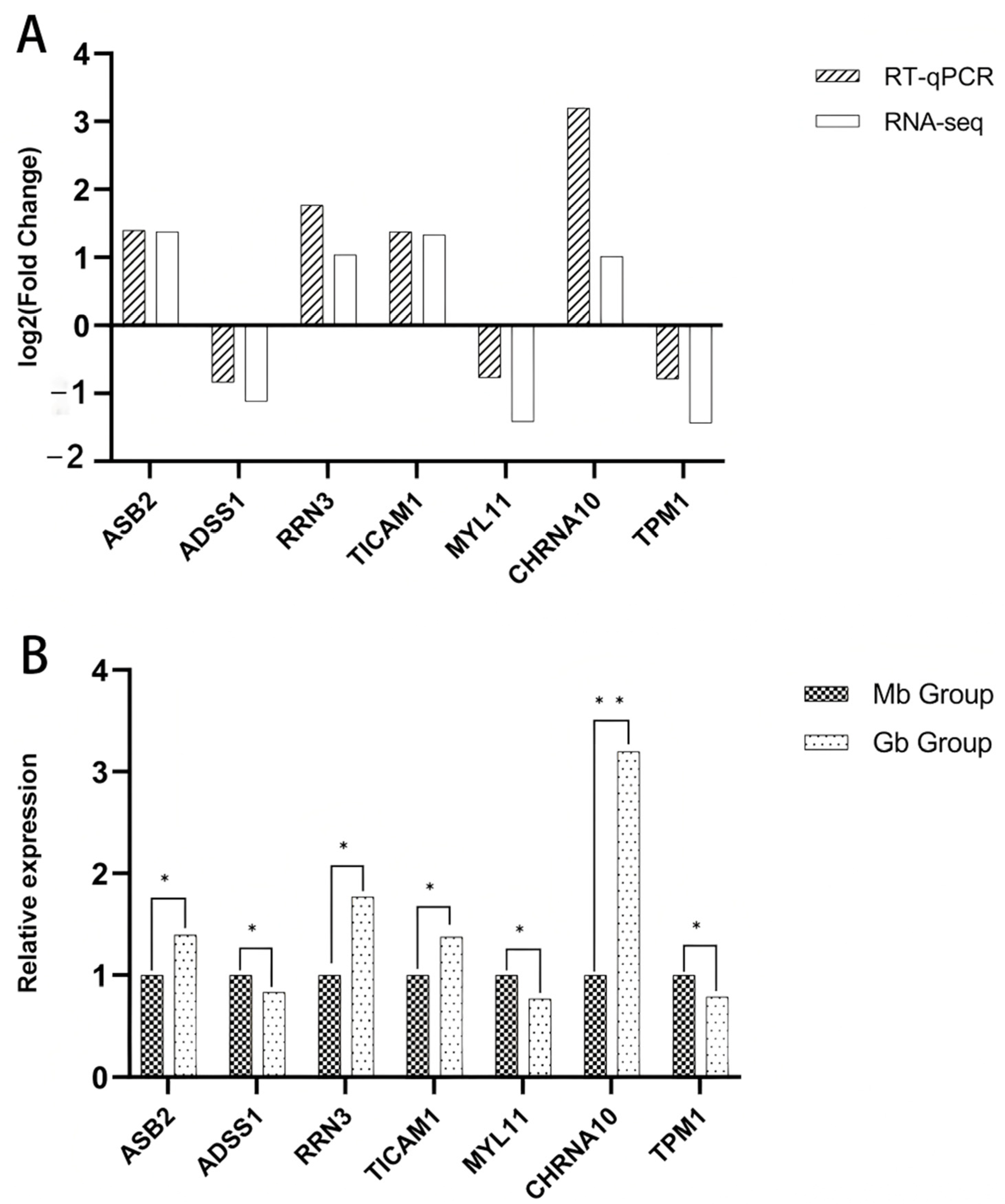
2.8. RT-qPCR Validation
In order to validate the expression of individual mRNAs, total RNA was reverse-transcribed into cDNA. RT-qPCR primer information is provided in
Table S1. RT-qPCR was conducted on a CFX Connect Fluorescence Quantitative PCR system (Bio-Rad Laboratories, Inc., Hercules, CA, USA) with three replicates per sample.
2.9. Ethical Statement
The experimental protocol and procedures were approved by the Animal Ethics Review Committee of Xinjiang Agricultural University (Approval No. 2023004).
4. Discussion
Skeletal muscle development in horses is regulated by multiple factors, including age and sex. Ren et al. [
1] investigated skeletal muscle development in Kazakh horses of varying ages and identified
BMP2,
MDH1, and
ATF3 as key genes involved in this process. Wang et al. [
2] found that
RYR3 and
MYH6 genes regulate fast and slow muscle function in stallion and mare Kazakh horses, potentially improving meat quality by altering muscle fiber composition. Despite these insights, studies specifically examining the effect of sex on equine skeletal muscle development remain limited [
8]. Therefore, this study selected eight three-year-old Kazakh horses (four stallions and four mares) and performed transcriptome sequencing analysis and histochemical analysis on samples from their longissimus dorsi, rectus abdominis, and diaphragm.
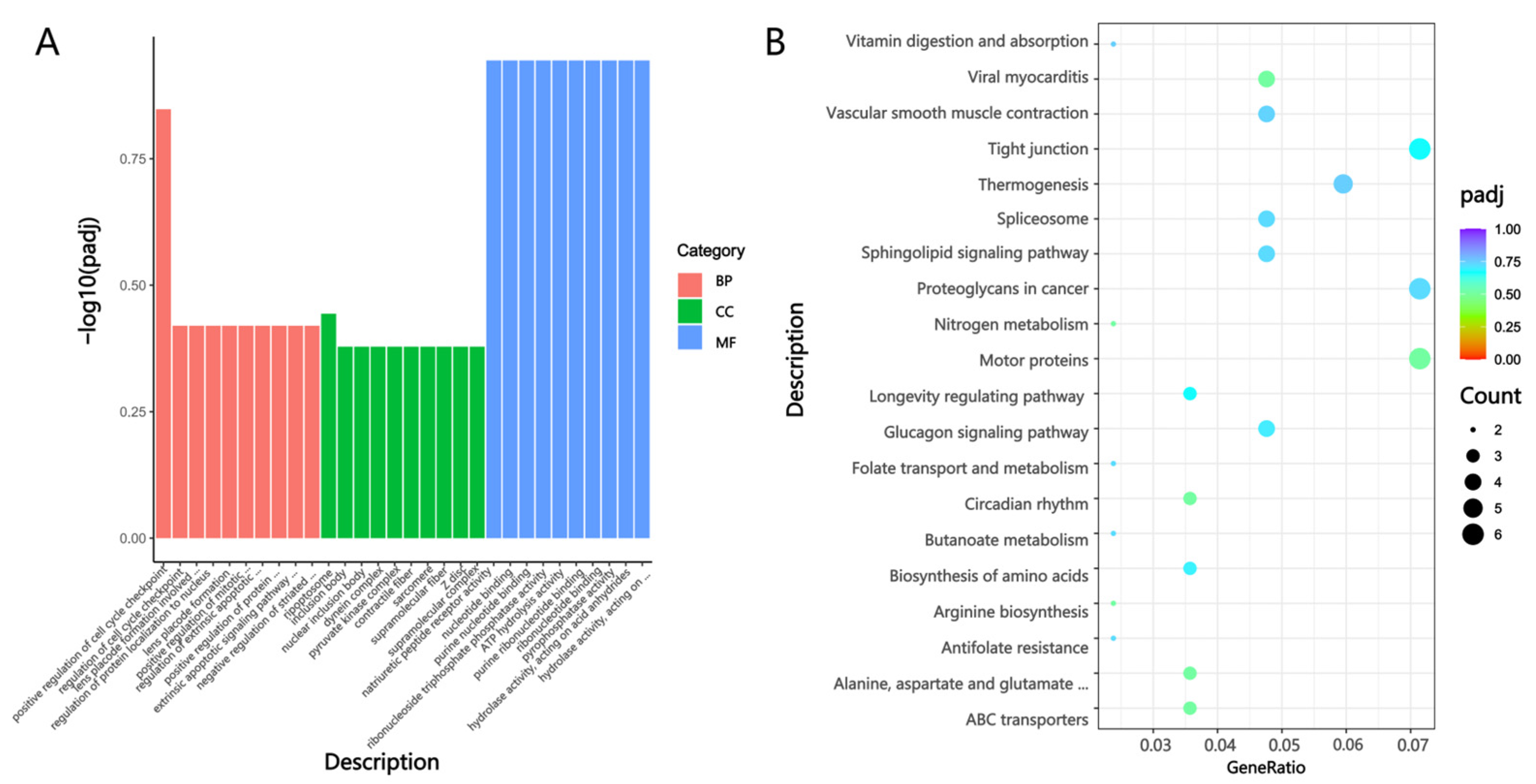
KEGG enrichment analysis revealed that DEGs were significantly enriched in pathways such as Cytoskeleton in muscle cells, the thyroid hormone signaling pathway, and the regulation of actin cytoskeleton. The cytoskeleton, primarily composed of microtubules, actin filaments, and intermediate filaments, plays an essential role in multiple fundamental cellular and BPs, such as cell migration, movement, division, and the establishment and maintenance of cellular and tissue structures [
23]. It forms a dynamic yet mechanically stable network of cytoskeletal filaments. Muscle contractile force is generated by actin and myosin filaments, among which the orderly arrangement of polar actin filaments is particularly crucial for striated muscle contraction [
24].
Jabre et al. [
25] reported that the cytoskeleton is anchored to the cell membrane, where surface-bound condensates exert and buffer mechanical forces, thereby conferring both stability and flexibility to the cytoskeletal structure. During skeletal muscle development,
TPM1 promotes sarcomere assembly by stabilizing actin filaments and regulates the differentiation of myocytes into functional fast-twitch (Type II) and slow-twitch (Type I) muscle fibers [
26]. Human studies indicate that the expression levels of
TPM1 directly influence the calcium sensitivity and contraction characteristics of muscle fibers [
27]. This study found that in the longissimus dorsi muscle group, the expression of
TPM1 was significantly higher in stallions than in mares, with marked enrichment in the cytoskeleton in muscle cells pathway. Muscle phenotype data further indicated that the average muscle fiber area in stallions exceeded that in mares. It has been reported that male animals generally exhibit larger average muscle fiber areas than females, which is consistent with the findings of this study. Sex hormones also play a pivotal role in maintaining skeletal muscle homeostasis. Among them, testosterone acts as a potent anabolic factor promoting protein synthesis and muscle regeneration [
28]. Therefore, it is hypothesized that muscle fiber thickness in horses may be sex-dependent.
MYL1, a member of the myosin light chain family, is essential for maintaining skeletal muscle function [
29]. Feng et al. [
30] discovered that aerobic and resistance exercise alleviate skeletal muscle atrophy in myocardial infarction (MI) models through the IGF-1/IGF-1R-PI3K/Akt signaling pathway by inhibiting apoptosis. Horses possess exceptional aerobic and muscular abilities, excelling in competitive sports. Murach KA et al. [
31] proposed a synergistic role for
TPM1 and
MYL1:
TPM1 exposes actin binding sites via calcium signaling, and
MYL1 enhances the myosin head’s motility, ensuring coordinated muscle contraction.
MYH3 is a member of the myosin family, which consists of two myosin heavy chains and four light chains [
32]. It functions as a motor protein in muscle contraction, cell migration, and cytokinesis. By hydrolyzing ATP,
MYH3 converts chemical energy into mechanical energy and contributes to muscle contraction, making it a critical component of muscle tissue [
33].
MYH3 impacts muscle development and regulates downstream protein phosphorylation via MAPK and TGF-β signaling pathways, thereby influencing muscle metabolism [
34]. In this study, diaphragm muscle expression of
MYL1 and
MYH3 was significantly higher in mares than in stallions, with both genes markedly enriched in the cytoskeleton in muscle cells pathway. Amino acid composition analysis revealed that glycine content in the diaphragm of mares significantly exceeded that of stallions. As a precursor of glutathione (GSH), glycine supplementation can indirectly enhance antioxidant capacity and reduce oxidative damage [
35]. Chemello et al. [
36] reported higher
MYH3 expression in the soleus muscle than in the extensor digitorum longus muscle. Estrogen has been proven to modulate muscle mass, function, and antioxidant capacity [
37]. Therefore, it is hypothesized that sex may influence the antioxidant capacity of equine muscle.
PPI network construction identified multiple DEGs, including
TPM1,
MYL1,
MYH3 and
PYGM.
PYGM catalyzes the degradation of glycogen to glucose-1-phosphate, providing energy by breaking down glycogen stored in muscle tissue [
38]. This study found that in the longissimus dorsi muscle group, the expression of
PYGM was significantly higher in stallions than in mares, suggesting superior energy-supplying capacity in stallions. Nam et al. [
39] examined the genomic structure and expression patterns of
PYGM in thoroughbred horses and found that
PYGM expression was highest in skeletal muscle compared to other tissues. Sun [
40] identified DEGs, including
TPM1,
MYL1,
MYH3 and
PYGM, co-expressed in the longissimus dorsi muscle of donkeys. These genes influence skeletal muscle development by modifying muscle fiber contraction and protein metabolism. This study demonstrated that the sex-specific differential expression of
TPM1,
MYL1,
MYH3 and
PYGM significantly impacts muscle structure, thereby providing a molecular-level explanation for observed sex-related differences. The differential expression of
TPM1,
MYL1 and
MYH3 suggests that the fiber type composition (i.e., the proportion of type I and type II fibers) may vary with Kazakh horse genders. However, given insufficient available data on fiber types in Kazakh horses, this study did not directly assess fiber type distribution. These transcriptomic findings imply potential sex-related differences in muscle fiber types, which warrant further validation through approaches such as immunohistochemistry of specific fiber types and single-fiber RNA sequencing.
In this study, the use of paraffin embedding and sectioning may introduce a certain degree of tissue shrinkage, potentially affecting the clarity of morphological observations. Although incomplete fibers were excluded during image analysis to ensure the reliability of quantitative data, future studies employing cryosectioning or alternative techniques that better preserve the native tissue architecture will provide further verification of these findings. Despite the lack of systematic multi-omics analyses, consistent patterns were observed between metabolic phenotypes and gene expression. For instance, sex-specific differences in fatty acid composition may reflect differential regulation of lipid metabolism genes. The elevated C18:2n6c content in stallion rectus abdominis may indicate an increased demand for glycine synthesis in the diaphragm. These variations may be associated with the expression of genes involved in muscle fiber remodeling. Future studies will integrate multiple data types to comprehensively analyze muscle fiber typing and multi-omics.