Synthesis, Characterization, and Screening Anticancer—Antibiofilm Activities of Theophylline Derivatives Containing CF3/OCF3 Moiety
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemistry
- General Synthesis of N(7)-Substituted Theophyllines (1a–1e)
- 1,3-Dimethyl-7-(2-(trifluoromethyl)benzyl)-3,7-dihydro-1H-purine-2,6-dione (1a)
- 1,3-Dimethyl-7-(3-(trifluoromethyl)benzyl)-3,7-dihydro-1H-purine-2,6-dione (1b)
- 1,3-Dimethyl-7-(4-(trifluoromethyl)benzyl)-3,7-dihydro-1H-purine-2,6-dione (1c)
- 1,3-Dimethyl-7-(3,5-bis(trifluoromethyl)benzyl)-3,7-dihydro-1H-purine-2,6-dione (1d)
- 1,3-Dimethyl-7-(4-(trifluoromethoxy)benzyl)-3,7-dihydro-1H-purine-2,6-dione (1e)
2.2. Molecular Docking Method
2.3. Evaluation of Anticancer Activity and Selectivity Index
2.4. Gene Expression Analysis by RT-qPCR
2.5. Quantification of Apoptosis-Related Proteins by ELISA
2.6. Determination of Antimicrobial Activity
2.7. Reduction in Biofilm Formation
3. Results and Discussion
3.1. Preparation and Characterization of N(7)-Substituted Theophyllines (1a–1e)
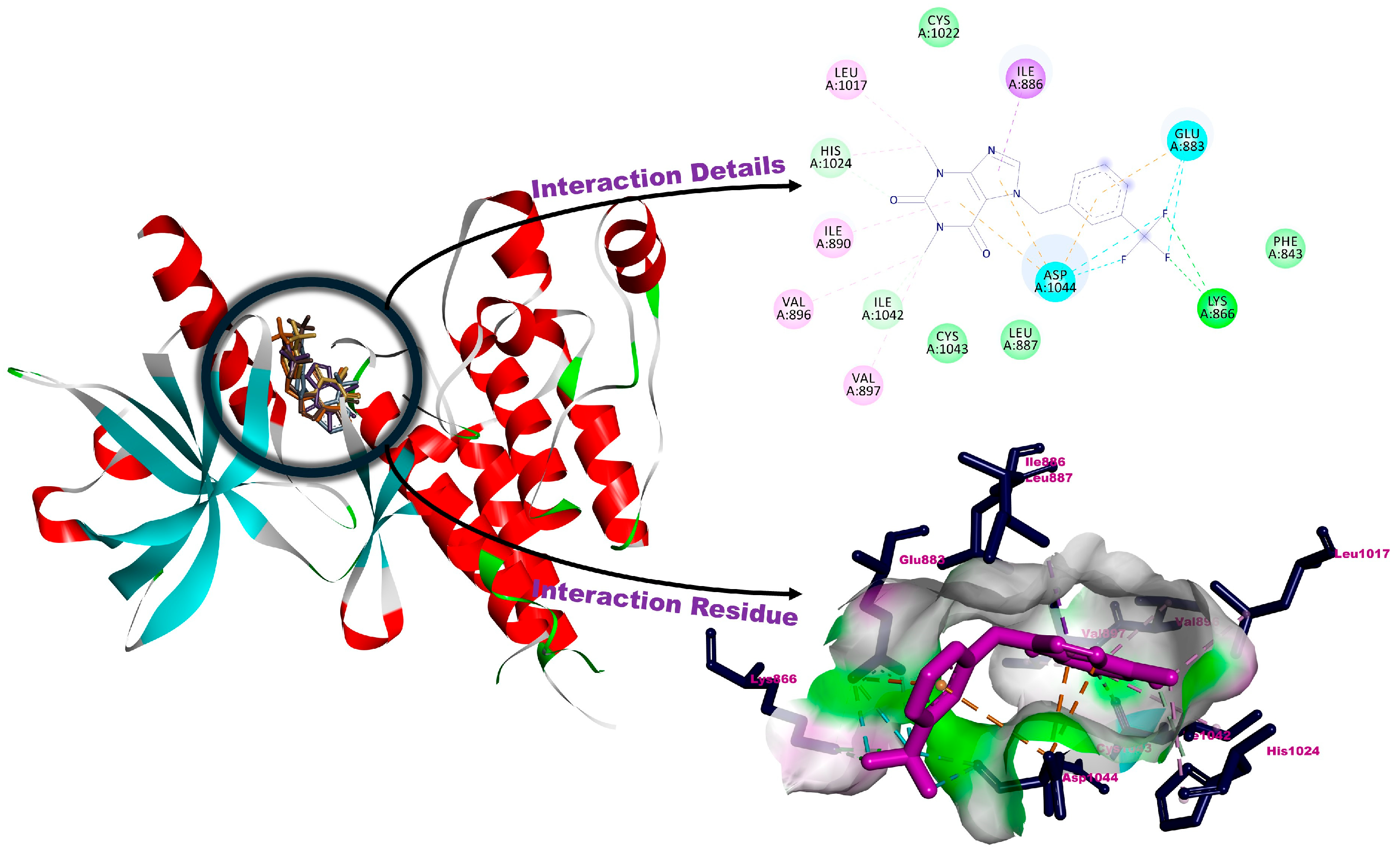
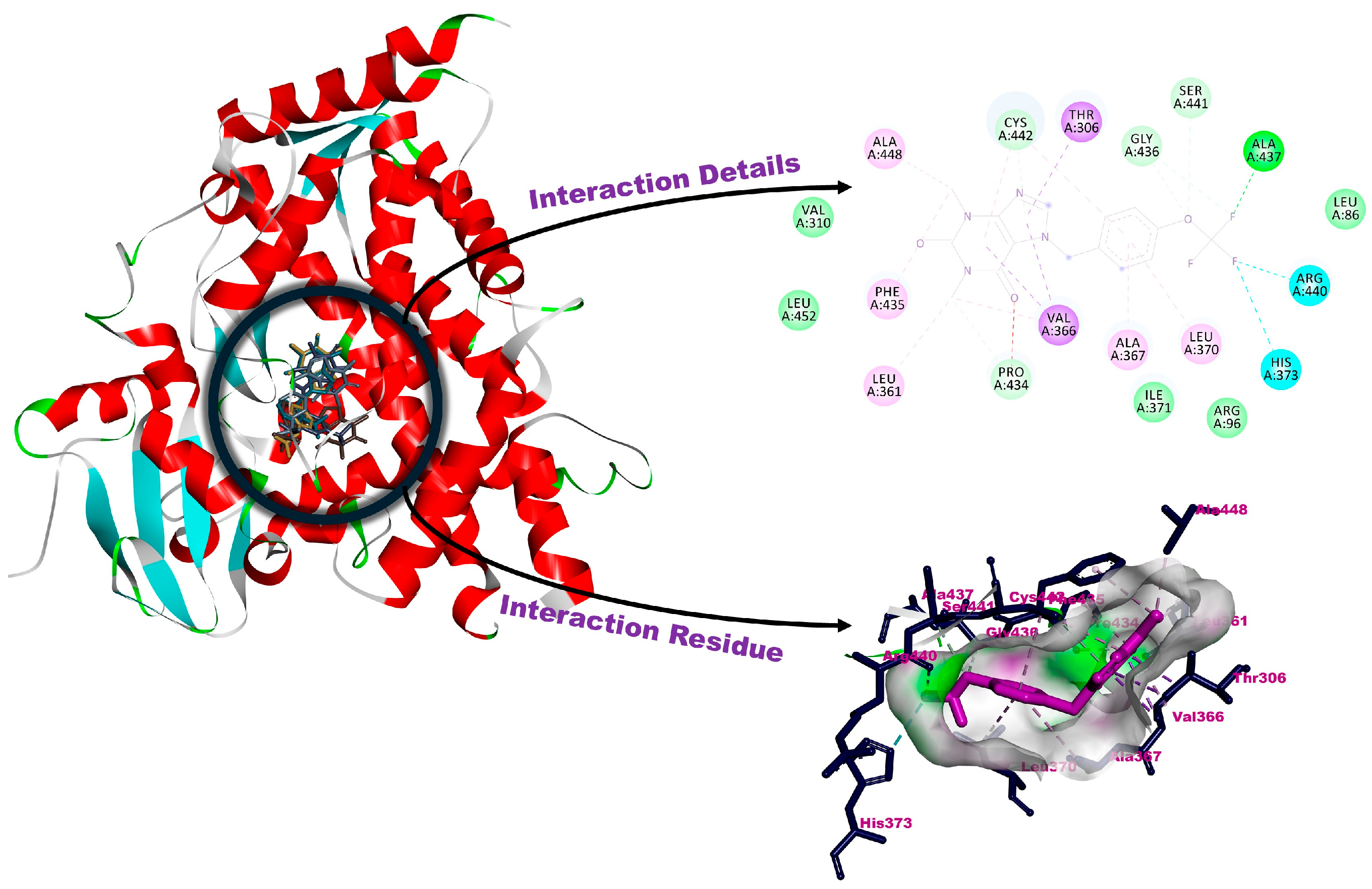
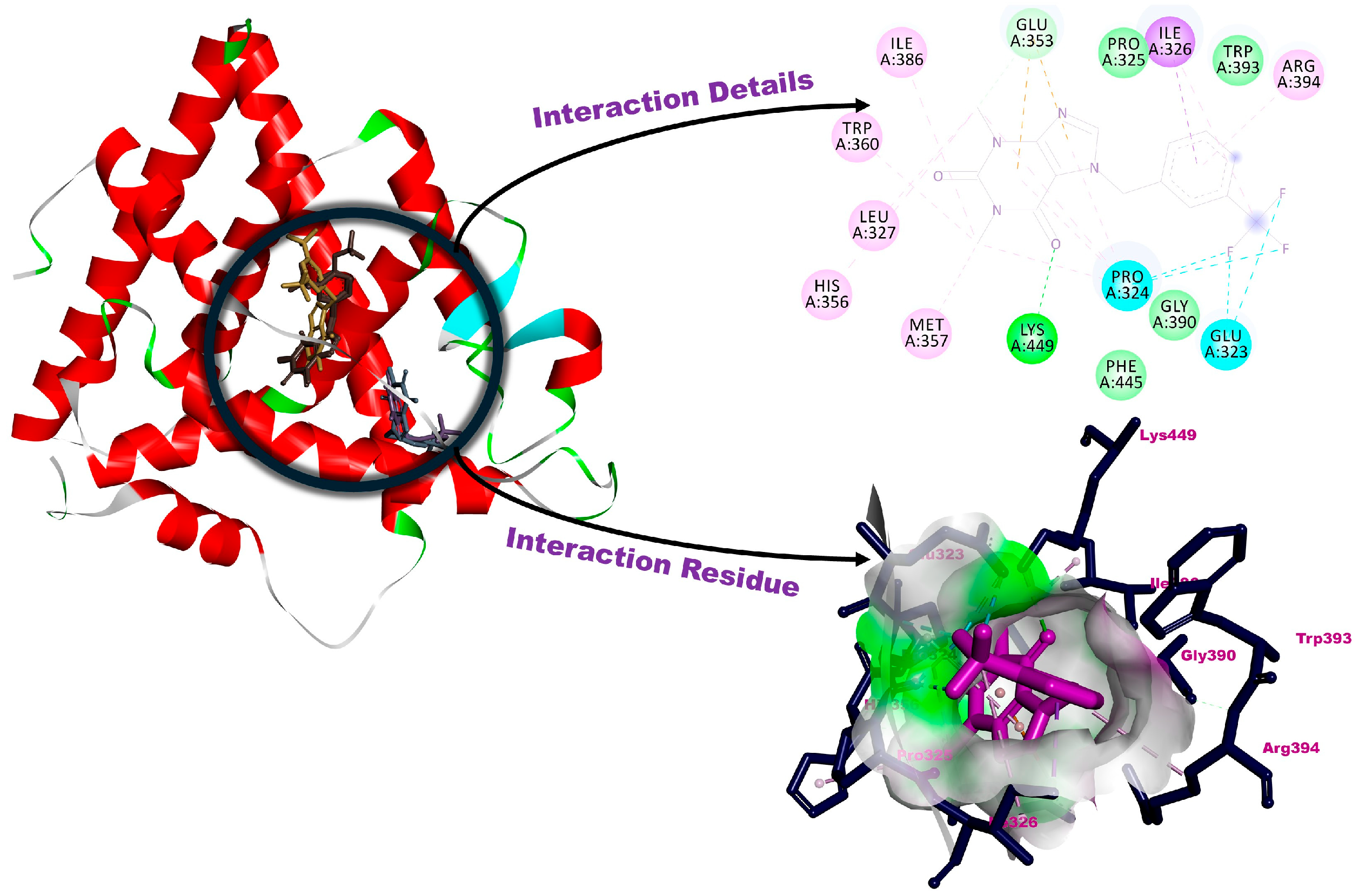
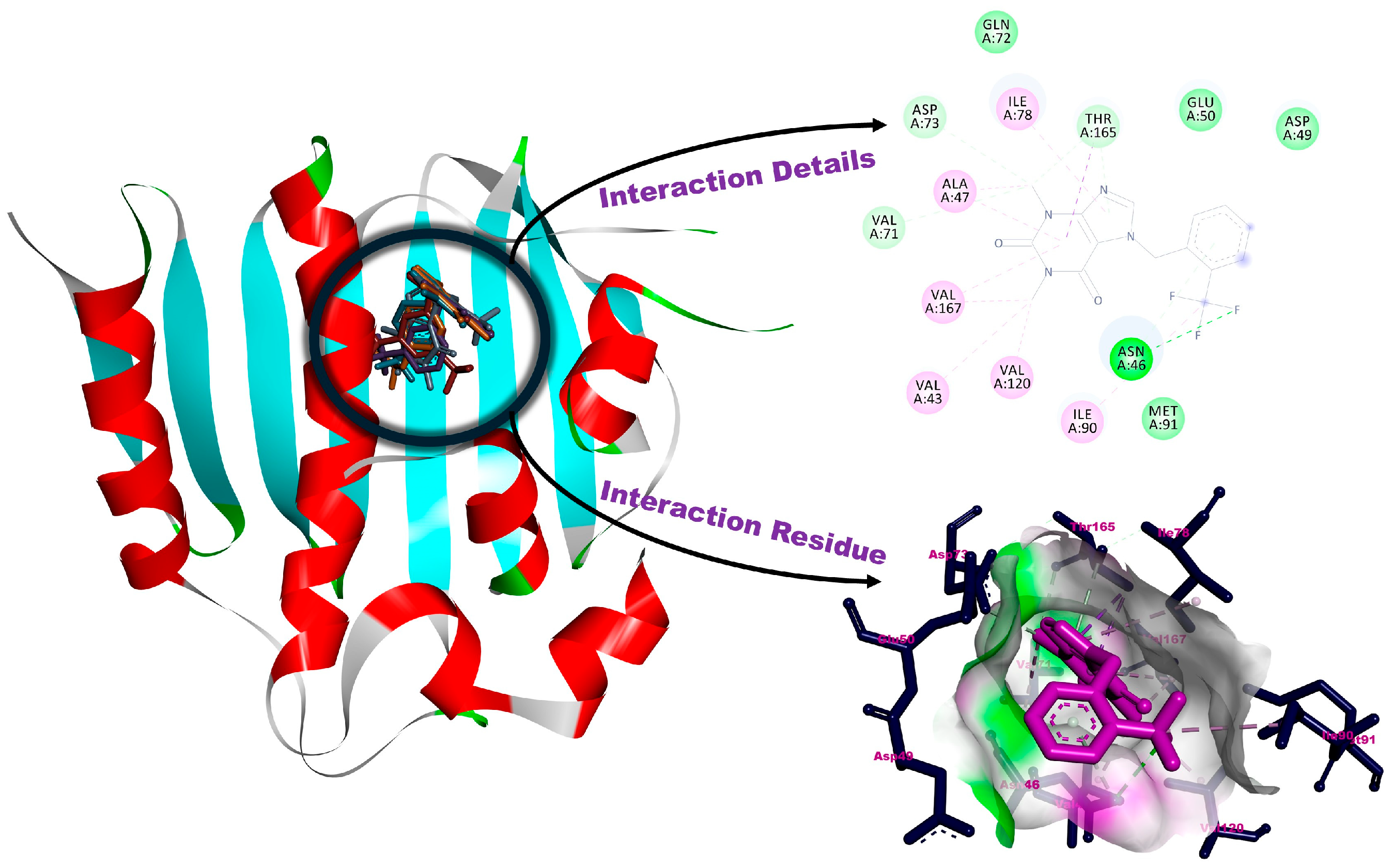
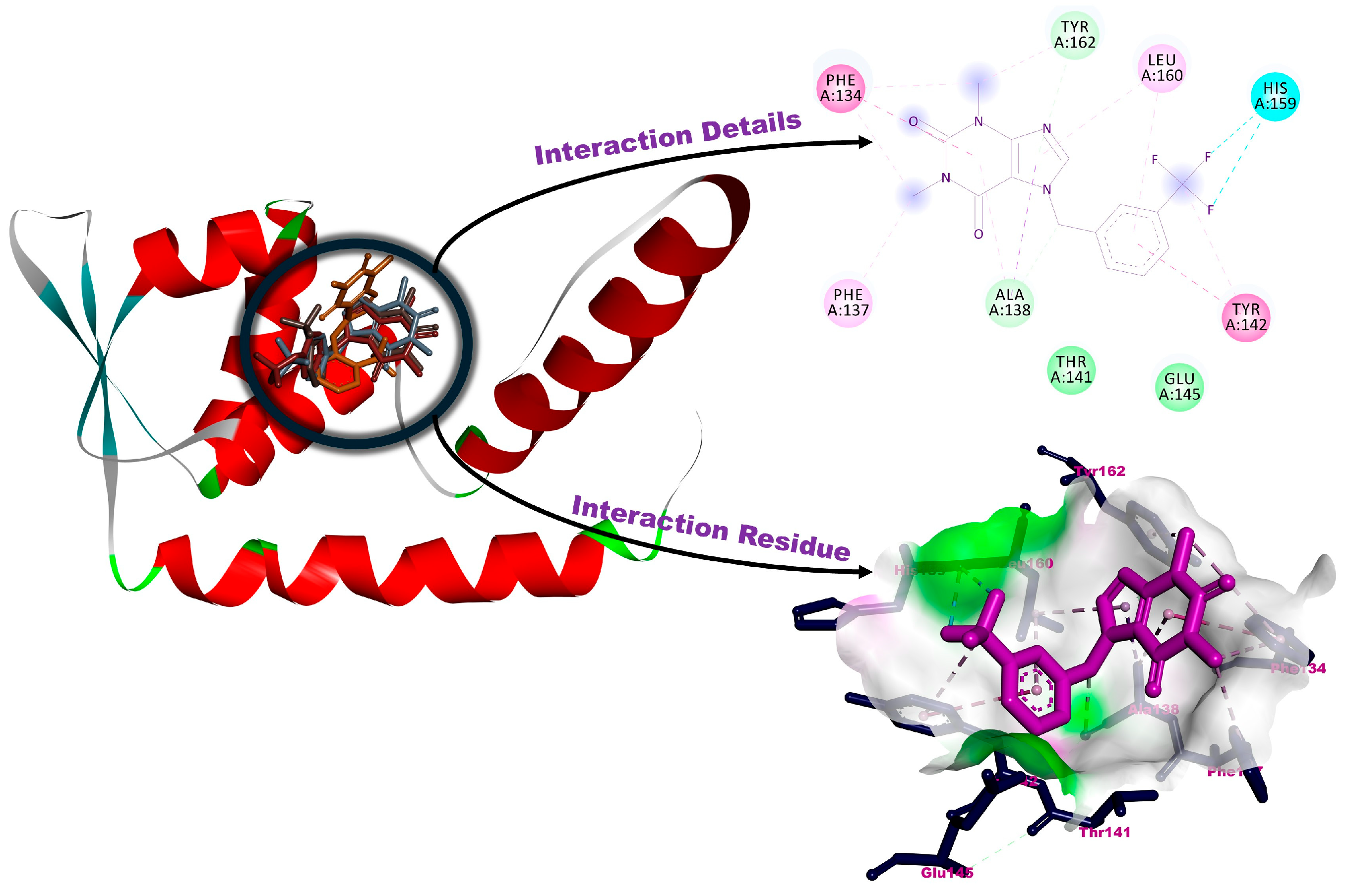
3.2. Molecular Docking
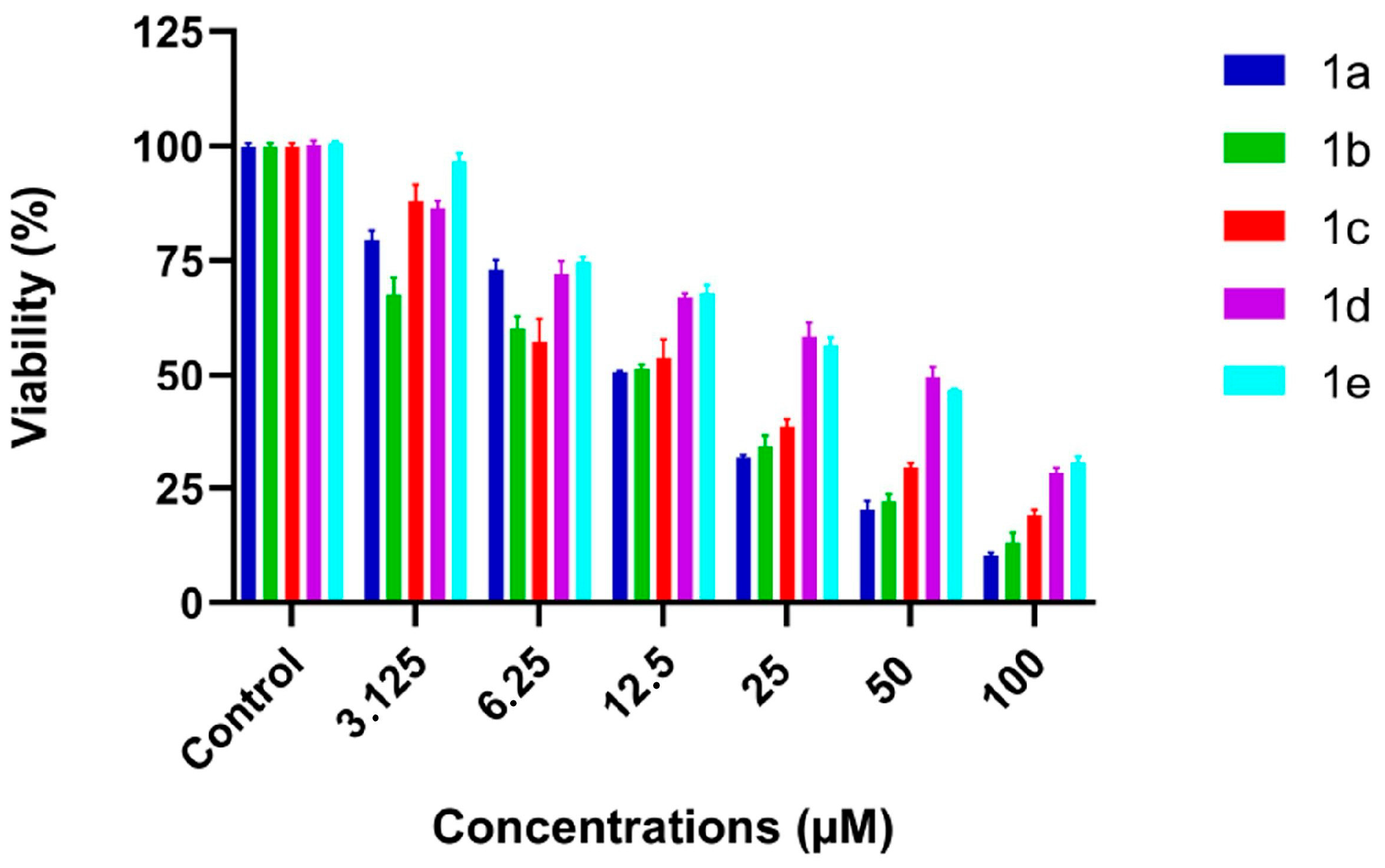
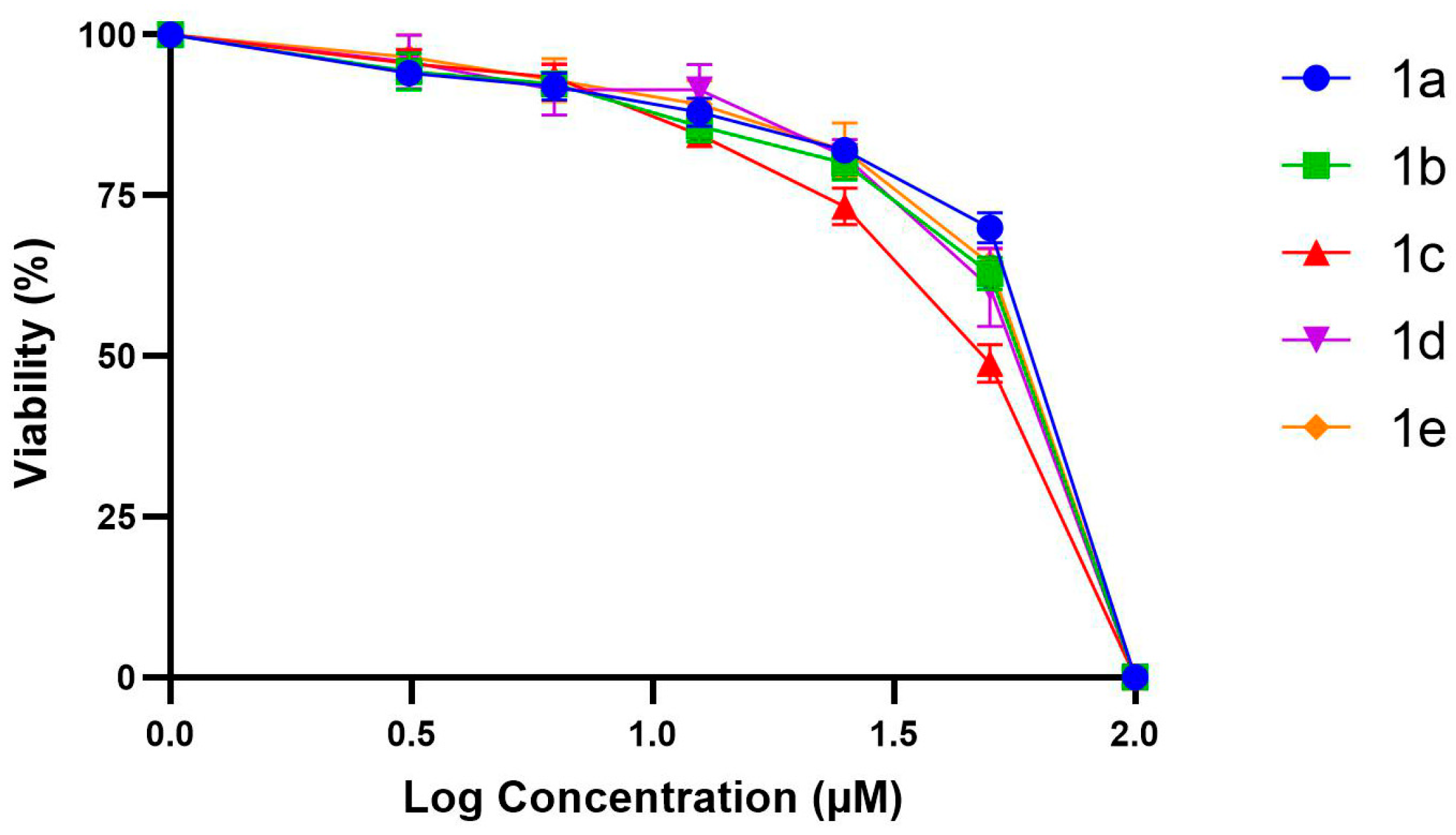
3.3. Anticancer Activity and Selectivity of Substituted Theophylline Derivatives
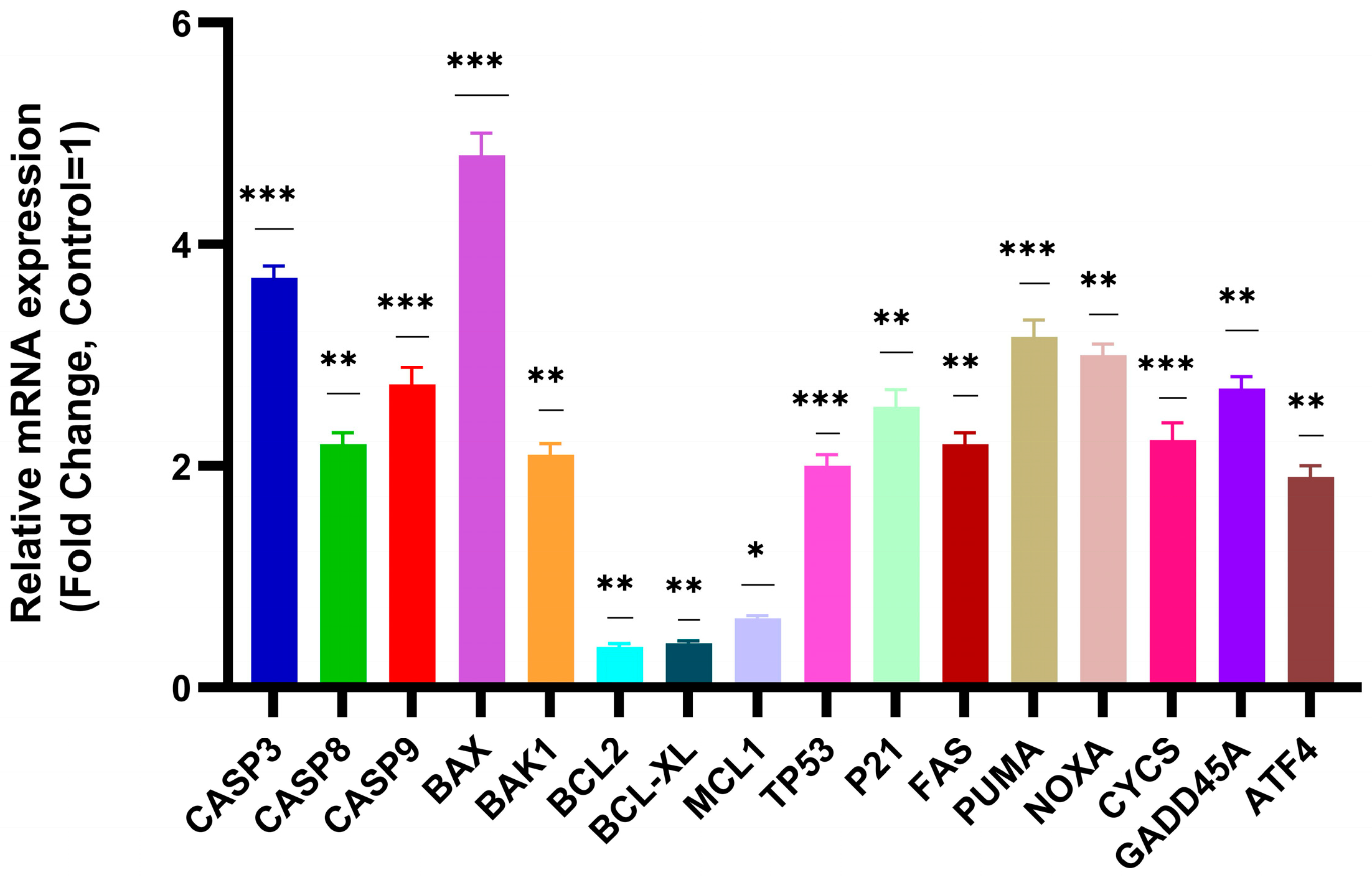
3.4. Apoptosis-Related Gene Expression Analysis by RT-qPCR
3.5. Protein-Level Characterization of Apoptotic Pathways
3.6. Antimicrobial Activity
3.7. Antibiofilm Activity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zuccolo, M.; Arrighetti, N.; Perego, P.; Colombo, D. Recent Progresses in Conjugation with Bioactive Ligands to Improve the Anticancer Activity of Platinum Compounds. Curr. Med. Chem. 2022, 29, 2566–2601. [Google Scholar] [CrossRef] [PubMed]
- Kangra, K.; Kakkar, S.; Mittal, V.; Kumar, V.; Aggarwal, N.; Chopra, H.; Malik, T.; Garg, V. Incredible use of plant-derived bioactives as anticancer agents. RSC Adv. 2025, 15, 1721–1746. [Google Scholar] [CrossRef]
- Singla, P.; Luxami, V.; Paul, K. Triazine as a promising scaffold for its versatile biological behavior. Eur. J. Med. Chem. 2015, 102, 39–57. [Google Scholar] [CrossRef]
- Kumar, A.; Singh, A.K.; Singh, H.; Vijayan, V.; Kumar, D.; Naik, J.; Thareja, S.; Yadav, J.P.; Pathak, P.; Grishina, M.; et al. Nitrogen Containing Heterocycles as Anticancer Agents: A Medicinal Chemistry Perspective. Pharmaceuticals 2023, 16, 299. [Google Scholar] [CrossRef] [PubMed]
- Tugrak, M.; Gul, H.I.; Bandow, K.; Sakagami, H.; Gulcin, I.; Ozkay, Y.; Supuran, C.T. Synthesis and biological evaluation of some new mono Mannich bases with piperazines as possible anticancer agents and carbonic anhydrase inhibitors. Bioorg. Chem. 2019, 90, 103095. [Google Scholar] [CrossRef]
- Belhi, Z.; Karci, H.; Dündar, M.; Gürbüz, N.; Özdemir, İ.; Koç, A.; Özdemir, İ. Novel benzimidazolium salts and their silver(I)-N-heterocyclic carbene complexes: Synthesis, characterization and their biological properties. J. Coord. Chem. 2023, 76, 120–133. [Google Scholar] [CrossRef]
- Kabir, E.; Uzzaman, M. A review on biological and medicinal impact of heterocyclic compounds. Results Chem. 2022, 4, 100606. [Google Scholar] [CrossRef]
- Habib, A.; Iqbal, M.A.; Bhatti, H.N.; Kamal, A.; Kamal, S. Synthesis of alkyl/aryl linked binuclear silver(I)-N-Heterocyclic carbene complexes and evaluation of their antimicrobial, hemolytic and thrombolytic potential. Inorg. Chem. 2020, 111, 107670. [Google Scholar] [CrossRef]
- Aatif, M.; Raza, M.A.; Javed, K.; Nashre-ul-Islam, S.M.; Farhan, M.; Alam, M.W. Potential Nitrogen-Based Heterocyclic Compounds for Treating Infectious Diseases: A Literature Review. Antibiotics 2022, 11, 1750. [Google Scholar] [CrossRef] [PubMed]
- Abd El-Karim, S.S.; Anwar, M.M.; Syam, Y.M.; Awad, H.M.; El-Dein, A.N.; El-Ashrey, M.K.; Alkahtani, H.M.; Abdelwahed, S.H. New Benzofuran–Pyrazole-Based Compounds as Promising Antimicrobial Agents: Design, Synthesis, DNA Gyrase B Inhibition, and In Silico Studies. Pharmaceuticals 2024, 17, 1664. [Google Scholar] [CrossRef]
- Düşünceli, S.D.; Ayaz, D.; Üstün, E.; Günal, S.; Özdemir, N.; Dinçer, M.; Özdemir, İ. Synthesis, antimicrobial properties, and theoreticalanalysis of benzimidazole-2-ylidene silver(I) complexes. J. Coord. Chem. 2020, 73, 1967–1986. [Google Scholar] [CrossRef]
- Özdemir, İ.; Demir, S.; Günal, S.; Özdemir, İ.; Arıcı, C.; Ülkü, D. Synthesis, characterization and antimicrobial activity of new silver complexes with N-heterocyclic carbene ligands. Inorg. Chim. Acta 2010, 363, 3803–3808. [Google Scholar] [CrossRef]
- Li, S.; Ren, Y.; He, Q.; Wei, Y.; Du, H. Synthesis of novel purine derivatives: Antiplateletaggregation activity evaluation and 3D-QSAR analysis. J. Heterocycl. Chem. 2022, 59, 2016–2024. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, H.; Wang, H.; Zheng, Z.; Meng, Z.; Xu, Z.; Li, J.; Xue, M. Design, Synthesis, and Biological Activity Studies of Istradefylline Derivatives Based on Adenine as A2A Receptor Antagonists. ACS Omega 2021, 6, 4386–4394. [Google Scholar] [CrossRef]
- Managutti, P.B.; Mangasuli, S.N.; Malaganvi, S.S. Synthesis, crystal structure, electronic structure, and anti-tubercular properties of two new coumarin derivatives bearing theophylline moiety. J. Mol.Struc. 2023, 1277, 134888. [Google Scholar] [CrossRef]
- Voynikov, Y.; Valcheva, V.; Momekov, G.; Peikov, P.; Stavrakov, G. Theophylline-7-acetic acid derivatives with amino acids as anti-tuberculosis agents. Bioorganic Med. Chem. Lett. 2014, 24, 3043–3045. [Google Scholar] [CrossRef]
- Horrigan, L.A.; Kelly, J.P.; Connor, T.J. Immunomodulatory effects of caffeine: Friend or foe? Pharmacol. Ther. 2006, 111, 877–892. [Google Scholar] [CrossRef] [PubMed]
- Zablocki, J.; Kalla, R.; Perry, T.; Palle, V.; Varkhedkar, V.; Xiao, D.; Piscopio, A.; Maa, T.; Gimbel, A.; Hao, J.; et al. The discovery of a selective, high affinity A2B adenosine receptor antagonist for the potential treatment of asthma. Bioorganic Med. Chem. Lett. 2005, 15, 609–612. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.J.; Wu, B.N.; Lo, Y.C.; An, L.M.; Dai, Z.K.; Lin, Y.T.; Tang, C.S.; Chen, I.J. A xanthine-based epithelium-dependent airway relaxant KMUP-3 (7-[2-[4-(4-nitrobenzene)piperazinyl]ethyl]-1,3-dimethylxanthine) increases respiratory performance and protects against tumor necrosis factor-alpha-induced tracheal contraction, involving nitric oxide release and expression of cGMP and protein kinase G. J. Pharmacol. Exp. Ther. 2006, 316, 709–717. [Google Scholar]
- Purser, S.; Moore, P.R.; Swallow, S.; Gouverneur, V. Fluorine in medicinal chemistry. Chem. Soc. Rev. 2008, 37, 320–330. [Google Scholar] [CrossRef]
- Sheppard, W.A. α-Fluorinated Ethers. I. Aryl Fluoroalkyl Ethers1. J. Org. Chem. 1964, 29, 1–11. [Google Scholar] [CrossRef]
- Aldrich, P.E.; Sheppard, W.A. α-Fluorinated Ethers. II. Alkyl Fluoroalkyl Ethers1. J. Org. Chem. 1964, 29, 11–15. [Google Scholar] [CrossRef]
- C.-Muniz, S.; Anthony, R.; Niemann, S.; Alffenaar, J.C. New Approaches and Therapeutic Options for Mycobacterium tuberculosis in a Dormant State. Clin. Microbiol. Rev. 2017, 31, 10–1128. [Google Scholar]
- Skripconoka, V.; Danilovits, M.; Pehme, L.; Tomson, T.; Skenders, G.; Kummik, T.; Cirule, A.; Leimane, V.; Kurve, A.; Levina, K.; et al. Delamanid improves outcomes and reduces mortality in multidrug-resistant tuberculosis. Eur. Respir. J. 2013, 41, 1393–1400. [Google Scholar] [CrossRef] [PubMed]
- Mathew, S.J.; Manji, H.K.; Charney, D.S. Novel Drugs and Therapeutic Targets for Severe Mood Disorders. Neuropsychopharmacology 2008, 33, 2080–2092. [Google Scholar] [CrossRef] [PubMed]
- Blumberg, D.; Doron, S.; Bitton, S. Effect Of Triflumuron On Two Species Of Nitidulid Beetles, Carpophilus Hemipterus And Urophor Us Humeralis. Phytoparasitica 1985, 13, 9–19. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, A.F.; Wang, W.X.; Zhang, Y.; Gao, T.C. Baseline sensitivity and efficacy of thifluzamide in Rhizoctoniasolani. Ann. Appl. Biol. 2012, 161, 247–254. [Google Scholar] [CrossRef]
- Whipker, B.E.; McCall, I.; Gibson, J.L.; Cavins, T.J. Efficacy Of Flurprimidol (Topflor) On Bedding Plants. Acta Hortic. 2003, 624, 413. [Google Scholar] [CrossRef]
- Jia, Y.; Yang, D.; Wang, W.; Hu, K.; Yan, M.; Zhang, L.; Gao, L.; Lu, Y. Recent advances in pharmaceutical cocrystals of theophylline. Nat. Prod. Bioprospect. 2024, 14, 53. [Google Scholar] [CrossRef]
- Slotkin, T.A.; Seidler, F.J. Antimitotic and cytotoxic effects of theophylline in MDA-MB-231 human breast cancer cells. Breast Cancer Res. Treat. 2000, 64, 259–267. [Google Scholar] [CrossRef]
- Wu, Z.; Liang, S.; Hu, S.; Wang, Y.; Mao, L.F.; Hou, X.; Yang, J. Synthesis, activity exploration, molecular docking, and affinity assessment of theophylline derivatives as inhibitors targeting IDO1. Heterocyc. Commun. 2025, 31, 20250180. [Google Scholar] [CrossRef]
- Gordon, A.T.; Abosede, O.O.; Ntsimango, S.; Hosten, E.C.; Myeza, N.; van Eyk, A.; Harmse, L.; Ogunlaja, A.S. Synthesis and anticancer evaluation of copper(II)- and manganese(II)- theophylline mixed ligand complexes. Polyhedron 2022, 214, 115649. [Google Scholar] [CrossRef]
- Ferreira, L.G.; Dos Santos, R.N.; Oliva, G.; Andricopulo, A.D. Molecular Docking and Structure-Based Drug Design Strategies. Molecules 2015, 20, 13384–13421. [Google Scholar] [CrossRef]
- Liu, X.J.; Zhao, H.C.; Hou, S.J.; Zhang, H.J.; Cheng, L.; Yuan, S.; Chen, S.W. Recent development of multi-target VEGFR-2 inhibitors for the cancer therapy. Bioorganic Chem. 2023, 133, 106425. [Google Scholar] [CrossRef]
- Yoshimoto, F.K.; Auchus, R.J. The diverse chemistry of cytochrome P450 17A1 (P450c17, CYP17A1). J. Steroid Biochem. Mol. Biol. 2015, 151, 52–65. [Google Scholar] [CrossRef]
- Duffy, M.J. Estrogen Receptors: Role in Breast Cancer. Crit. Rev. Clin. Lab. Sci. 2006, 43, 325–347. [Google Scholar] [CrossRef]
- Ferrara, F.; Castagna, T.; Pantolini, B.; Campanardi, M.C.; Roperti, M.; Grotto, A.; Fattori, M.; Maso, L.D.; Carrara, F.; Zambarbieri, G.; et al. The challenge of antimicrobial resistance (AMR): Current status and future prospects. Naunyn Schmiedebergs Arch. Pharmacol. 2024, 397, 9603–9615. [Google Scholar] [CrossRef]
- Samrot, A.V.; Mohamed, A.A.; Faradjeva, E.; Jie, L.S.; Sze, C.H.; Arif, A.; Sean, T.C.; Michael, E.N.; Mun, C.Y.; Qi, N.X.; et al. Mechanisms and Impact of Biofilms and Targeting of Biofilms Using Bioactive Compounds—A Review. Medicina 2021, 57, 839. [Google Scholar] [CrossRef] [PubMed]
- Cozzarelli, N.R. DNA Gyrase and the Supercoiling of DNA. Science 1980, 207, 953–960. [Google Scholar] [CrossRef] [PubMed]
- Painter, K.L.; Krishna, A.; Wigneshweraraj, S.; Edwards, A.M. What role does the quorum-sensing accessory gene regulator system play during Staphylococcus aureus bacteremia? Trends Microbiol. 2014, 22, 676–685. [Google Scholar] [CrossRef]
- Ginger, R.D.; Samour, C.M. Trifluoroalkyl, fluorobenzyl, pentafluorobenzyl, fluorobenzenesulfonyl, and pentafluorobenzenesulfonyl theophyllines United States. Patent. Patent No: US3900474, 19 August 1975. [Google Scholar]
- Bertrand, B.; Stefan, L.; Pirrotta, M.; Monchaud, D.; Bodio, E.; Richard, P.; Le Gendre, P.; Warmerdam, E.; de Jager, M.H.; Groothuis, G.M.M.; et al. Caffeine-Based Gold(I) N-Heterocyclic Carbenes as Possible Anticancer Agents: Synthesis and Biological Properties. Inorg. Chem. 2014, 53, 2296–2303. [Google Scholar] [CrossRef] [PubMed]
- Neese, F. The ORCA Program System. Wires Comput. Mol. Sci. 2012, 2, 73–78. [Google Scholar] [CrossRef]
- Neese, F. Software update: The ORCA program system, version 4.0. Wires Comput. Mol. Sci. 2018, 8, e1327. [Google Scholar] [CrossRef]
- Miyazaki, Y.; Matsunaga, S.; Tang, J.; Maeda, Y.; Nakano, M.; Philippe, R.J.; Shibahara, M.; Liu, W.; Sato, H.; Wang, L.; et al. Novel 4-amino-furo[2,3-d]pyrimidines as Tie-2 and VEGFR2 dual inhibitors. Bioorganic Med. Chem. Lett. 2005, 15, 2203–2207. [Google Scholar] [CrossRef]
- DeVore, N.M.; Scott, E.E. Structures of cytochrome P450 17A1 with prostate cancer drugs abiraterone and TOK-001. Nature 2012, 482, 116–119. [Google Scholar] [CrossRef] [PubMed]
- Shiau, A.K.; Barstad, D.; Loria, P.M.; Cheng, L.; Kushner, P.J.; Agard, D.A.; Greene, G.L. The Structural Basis of Estrogen Receptor/Coactivator Recognition and the Antagonism of This Interaction by Tamoxifen. Cell 1998, 95, 927–937. [Google Scholar] [CrossRef] [PubMed]
- Lafitte, D.; Lamour, V.; Tsvetkov, P.O.; Makarov, A.A.; Klich, M.; Deprez, P.; Moras, D.; Briand, C.; Gilli, R. DNA Gyrase Interaction with Coumarin-Based Inhibitors: The Role of the Hydroxybenzoate Isopentenyl Moiety and the 5′-Methyl Group of the Noviose. Biochemistry 2002, 41, 7217–7223. [Google Scholar] [CrossRef]
- Liu, Y.; Manna, A.C.; Pan, C.H.; Kriksunov, I.A.; Thiel, D.J.; Cheung, A.L.; Zhang, G. Structural and function analyses of the global regulatory protein SarA from Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 2006, 103, 2392–2397. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. Software News and Updates Auto Dock4 and Auto Dock Tools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Cosconati, S.; Forli, S.; Perryman, A.L.; Harris, R.; Goodsell, D.S.; Olson, A.J. Virtual screening with AutoDock: Theory and practice. Expert. Opin. Drug Discov. 2010, 5, 597–607. [Google Scholar] [CrossRef]
- Türker, D.; Üstün, E.; Günal, S.; Yıldız, H.; Düşünceli, S.D.; Özdemir, İ. Cyanopropyl functionalized benzimidazolium salts and theirsilver N-heterocyclic carbene complexes: Synthesis, antimicrobial activity, and theoretical analysis. Arch. Pharm. 2022, 355, e2200041. [Google Scholar] [CrossRef]
- Tutar, U.; Çelik, C.; Üstün, E.; Özdemir, N.; Şahin, N.; Sémeril, D.; Gürbüz, N.; Özdemir, İ. Benzimidazol-2-ylidene Silver Complexes: Synthesis, Characterization, Antimicrobial and Antibiofilm Activities, Molecular Docking and Theoretical Investigations. Inorganics 2023, 11, 385. [Google Scholar] [CrossRef]
- Çalışkan, R.; Sarı, S.P.; Altınbaşak, B.B.; Dinç, H.Ö.; Balekoğlu, A.; Issa, G.; Mayda, P.Y. Bioactive Components and Antioxidant and Antimicrobial Activities of Rhus coriaria, a Sumac Species found in Turkey. Bezmiâlem Sci. 2022, 10, 796–804. [Google Scholar] [CrossRef]
- Gschwend, D.A.; Good, A.C.; Kuntz, I.D. Molecular docking towards drug discovery. J. Mol. Recognit. 1996, 9, 175–186. [Google Scholar] [CrossRef]
- Serdaroğlu, G.; Uludag, N.; Üstün, E. Efficient synthesis of chromeno[2,3-b]pyridine derivatives using Zn(OTf)2 as a catalyst: DFT computations, molecular docking and ADME studies. J. Mol. Liq. 2023, 375, 121364. [Google Scholar] [CrossRef]
- Modi, S.J.; Kulkarni, V.M. Vascular Endothelial Growth Factor Receptor (VEGFR-2)/KDR Inhibitors: Medicinal Chemistry Perspective. Med. Drug Discov. 2019, 2, 100009. [Google Scholar] [CrossRef]
- Joukov, V.; Pajusola, K.; Kaipainen, A.; Chilov, D.; Lahtinen, I.; Kukk, E.; Saksela, O.; Kalkkinen, N.; Alitalo, K. A novel vascular endothelial growth factor, VEGF-C, is a ligand for the Flt4 (VEGFR-3) and KDR (VEGFR-2) receptor tyrosine kinases. EMBO J. 1996, 15, 290. [Google Scholar] [CrossRef] [PubMed]
- Al-Hazmy, S.M.; Zouaghi, M.O.; Al-Johani, J.N.; Arfaoui, Y.; Al-Ashwal, R.; Hammami, B.; Alhagri, I.A.; Alhemiary, N.A.; Hamdi, N. Chemosensing Properties of Coumarin Derivatives: Promising Agents with Diverse Pharmacological Properties, Docking and DFT Investigation. Molecules 2022, 27, 5921. [Google Scholar] [CrossRef] [PubMed]
- Serdaroğlu, G.; Şahin, N.; Bölükbaşı, S.Ş.; Üstün, E. Novel Ag(I)-NHC complex: Synthesis, in vitro cytotoxic activity, molecular docking, and quantum chemical studies. Z. Für Naturforschung C 2022, 77, 21–36. [Google Scholar] [CrossRef]
- Mamta; Pinki; Chaudhary, A. Synthesis, spectroscopic elucidation, density functionaltheory calculation, and molecular docking studies of a novelseries of tetradentate macrocyclic Schiff base ligands andtheir Zn(II) complexes and investigations of theirantimicrobial, anti-inflammatory, and anticancer activities. Appl. Organomet. Chem. 2024, 38, e7330. [Google Scholar]
- Moitessier, N.; Englebienne, P.; Lee, D.; Lawandi, J.; Corbeil, C.R. Towards the development of universal, fast and highly accurate docking/scoring methods: A long way to go. Br. J. Pharmacol. 2008, 153, S7. [Google Scholar] [CrossRef]
- Guengerich, F.P. Reactions and significance of cytochrome P-450 enzymes. J. Biol. Chem. 1991, 266, 10019–10022. [Google Scholar] [CrossRef]
- Haidar, S.; Ehmer, P.B.; Barassin, S.; Batzl-Hartmann, C.; Hartmann, R.W. Effects of novel 17-hydroxylase/C17, 20-lyase (P450 17, CYP 17) inhibitors on androgen biosynthesis in vitro and in vivo. J. Steroid Biochem. Mol. Biol. 2003, 84, 555–562. [Google Scholar] [CrossRef]
- Dhawale, S.A.; Bhosle, P.; Mahajan, S.; Patil, G.; Gawale, S.; Ghodke, M.; Tapadiya, G.; Ansari, A. Dual targeting in prostate cancer with phytoconstituents as a potent lead: Acomputational approach for novel drug discovery. J. Biomol. Struct. Dyn. 2024, 42, 8906–8919. [Google Scholar] [CrossRef]
- Behzad, M.; Ghasemi, L.; Dusek, M.; Kučeráková, M. Computer-Aided Investigation of Anticancer Properties ofNew Mixed-Ligand Cu (II) Complexes of an UnsymmetricalSchiff Base Ligand by ADMET and Molecular Docking:Comparison With Chemical Drugs and Curcumin. Appl. Organomet. Chem. 2025, 39, e7918. [Google Scholar] [CrossRef]
- Tajiani, F.; Ahmadi, S.; Lotfi, S.; Kumar, P.; Almasirad, A. In-silico activity prediction and docking studies of some favonol derivatives as anti-prostate cancer agents based on Monte Carlo optimization. BMC Chem. 2023, 17, 87. [Google Scholar] [CrossRef] [PubMed]
- Yue, W.; Wang, J.P.; Li, Y.; Fan, P.; Liu, G.; Zhang, N.; Conaway, M.; Wang, H.; Korach, K.S.; Bocchinfuso, W.; et al. Effects of estrogen on breast cancer development: Role ofestrogen receptor independent mechanisms. Int. J. Cancer 2010, 127, 1748–1757. [Google Scholar] [CrossRef]
- Kumar, P.H.; Rambabu, M.; Vijayakumar, V.; Sarveswari, S. Palladium-Mediated Synthesis of 2-([Biphenyl]-4-yloxy)quinolin-3carbaldehydes through Suzuki−Miyaura Cross-Coupling and Their in Silico Breast Cancer Studies on the 3ERT Protein. ACS omega 2023, 8, 11806–11812. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, D.; Maiti, A.K. Virtual Screening and Molecular Docking Studies with Organosulfur and Flavonoid Compounds of Garlic Targeting the Estrogen Receptor Protein for the Therapy of Breast Cancer. Biointerface Res. Appl. Chem. 2023, 13, 49. [Google Scholar]
- Sehrawat, R.; Rathee, P.; Rathee, P.; Khatkar, S.; Akkol, E.K.; Khatkar, A.; S.-Sánchez, E. In silico design of novel bioactive molecules to treat breast cancer with chlorogenic acid derivatives: A computational and SAR approach. Front. Pharmacol. 2023, 14, 1266833. [Google Scholar] [CrossRef]
- Khan, T.; Sankhe, K.; Suvarna, V.; Sherje, A.; Patel, K.; Dravyakar, B. DNA gyrase inhibitors: Progress and synthesis of potent compounds as antibacterial agents. Biomed. Pharmacotherp. 2018, 103, 923–938. [Google Scholar] [CrossRef]
- Bhukal, A.; Kumar, V.; Raman, A.P.S.; Kumar, A.; Singh, P.; Lal, K. Pyrazoline Spiro-oxindole tethered 1,2,3-triazole hybrids: Design, synthesis, antimicrobial efcacy and molecular modelling studies. Mol. Divers. 2025, 29, 1479–1492. [Google Scholar] [CrossRef]
- Üstün, E.; Şahin, N.; Özdemir, İ.; Günal, S.; Gürbüz, N.; Özdemir, İ.; Sémeril, D. Design, synthesis, antimicrobial activity and moleculardocking study of cationic bis-benzimidazole-silver(I)complexes. Arch. Pharm. 2023, 356, 2300302. [Google Scholar] [CrossRef]
- Gowthami, P.; Kosiha, A.; Kalaiarasi, G.; Sangeetha, N.S.; Shaik, B.; Jerome, P.; Oh, T.H. Phyto-mediated fabrication of cerium oxide nanoparticles using Mollugo oppositifolia L aqueous leaf extract: Antibacterial, antitonicity, and molecular docking studies. J. Indian. Chem. Soc. 2024, 101, 101399. [Google Scholar] [CrossRef]
- Arya, R.; Ravikumar, R.; Santhosh, R.S.; Princy, S.A. SarA based novel therapeutic candidate against Staphylococcus aureus associated with vascular graft infections. Front. Microbiol. 2015, 6, 416. [Google Scholar] [CrossRef]
- Cetinkaya, E.A.; Koc, A.; Koç, H.K.; Karabıyık, H.; Karabıyık, H.; Üstün, E.; Özdemir, İ. Synthesis, characterization and antimicrobial properties of silver complexes derived from 5,6-Dimethylbenzimidazol-2-ylidene. Polyhedron 2023, 237, 116383. [Google Scholar] [CrossRef]
- Kathiresan, N.; Prabu, D.; Kasilingam, H.; Sangavi, P.; ravi Arumugam, V.; Kulanthaivel, L. Anti-quorum sensing mediated anti-infective efficacy of pentadecanoic acid against Staphylococcus aureus infections through in vitro, and in silico analyses. Biocatal. Agric. Biotechnol. 2024, 62, 103442. [Google Scholar] [CrossRef]
- Beniaich, G.; Mabchour, I.; Mssillou, I.; Lfitat, A.; El Kamari, F.; Allali, A.; Hosen, M.E.; Supti, S.J.; Bin Jardan, Y.A.; Shazly, G.A.; et al. Antioxidant, antimicrobial, and insecticidal properties of chemicallycharacterized essential oils isolated from Artemisia herba-alba: In vivo, in vitro,and in silico approaches. Plant Biosyst. 2025, 159, 275–288. [Google Scholar] [CrossRef]
- Henary, E.; Casa, S.; Dost, T.L.; Sloop, J.C.; Henary, M. The Role of Small Molecules Containing Fluorine Atoms in Medicine and Imaging Applications. Pharmaceuticals 2024, 17, 281. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Elmore, S. Apoptosis: A review of programmed cell death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef] [PubMed]
- Vousden, K.H.; Lane, D.P. p53 in health and disease. Nat. Rev. Mol. Cell Biol. 2007, 8, 275–283. [Google Scholar] [CrossRef] [PubMed]







| Compounds | Molecular Formula | Yield (%) | M.p. (°C) | IR (vC(2)=O, vC(6)=O) (cm−1) | IR (vC(8)=N) (cm−1) | 1H NMR (C(8)-H) (ppm) | 13C NMR (C(8)) (ppm) |
|---|---|---|---|---|---|---|---|
| 1a | C15H13F3N4O2 | 92 | 214–215 | 1704, 1650 | 1550 | 7.53 | 141.50 |
| 1b | C15H13F3N4O2 | 81 | 128–129 | 1694, 1644 | 1547 | 7.58 | 139.78 |
| 1c | C15H13F3N4O2 | 87 | 196–197 | 1703, 1654 | 1548 | 7.56 | 140.88 |
| 1d | C16H12F6N4O2 | 93 | 157–158 | 1703, 1645 | 1551 | 7.69 | 140.71 |
| 1e | C15H13F3N4O3 | 90 | 177–178 | 1703, 1656 | 1547 | 7.54 | 140.74 |
| Molecules | BA * | Amino Acids Residue |
|---|---|---|
| VEGFR-2 | ||
| 1a | −5.07 | Glu883, His1024 (H-bonds), Asp1044 (halogenic interaction), Ile886, Leu887, Ile890, Val896, Leu1017 (alkylic interactions), Lys866, Val897, Cys1022, Ile1042, Cys1043 (van der Waals interactions) |
| 1b | −5.69 | Lys866, His1024, Ile1042 (H-bonds), Glu883, Asp1044 (halohenic interactions), Ile886 (pi-interactions), Ile890, Val896, Val897, Leu1017 (alkylic interacttions), Phe843, Leu887, Cys1022, Cys1043 (van der Waals interactions) |
| 1c | −5.27 | Ile1023, His1024, Ile1042 (H-bonds), Glu883, Asp1044 (pi-interactionss), val896, Leu1017 (alkylic interactions), Phe843, Lys866, Ile886, Ile890, Val897, Cys1022, Cys1043 (van der Waals interactions) |
| 1d | −5.17 | Lys866, His1024, Ile1042 (H-bonds), Glu883, Asp1044 (halogenic interactions), Leu887 (pi-interactions), Ile886, Ile890, Val896, Leu1017 (alkylic interactions), Ser882, Phe843, Val897, Cys1022, Ile1023, Cys1043 (van der Waals interactions) |
| 1e | −5.34 | Lys866, His1024 (H-bonds), Glu883 (halogenic interaction), Leu887, Asp1044 (pi-interactions), Ile886, Ile890, Val896, Leu1017, Ile1042 (alkylic interactions) Phe843, Ala879, Val897, Cys1022, Ile1023, Cys1043 (van der Waals interactions) |
| AAFP | −5.61 | Glu883, Glu915, Cys917, Asp1044 (H-bonds), Val846 (pi-interaction), Ile1042, Cys1043 (halogenic interactions), Leu838, Ala864, Leu887, Ile890, Val896, Val897, Val914, Leu1017, Leu1033 (alkylic interactions), Gly839, Arg840, Gly841, Lys866, Ile886, Phe916, His1024 (van der Waals interactions) |
| Human Cytochrome P450 | ||
| 1a | −6.37 | Ala367, Cys442 (H-bonds), Arg440 (halogenic interaction), Val366 (pi-interaction), Leu361, leu370, Ile371, Phe435, Ala448 (alkylic interactions), Arg96, Thr306, Val310, Gly436, Ser441 (van der Waals interactions) |
| 1b | −6.42 | Gly436, Arg440, Ser441, Cys442, Ile443 (H-bonds), His373 (halogenic interactions), Ala302, Ala367, Leu370 (alkylic interactions), Leu86, Arg96, Ile112, Arg125, Thr306, Ile371, Pro434, Phe435, Ala437, Gly444 (van der Waals interactions) |
| 1c | −6.60 | Gly436, Ser441, Cys442 (H-bonds), Phe435, Arg440 (halogenic interactions), Thr306, Val366 (pi-interactions), Ala302, Ala367, Leu370, Ala448 (alkylic interactions), Arg96, Pro434, Ala437 (van der Waals interactions) |
| 1d | −6.28 | Gly436, Ser441, Cys442, Ile443 (H-bonds), Phe435, Arg440 (halogenic interactions), Val366 (pi-interaction), Leu361, Ala367, Leu370, Ala448 (alkylic interactions), Arg96, Ala113, Arg125, Thr306, Ile371, Ala437, Gly444 (van der Waals interactions) |
| 1e | −6.69 | Pro434, Gly436, Ala437, Ser441, Cys442 (H-bonds), His373, Arg440 (halogenic interactions), Thr306, Val366 (pi-interactions), Leu361, Ala367, Leu370, Phe435, Ala448 (alkylic interactions), Leu86, Arg96, Val310, Ile371, Leu452 (van der Waals interactions) |
| Abiraterone | −9.01 | Asn202 (H-bond), Ala113, Phe114, Ile205, Leu209, Ala302, Val482 (alkylic interactions), Tyr201, Ile206, Arg239, Gly297, Asp298, Gly301, Glu305, Thr306, Val366, Ala367, Ile371, Cys442, Val483 (van der Waals interactions) |
| Estrogen Receptor | ||
| 1a | −6.64 | Leu346, Glu353 (H-bonds), Leu384, Met388 (pi interactions), Met343, Leu349, Ala350, Leu387, Leu391, Met421, Leu525 (alkylic interactions), Thr347, Arg394, Phe404, Glu419, Gly420, Ile424, Gly51-21, His524 (van der Waals interactions) |
| 1b | −6.70 | Glu353, Lys449 (H-bonds), Glu323, Pro324 (halogenic interactions), Ile326 (pi-interaction), Leu327, His356, Met357, Trp360, Ile386, Arg394 (alkylic interactions), Pro325, Gly390, Trp393, Phe445 (van der Waals interactions) |
| 1c | −6.36 | Leu387, Arg394, Gly521, His524, Leu525 (H-bonds), Glu419, Gly420 (halogenic interactions), Ala350, Glu353 (pi-interactions), Leu346, Leu349, Leu384, Met388, Leu391, Phe404 (alkylic interactions), Met343, Thr347, Trp383, Met421, Ile424 (van der Waals interactions) |
| 1d | −6.40 | Pro325, Ile326, Ile386, Trp393, Lys449 (H-bonds), Glu323, Pro324 (halogenic interactions), Glu353, Arg394 (pi-interactions), Leu327, Phe445 (alkylic interactions), Met357, Leu387, Gly390, Val446 (van der Waals interactions) |
| 1e | −6.54 | Pro325, Glu353, Lys449 (H-bonds), Trp393, Phe445 (pi-interactions), Pro324, Ile326, Leu327, His356, Met357, Trp360, Ile386, Arg394 (alkylic interactions), Glu323, Gly390 (van der Waals interactions) |
| Hat | −10.35 | Asp351, Glu353, Arg394 (H-bonds), Met343, Leu346 (pi-interactions), Ala350, Leu387, Met388, Leu391, Ile424, Leu428, Leu525 (alkylic interactions), Thr347, Leu349, Trp383, Leu384, Phe404, Glu419, Gly420, Gly521, His524 (van der Waals interactions) |
| DNA Gyrase | ||
| 1a | −5.56 | Asn46, Val71, Asp73, Thr165 (H-bonds), Val43, Ala47, Ile78, Ile90, Val120, Val167 (alkylic interactions), Asp49, Glu50, Gln72, Met91 (van der Waals interactions) |
| 1b | −5.06 | Asn46, Asp (H-bonds), Val43, Val71, Asp73, Thr165 (halogenic interactions), Ala47, Ile90, Val120, Val167 (alkylic interactions), Val44, Glu50, Gln72, Ile78, Met91, Met166 (van der Waals interactions) |
| 1c | −4.62 | Asn46, Asp49, Val167 (H-bonds), Val43, Val71, Thr165 (halogenic interactions), Ala47, Ile78, Ile90 (alkylic interactions), Glu50, Gln72, Asp73, Val120, Met166 (van der Waals interactions) |
| 1d | −4.59 | Asn46, Asp49 (H-bonds), Val43, Val71, Asp73, Thr165 (halogenic interactions), Ala47, Ile78, Ile90, Met91, Val120 (alkylic interactions), Val44, Glu50, Ile59, Gln72, Leu132, Met166 (van der Waals interactions) |
| 1e | −5.11 | Asn46, Asp49, Val167 (H-bonds), Val71, Thr165 (halogenic interactions), Ala47, Ile78, Ile90 (alkylic interactions),Val43, Glu50, Gln72, Asp73, Val120, Met166 (van der Waals interactions) |
| Clorobiocin | −7.15 | Asn46, Asp73, Arg136 (H-bonds), Glu50, Arg76, Thr165 (pi-interactions), Gly77 (halogenic interaction), Ala47, Ile78, Pro79, Ile90 (alkylic interactions), Val43, Asp49, Val71, Gln72, Ala86, Met91, Val120, Met166, Val167 (van der Waals interactions) |
| SarA | ||
| 1a | −4.56 | His 159 (H-bond), Leu160 (pi-interaction), Phe134, Phe137, Ala138 (alkylic interactions), Thr141, Tyr142, Glu145, Asn161, Tyr162 (van der Waals interactions) |
| 1b | −4.90 | Ala138, Tyr162 (H-bonds), His159 (halogenic interaction), Phe134, Tyr142 (pi-interactions), Phe137, Leu160 (alkylic interactions), Thr141, Glu145 (van der Waals interactions) |
| 1c | −4.68 | Ala138, Tyr162 (H-bonds), Glu145 (halogenic interaction), Phe134, Tyr142 (pi-interactions), Leu160 (alkylic interaction), Phe137, Thr141, His159 (van der Waals interactions) |
| 1d | −4.64 | Ala138, Tyr162 (H-bonds), Leu122 (halogenic interaction), Phe137 (pi-interaction), Lys123, Ile126, Phe134, Leu160, Ile215 (alkylic interactions), Thr141, Tyr142 (van der Waals interactions) |
| 1e | −4.53 | Ala138, Tyr162 (H-bonds), Glu145 (halogenic interaction), Phe134, Tyr142 (pi-interactions), Phe137, Leu160 (alkylic interactions), Thr141, Asn146, His159 (van der Waals interactions) |
| Molecules | BEAS-2B (μM) | A549 (μM) | SI (A549) | HeLa (μM) | SI (HeLa) |
|---|---|---|---|---|---|
| 1a | 100 ± 2 | 12.4 ± 0.7 | 8.06 ± 0.10 | 11.5 ± 1.2 | 8.70 ± 0.20 |
| 1b | 120 ± 3 | 13.6 ± 0.9 | 8.82 ± 0.20 | 14.7 ± 0.7 | 8.16 ± 0.16 |
| 1c | 90 ± 3 | 15.7 ± 0.6 | 5.73 ± 0.15 | 11.5 ± 1.2 | 7.83 ± 0.22 |
| 1d | 150 ± 4 | 31.4 ± 1.0 | 4.78 ± 0.14 | 34.5 ± 1.0 | 4.35 ± 0.13 |
| 1e | 140 ± 3 | 29.1 ± 0.5 | 4.81 ± 0.09 | 32.4 ± 0.8 | 4.32 ± 0.11 |
| Compound | Concentration Range (µg/mL or %) | Staphylococcus aureus (%) | Enterococcus faecalis (%) | Escherichia coli (%) | Pseudomonas aeruginosa (%) | Candida albicans (%) |
|---|---|---|---|---|---|---|
| 1a | 0.488–1000 | – | 1.08–14.7 | – | – | 0.5–13.7 |
| 1b | 0.97–1000 | – | 5.7–23.1 | – | – | 0.8–23.9 |
| 1c | 0.488–1000 | 20.1 | 0.7–16.0 | – | – | 0.9–12.3 |
| 1d | 0.244–1000 | – | 0.7–23.9 | – | – | 1.8–11.8 |
| 1e | 0.244–1000 | 13.6 | 5.1–13.9 | – | – | 0.4–23.4 |
| Brucella broth (NC) | – | – | – | – | – | – |
| Ciprofloxacin (PC) | 0.03125–64 | 50.56 | 18.11 | 36.67 | 35.79 | NT |
| Fluconazole (PC) | 0.3125–640 | NT | NT | NT | NT | 29.10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Düşünceli, S.D.; Coşkun, K.A.; Kaloğlu, M.; Üstün, E.; Çalışkan, R.; Tutar, Y. Synthesis, Characterization, and Screening Anticancer—Antibiofilm Activities of Theophylline Derivatives Containing CF3/OCF3 Moiety. Biology 2025, 14, 1180. https://doi.org/10.3390/biology14091180
Düşünceli SD, Coşkun KA, Kaloğlu M, Üstün E, Çalışkan R, Tutar Y. Synthesis, Characterization, and Screening Anticancer—Antibiofilm Activities of Theophylline Derivatives Containing CF3/OCF3 Moiety. Biology. 2025; 14(9):1180. https://doi.org/10.3390/biology14091180
Chicago/Turabian StyleDüşünceli, Serpil Demir, Kübra Açıkalın Coşkun, Murat Kaloğlu, Elvan Üstün, Reyhan Çalışkan, and Yusuf Tutar. 2025. "Synthesis, Characterization, and Screening Anticancer—Antibiofilm Activities of Theophylline Derivatives Containing CF3/OCF3 Moiety" Biology 14, no. 9: 1180. https://doi.org/10.3390/biology14091180
APA StyleDüşünceli, S. D., Coşkun, K. A., Kaloğlu, M., Üstün, E., Çalışkan, R., & Tutar, Y. (2025). Synthesis, Characterization, and Screening Anticancer—Antibiofilm Activities of Theophylline Derivatives Containing CF3/OCF3 Moiety. Biology, 14(9), 1180. https://doi.org/10.3390/biology14091180










