Mitogenomic Insights into Orthocladiinae (Diptera: Chironomidae): Structural Diversity and Phylogenetic Implications
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
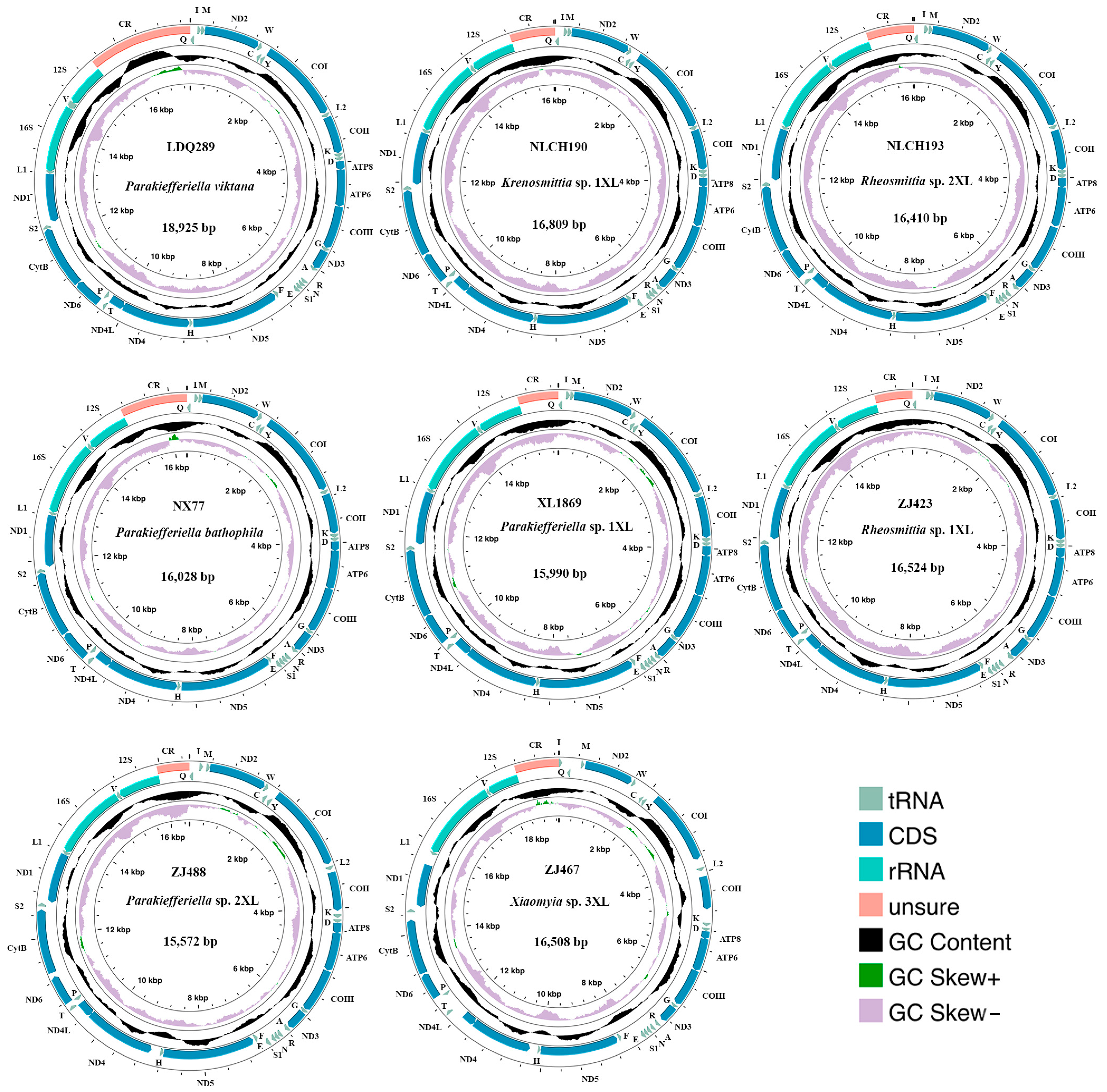
2.2. Sequencing, Assembly, and Annotation
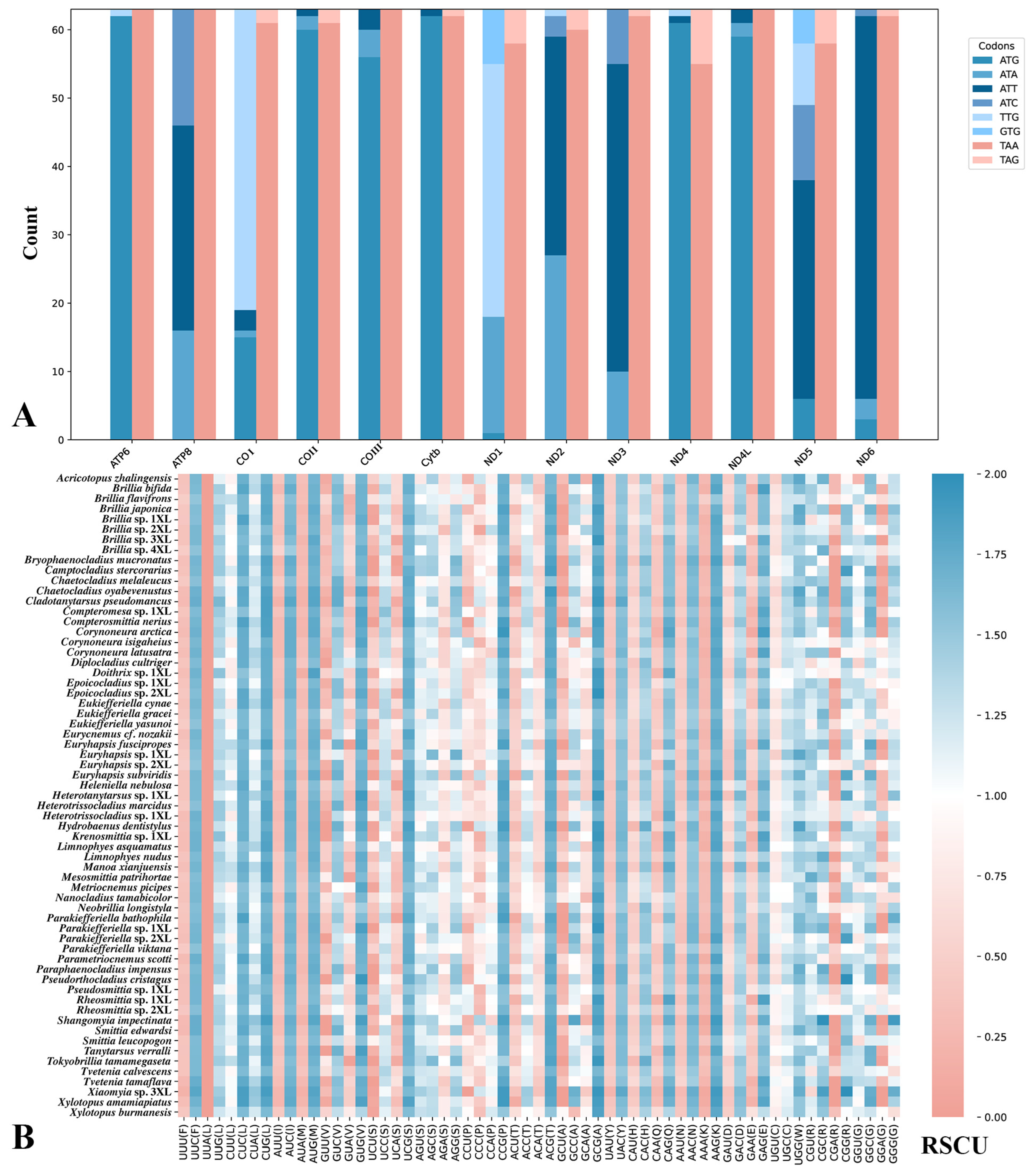
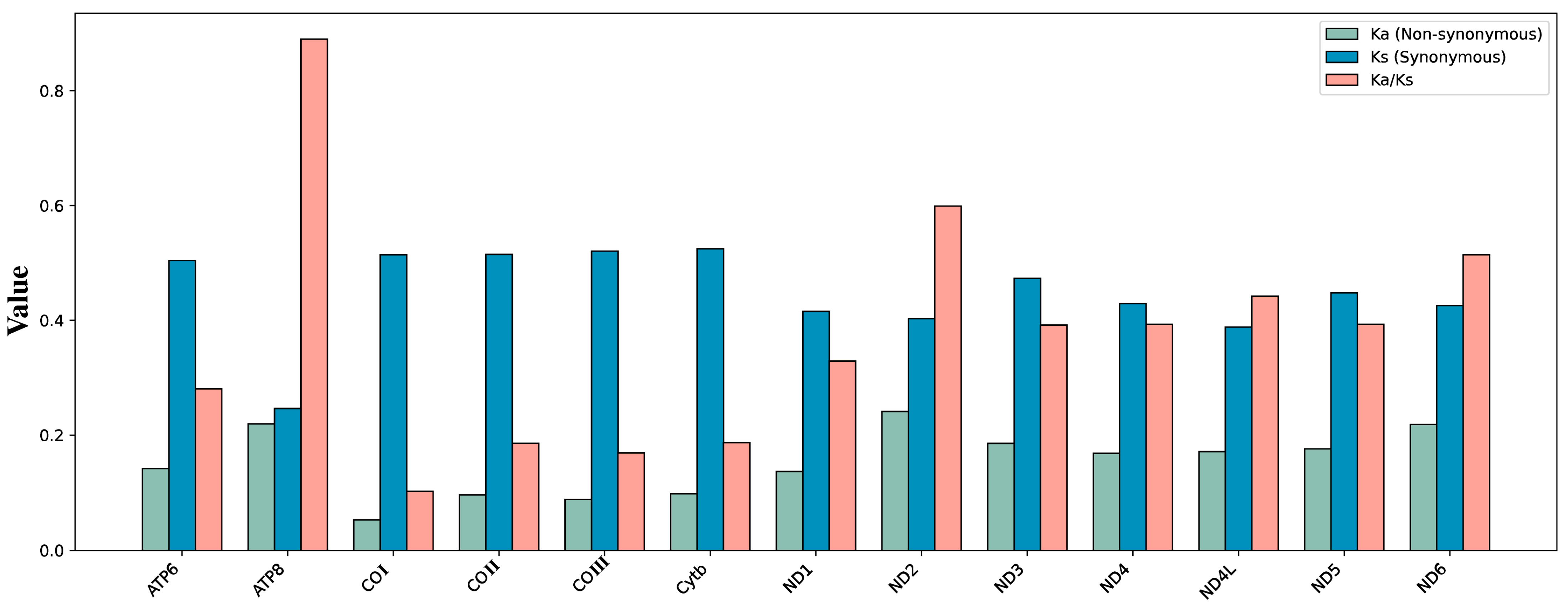
2.3. Composition Analyses, RSCU, and Evolutionary Rate
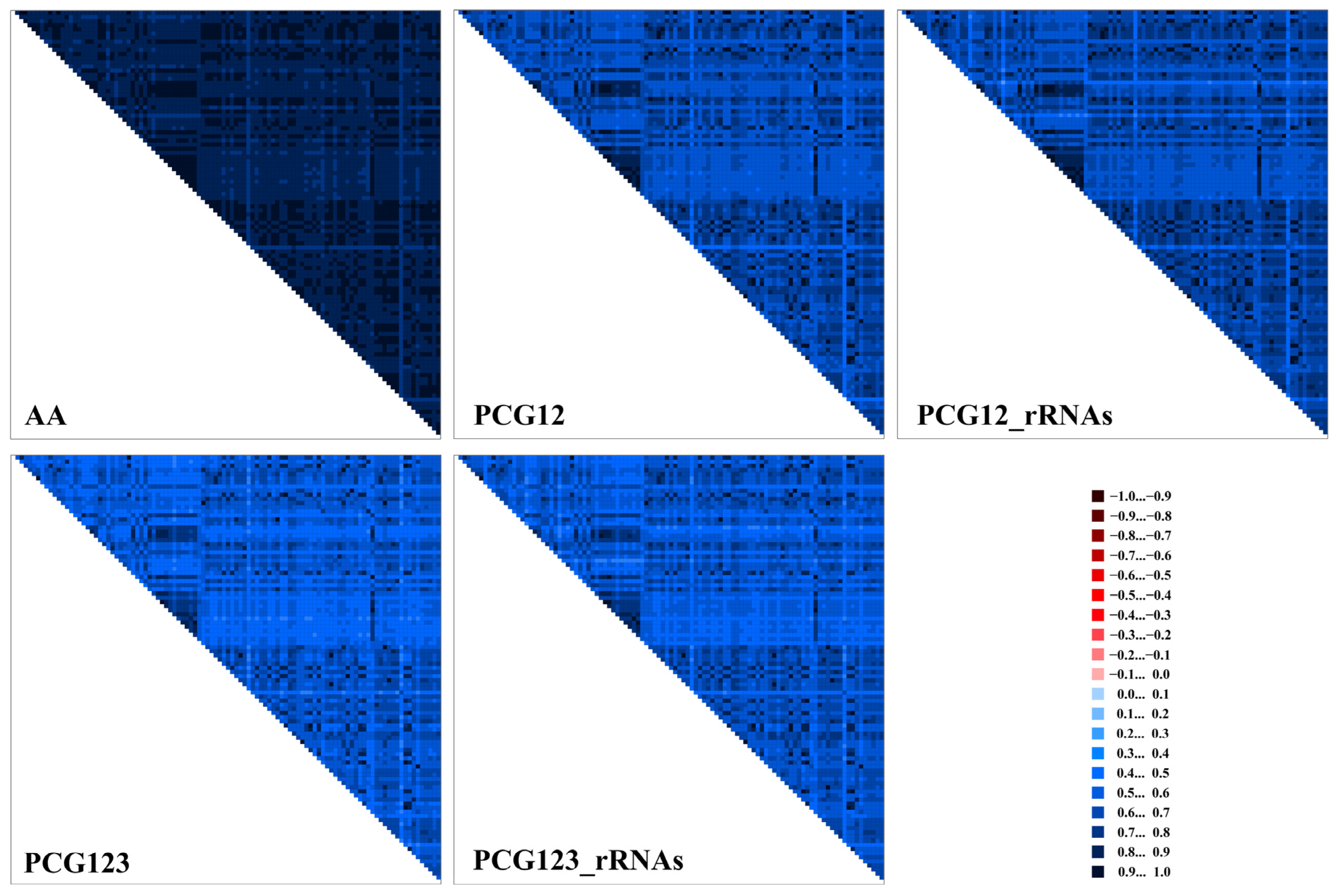
2.4. Phylogenetic Analyses
3. Results and Discussion
3.1. Nucleotide Composition
3.2. Codon Usage
3.3. Substitution Rates and Nucleotide Diversity
3.4. Heterogeneity Analyses of Mitogenomes
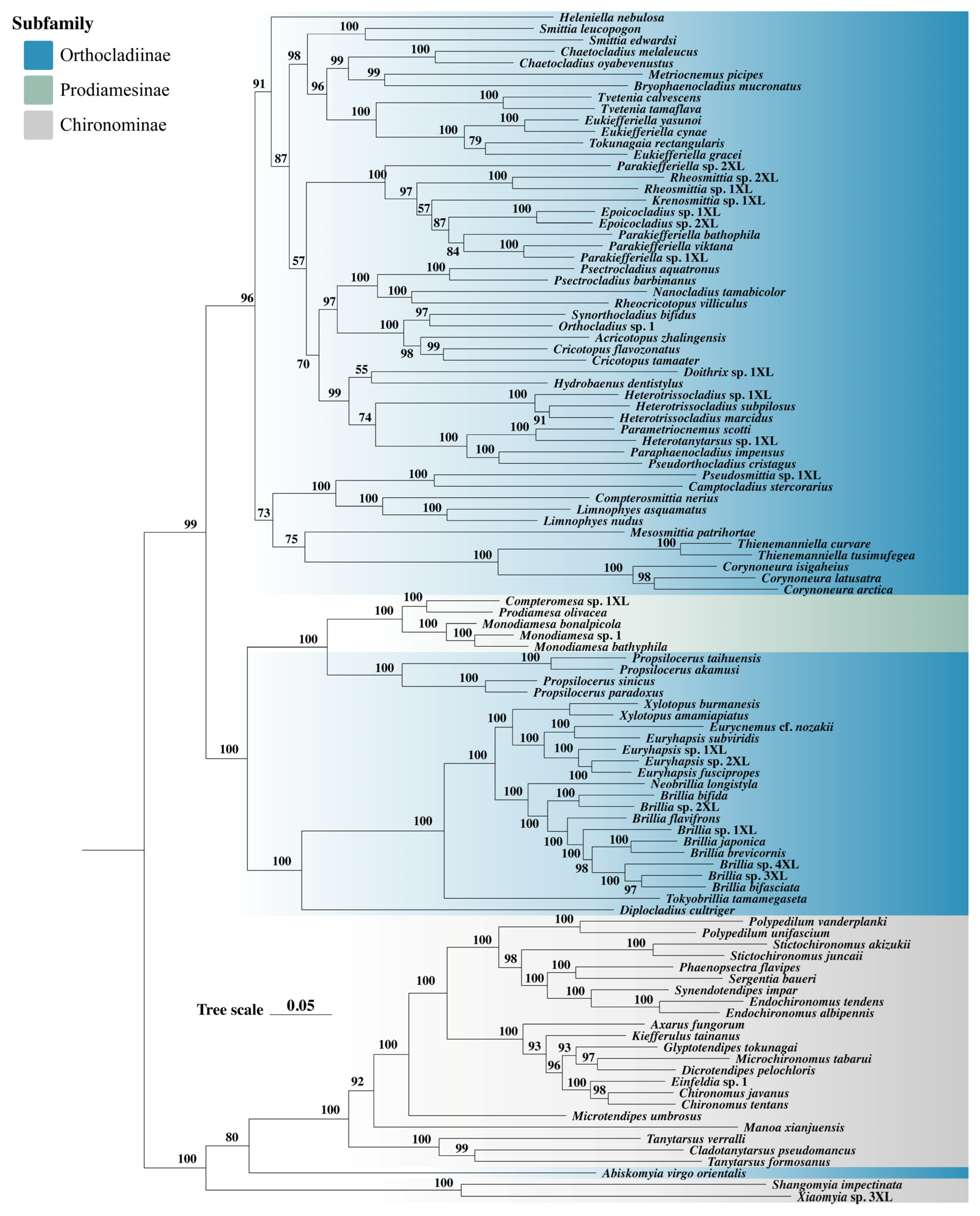
3.5. Phylogenetic Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cameron, S.L. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef]
- Curole, J.P.; Kocher, T.D. Mitogenomics: Digging deeper with complete mitochondrial genomes. Trends Ecol. Evol. 1999, 14, 394–398. [Google Scholar] [CrossRef] [PubMed]
- Munasinghe, M.; Ågren, J.A. When and why are mitochondria paternally inherited? Curr. Opin. Genet. Dev. 2023, 80, 102053. [Google Scholar] [CrossRef]
- Hu, W.; Zhang, J.; Wu, Z.; Wu, Y.; Hu, Y.; Hu, X.; Cao, J. Research progress on paternal mitochondrial inheritance: An overview. Mitochondrion 2025, 82, 102019. [Google Scholar] [CrossRef]
- Cao, J.; Luo, Y.; Chen, Y.; Wu, Z.; Zhang, J.; Wu, Y.; Hu, W. Maternal mitochondrial function affects paternal mitochondrial inheritance in Drosophila. Genetics 2024, 226, iyae014. [Google Scholar] [CrossRef]
- Wolstenholme, D.R. Animal mitochondrial DNA: Structure and evolution. Int. Rev. Cytol. 1992, 141, 173–216. [Google Scholar] [CrossRef] [PubMed]
- Talavera, G.; Vila, R. What is the phylogenetic signal limit from mitogenomes? The reconciliation between mitochondrial and nuclear data in the Insecta class phylogeny. BMC Evol. Biol. 2011, 11, 315. [Google Scholar] [CrossRef] [PubMed]
- Baker, R.H.; Wilkinson, G.S.; DeSalle, R. Phylogenetic Utility of Different Types of Molecular Data Used to Infer Evolutionary Relationships among Stalk-Eyed Flies (Diopsidae). Syst. Biol. 2001, 50, 87–105. [Google Scholar] [CrossRef]
- Cao, J.-J.; Wang, Y.; Murányi, D.; Cui, J.-X.; Li, W.-H. Mitochondrial genomes provide insights into the Euholognatha (Insecta: Plecoptera). BMC Evol. Biol. 2024, 24, 16. [Google Scholar] [CrossRef]
- Li, R.; Ma, Z.; Zhou, C. The first two complete mitochondrial genomes of Neoephemeridae (Ephemeroptera): Comparative analysis and phylogenetic implication for Furcatergalia. Genes 2021, 12, 1875. [Google Scholar] [CrossRef]
- Zhao, W.; Zhao, Q.; Li, M.; Wei, J.; Zhang, X.; Zhang, H. Comparative Mitogenomic Analysis of the Eurydema Genus in the Context of Representative Pentatomidae (Hemiptera: Heteroptera) Taxa. J. Insect Sci. 2019, 19, 20. [Google Scholar] [CrossRef]
- Dettai, A.; Gallut, C.; Brouillet, S.; Pothier, J.; Lecointre, G.; Debruyne, R. Conveniently pre-tagged and pre-packaged: Extended molecular identification and metagenomics using complete metazoan mitochondrial genomes. PLoS ONE 2012, 7, e51263. [Google Scholar] [CrossRef] [PubMed]
- Ge, X.; Zang, H.; Ye, X.; Peng, L.; Wang, B.; Lian, G.; Sun, C. Comparative Mitogenomic Analyses of Hydropsychidae Revealing the Novel Rearrangement of Protein-Coding Gene and tRNA (Trichoptera: Annulipalpia). Insects 2022, 13, 759. [Google Scholar] [CrossRef] [PubMed]
- Ge, X.; Peng, L.; Vogler, A.P.; Morse, J.C.; Yang, L.; Sun, C.; Wang, B. Massive gene rearrangements of mitochondrial genomes and implications for the phylogeny of Trichoptera (Insecta). Syst. Entomol. 2022, 48, 278–295. [Google Scholar] [CrossRef]
- Zhang, X.; Yang, D.; Kang, Z.-H. New data on the mitochondrial genome of Nematocera (lower Diptera): Features, structures and phylogenetic implications. Zool. J. Linn. Soc. 2022, 197, 229–245. [Google Scholar] [CrossRef]
- Zhang, X.; Kang, Z.; Ding, S.; Wang, Y.; Borkent, C.; Saigusa, T.; Yang, D. Mitochondrial genomes provide insights into the phylogeny of Culicomorpha (Insecta: Diptera). Int. J. Mol. Sci. 2019, 20, 747. [Google Scholar] [CrossRef]
- Ashe, P.; O’Connor, J.P. A World Catalogue of Chironomidae (Diptera), Part 1: Buchonomyiinae, Chilenomyiinae, Podonominae, Aphroteniinae, Tanypodinae, Usambaromyiinae, Diamesinae, Prodiamesinae and Telmatogetoninae, 1st ed.; Irish Biogeographical Society: Dublin, Ireland, 2009; p. 445. [Google Scholar]
- Ferrington, L.C. Global diversity of non-biting midges (Chironomidae; Insecta-Diptera) in freshwater. Hydrobiologia 2008, 595, 447–455. [Google Scholar] [CrossRef]
- Pape, T.; Evenhuis, N. Systema Dipterorum. Biodivers. Inf. Sci. Stand. 2023, 7, e111959. [Google Scholar] [CrossRef]
- Armitage, P.D.; Pinder, L.; Cranston, P. The Chironomidae: Biology and Ecology of Non-Biting Midges, 1st ed.; Springer Science & Business Media: Dordrecht, The Netherlands, 2012; p. 572. [Google Scholar]
- Sæther, O.A. Some Nearctic Podonominae, Diamesinae, and Orthocladiinae (Diptera: Chironomidae); Fisheries Research Board of Canada: Nanaimo, BC, Canada, 1969; Volume 170, pp. 1–154. [Google Scholar]
- Brodin, Y.; Ejdung, G.; Strandberg, J.; Lyrholm, T. Improving environmental and biodiversity monitoring in the Baltic Sea using DNA barcoding of Chironomidae (Diptera). Mol. Ecol. Resour. 2013, 13, 996–1004. [Google Scholar] [CrossRef] [PubMed]
- Sæther, O.A. Female genitalia in Chironomidae and other Nematocera: Morphology, Phylogenies, Keys; Fisheries Research Board of Canada: Nanaimo, BC, Canada, 1977; Volume 197, pp. 1–209. [Google Scholar]
- Cranston, P.S.; Oliver, D.R.; Sæther, O.A. The Larvae of Orthocladiinae (Diptera: Chironomidae) of the Holarctic Region: Keys and Diagnoses, 1st ed.; Scandinavian Society of Entomology: Sandby, Sweden, 1983; pp. 149–291. [Google Scholar]
- Lin, X.-L.; Liu, Z.; Yan, L.-P.; Duan, X.; Bu, W.-J.; Wang, X.-H.; Zheng, C.-G. Mitogenomes provide new insights of evolutionary history of Boreheptagyiini and Diamesini (Diptera: Chironomidae: Diamesinae). Ecol. Evol. 2022, 12, e8957. [Google Scholar] [CrossRef] [PubMed]
- Ekrem, T.; Willassen, E.; Stur, E. Phylogenetic utility of five genes for dipteran phylogeny: A test case in the Chironomidae leads to generic synonymies. Mol. Phylogenet. Evol. 2010, 57, 561–571. [Google Scholar] [CrossRef]
- Cranston, P.S.; Hardy, N.B.; Morse, G.E. A dated molecular phylogeny for the Chironomidae (Diptera). Syst. Entomol. 2012, 37, 172–188. [Google Scholar] [CrossRef]
- Cameron, S.L.; Miller, K.B.; D’Haese, C.A.; Whiting, M.F.; Barker, S.C. Mitochondrial genome data alone are not enough to unambiguously resolve the relationships of Entognatha, Insecta and Crustacea sensu lato (Arthropoda). Cladistics 2004, 20, 534–557. [Google Scholar] [CrossRef] [PubMed]
- Oliver, D.R.; Roussel, M.E. Redescription of Brillia Kieffer (Diptera, Chironomidae) with Descriptions of Nearctic Species. Can. Entomol. 1983, 115, 257–279. [Google Scholar] [CrossRef]
- Wang, X.-H.; Zheng, L.-Y.; Ji, B.-C. A taxonomic study on orthodadiinae (Diptera: Chironomidae) of China. Ⅱ. genus Brillia kieffer. Acta Ent. Sin. 1994, 37, 359–363. [Google Scholar]
- Cobo, F.; Gonzalez, M.; Vieira-Lanero, R. Notes on some taxonomic problems in the Iberian species of Brillia Kieffer, 1913 (Diptera: Chironomidae), with a description of B. pudorosa sp.n. Ann. Limnol.-Int. J. Lim 1995, 31, 245–252. [Google Scholar] [CrossRef]
- Kawai, K. Seven new chironomid species (Diptera, Chironomidae) from Japan. Jpn. J. Limnol. 1991, 52, 161–171. [Google Scholar] [CrossRef]
- Sæther, O.A.; Wang, X.H. Euryhapsis fuscipropes sp. n. from China and Tokyobrillia anderseni sp.n. from Tanzania, with a review of genera near Irisobrillia Oliver (Diptera: Chironomidae). Ann. Limnol.-Int. J. Lim 1992, 28, 209–223. [Google Scholar] [CrossRef]
- Li, L.; Tang, H. Eurycnemus vd Wulp (Diptera, Chironomidae) newly recorded in China. J. Entomol. Acarol. Res. 2017, 49, 144–149. [Google Scholar] [CrossRef]
- Liu, W.-B.; Yao, Y.; Duan, X.; Lin, X.-L.; Yan, C.-C. New record, COI barcode and redescription of Xylotopus amamiapiatus (Sasa, 1990) (Diptera, Chironomidae) from China. Ann. Zool. Fennici. 2021, 58, 69–74. [Google Scholar] [CrossRef]
- Li, S.Y.; Chen, M.H.; Sun, L.; Wang, R.H.; Li, C.H.; Gresens, S.; Li, Z.; Lin, X.L. New mitogenomes from the genus Cricotopus (Diptera: Chironomidae, Orthocladiinae): Characterization and phylogenetic implications. Arch. Insect Biochem. Physiol. 2024, 115, e22067. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.-G.; Zhu, X.-X.; Yan, L.-P.; Yao, Y.; Bu, W.-J.; Wang, X.-H.; Lin, X.-L. First complete mitogenomes of Diamesinae, Orthocladiinae, Prodiamesinae, Tanypodinae (Diptera: Chironomidae) and their implication in phylogenetics. PeerJ 2021, 9, e11294. [Google Scholar] [CrossRef]
- Li, S.-Y.; Zhao, Y.-M.; Guo, B.-X.; Li, C.-H.; Sun, B.-J.; Lin, X.-L. Comparative Analysis of Mitogenomes of Chironomus (Diptera: Chironomidae). Insects 2022, 13, 1164. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Bu, W.J.; Zheng, C.G.; Lin, X.L.; Jiao, K.L. New data on mitogenomes of Thienemanniella Kieffer, 1911 (Diptera: Chironomidae, Orthocladiinae). Arch. Insect Biochem. Physiol. 2023, 114, 1–9. [Google Scholar] [CrossRef]
- Semenchenko, A.A.; Krasheninnikov, A.B.; Seliverstov, N.A.; Khamenkova, E.V.; Vinnikov, K.A. How reliable is mitochondrial DNA for recovering the phylogeny of Chironomidae (Culicomorpha: Diptera)? J. Insect Sci. 2025, 25, 1. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Wang, X.; Mao, B.; Xiao, Y.; Shen, M.; Fu, Y. Comparative mitogenome analyses of twelve non-biting flies and provide insights into the phylogeny of Chironomidae (Diptera: Culicomorpha). Sci. Rep. 2023, 13, 9200. [Google Scholar] [CrossRef] [PubMed]
- Kong, F.-Q.; Zhao, Y.-C.; Chen, J.-L.; Lin, X.-L. First report of the complete mitogenome of Microchironomus tabarui Sasa, 1987 (Diptera, Chironomidae) from Hebei Province, China. Mitochondrial DNA Part B 2021, 6, 2845–2846. [Google Scholar] [CrossRef]
- Lei, T.; Song, C.; Zhu, X.-D.; Xu, B.-Y.; Qi, X. The complete mitochondrial genome of a non-biting midge Polypedilum unifascium (Tokunaga, 1938) (Diptera: Chironomidae). Mitochondrial DNA Part B 2021, 6, 2212–2213. [Google Scholar] [CrossRef]
- Deviatiiarov, R.; Kikawada, T.; Gusev, O. The complete mitochondrial genome of an anhydrobiotic midge Polypedilum vanderplanki (Chironomidae, Diptera). Mitochondrial DNA Part A 2017, 28, 218–220. [Google Scholar] [CrossRef]
- Zhang, D.; He, F.-X.; Li, X.-B.; Aishan, Z.; Lin, X.-L. New Mitogenomes of the Polypedilum Generic Complex (Diptera: Chironomidae): Characterization and Phylogenetic Implications. Insects 2023, 14, 238. [Google Scholar] [CrossRef]
- Cao, J.-K.; Lei, T.; Gu, J.-J.; Song, C.; Xin, Q. Codon bias analysis of the mitochondrial genome reveals natural selection in the nonbiting midge Microtendipes umbrosus Freeman, 1955 (Diptera: Chironomidae). Pan-Pac. Entomol. 2023, 99, 217–225. [Google Scholar] [CrossRef]
- Qi, Y.; Duan, X.; Jiao, K.-L.; Lin, X.-L. First complete mitogenome of Axarus fungorum (Albu, 1980) from Guizhou Province, China (Diptera, Chironomidae). Mitochondrial DNA Part B 2022, 7, 1807–1809. [Google Scholar] [CrossRef]
- Chen, X.-Y.; Xiao, X.-R.; Zhang, Y.; Zhang, Z.-C.; Zhang, D.-S.; Liu, Z.; Lin, X.-L. Comparative Mitogenomic Analyses of Psectrocladius (Diptera: Chironomidae). Insects 2025, 16, 420. [Google Scholar] [CrossRef]
- Lin, X.L.; Zhao, Y.M.; Yan, L.P.; Liu, W.B.; Bu, W.J.; Wang, X.H.; Zheng, C.G. Mitogenomes provide new insights into the evolutionary history of Prodiamesinae (Diptera: Chironomidae). Zool. Scr. 2022, 51, 119–132. [Google Scholar] [CrossRef]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Lin, X.-L.; Stur, E.; Ekrem, T. DNA barcodes and morphology reveal unrecognized species in Chironomidae (Diptera). Insect Syst. Evol. 2018, 47, 329–398. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017, 45, e18. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Leung, H.C.; Yiu, S.-M.; Chin, F.Y. IDBA-UD: A de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 2012, 28, 1420–1428. [Google Scholar] [CrossRef] [PubMed]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Shen, W.; Le, S.; Li, Y.; Hu, F. SeqKit: A cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS ONE 2016, 11, e0163962. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Suleski, M.; Sanderford, M.; Sharma, S.; Tamura, K. MEGA12: Molecular Evolutionary Genetic Analysis Version 12 for Adaptive and Green Computing. Mol. Biol. Evol. 2024, 41, msae263. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.-y.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Kück, P.; Longo, G.C. FASconCAT-G: Extensive functions for multiple sequence alignment preparations concerning phylogenetic studies. Front. Zool. 2014, 11, 81. [Google Scholar] [CrossRef]
- Kück, P.; Meid, S.A.; Groß, C.; Wägele, J.W.; Misof, B. AliGROOVE–visualization of heterogeneous sequence divergence within multiple sequence alignments and detection of inflated branch support. BMC Bioinform. 2014, 15, 294. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Sun, X.; Cheng, J. Comparative mitogenomic analyses and new insights into the phylogeny of Thamnocephalidae (Branchiopoda: Anostraca). Genes 2022, 13, 1765. [Google Scholar] [CrossRef]
- Xu, S.; Miao, P.; Liu, Q.; Song, F.; Yang, H.; Xia, Z.; Huang, W. Comparative analysis of five mitochondrial genomes of the subfamily Galerucinae (Coleoptera: Chrysomelidae) and evolution of control regions inferred from phylogeny. Front. Ecol. Evol. 2025, 13, 1495703. [Google Scholar] [CrossRef]
- Dowton, M.; Cameron, S.L.; Dowavic, J.I.; Austin, A.D.; Whiting, M.F. Characterization of 67 Mitochondrial tRNA Gene Rearrangements in the Hymenoptera Suggests That Mitochondrial tRNA Gene Position Is Selectively Neutral. Mol. Biol. Evol. 2009, 26, 1607–1617. [Google Scholar] [CrossRef]
- Beckenbach, A.T.; Joy, J.B. Evolution of the mitochondrial genomes of gall midges (Diptera: Cecidomyiidae): Rearrangement and severe truncation of tRNA genes. Genome Biol. Evol. 2009, 1, 278–287. [Google Scholar] [CrossRef] [PubMed]
- Lei, T.; Zheng, X.; Song, C.; Jin, H.; Chen, L.; Qi, X. Limited variation in codon usage across mitochondrial genomes of non-biting midges (Diptera: Chironomidae). Insects 2024, 15, 752. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Wang, J.; Li, H.; Yu, K.; Yang, Y.; Li, M.; Smagghe, G.; Dai, R. Mitochondrial genomes of Macropsini (Hemiptera: Cicadellidae: Eurymelinae): Structural features, codon usage patterns, and phylogenetic implications. Ecol. Evol. 2024, 14, e70268. [Google Scholar] [CrossRef] [PubMed]
- Parvathy, S.T.; Udayasuriyan, V.; Bhadana, V. Codon usage bias. Mol. Biol. Rep. 2022, 49, 539–565. [Google Scholar] [CrossRef]
- Plotkin, J.B.; Kudla, G. Synonymous but not the same: The causes and consequences of codon bias. Nat. Rev. Genet. 2011, 12, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Kondrashov, F.A.; Rogozin, I.B.; Wolf, Y.I.; Koonin, E.V. Selection in the evolution of gene duplications. Genome Biol. 2002, 3, research0008.0001. [Google Scholar] [CrossRef]
- Xie, B.-J.; Chen, P.-P.; Damgaard, J.; Xie, J.-Y.; Xie, Q.; Wang, Y.-H. Paraphyly of the subgenus Micronecta (Micronecta) Kirkaldy, 1897 (Hemiptera: Heteroptera: Micronectidae) based on mitochondrial genomes and nuclear rDNAs. Arthropod Syst. Phylogeny 2024, 82, 77–87. [Google Scholar] [CrossRef]
- Cheng, M.; Liu, Y.; Zheng, X.; Zhang, R.; Feng, K.; Yue, B.; Du, C.; Zhou, C. Characterization of seventeen complete mitochondrial genomes: Structural features and phylogenetic implications of the lepidopteran insects. Insects 2022, 13, 998. [Google Scholar] [CrossRef]
- Castro-Chavez, F. The rules of variation: Amino acid exchange according to the rotating circular genetic code. J. Theor. Biol. 2010, 264, 711–721. [Google Scholar] [CrossRef][Green Version]
- Castlejohn, C. Multi-Gene Phylogenetic Analysis of the Supergroup Excavata. Master’s Thesis, University of Georgia, Athens, GA, USA, 2009. [Google Scholar]
- Castle, J.C. SNPs occur in regions with less genomic sequence conservation. PLoS ONE 2011, 6, e20660. [Google Scholar] [CrossRef] [PubMed]
- Bast, F. Tutorial on phylogenetic inference—2. Resonance 2015, 20, 445–457. [Google Scholar] [CrossRef]
- Svanishvili, G. DIMT1 and Ribosomal Function: How Ribosomal Biogenesis Influences Lifespan and Energy Homeostasis. Science 2024, 1, 100027. [Google Scholar] [CrossRef]
- Sæther, O.A. Revision of Hydrobaenus, Trissocladius, Zalutschia, Paratrissocladius and Some Related Genera (Diptera, Chironomidae); Fisheries Research Board of Canada: Nanaimo, BC, Canada, 1976; Volume 195, 287p. [Google Scholar]
- Wang, X.-H.; Sæther, O.A. Xiaomyia, Shangomyia and Zhouomyia, three new and unusual genera of Chironomini from Oriental China (Diptera: Chironomidae). Insect Syst. Evol. 1993, 24, 185–195. [Google Scholar] [CrossRef]
- Tang, H.; Cranston, P.S. A new tribe in the Chironominae (Diptera: Chironomidae) validated by first immature stages of Xiaomyia Sæther & Wang and a phylogenetic review. Raffles Bull. Zool. 2019, 67, 684–693. [Google Scholar] [CrossRef]
- Rubinoff, D.; Holland, B.S. Between Two Extremes: Mitochondrial DNA is neither the Panacea nor the Nemesis of Phylogenetic and Taxonomic Inference. Syst. Biol. 2005, 54, 952–961. [Google Scholar] [CrossRef]
- Funk, D.J.; Omland, K.E. Species-level paraphyly and polyphyly: Frequency, causes, and consequences, with insights from animal mitochondrial DNA. Annu. Rev. Ecol. Evol. Syst. 2003, 34, 397–423. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, H.-F.; Xiao, X.-R.; Zhang, Z.-C.; Li, Y.-F.; Lin, X.-L. Mitogenomic Insights into Orthocladiinae (Diptera: Chironomidae): Structural Diversity and Phylogenetic Implications. Biology 2025, 14, 1178. https://doi.org/10.3390/biology14091178
Xu H-F, Xiao X-R, Zhang Z-C, Li Y-F, Lin X-L. Mitogenomic Insights into Orthocladiinae (Diptera: Chironomidae): Structural Diversity and Phylogenetic Implications. Biology. 2025; 14(9):1178. https://doi.org/10.3390/biology14091178
Chicago/Turabian StyleXu, Hai-Feng, Xiu-Ru Xiao, Zhi-Chao Zhang, Yu-Fan Li, and Xiao-Long Lin. 2025. "Mitogenomic Insights into Orthocladiinae (Diptera: Chironomidae): Structural Diversity and Phylogenetic Implications" Biology 14, no. 9: 1178. https://doi.org/10.3390/biology14091178
APA StyleXu, H.-F., Xiao, X.-R., Zhang, Z.-C., Li, Y.-F., & Lin, X.-L. (2025). Mitogenomic Insights into Orthocladiinae (Diptera: Chironomidae): Structural Diversity and Phylogenetic Implications. Biology, 14(9), 1178. https://doi.org/10.3390/biology14091178






