Systematic Survey and Expression Analysis of the Glutaredoxin Gene Family in Capsicum annuum Under Hypoxia Stress
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of GRX Genes in Pepper, Conserved Domain Organization, and Promoter Analysis
2.2. GRX Gene Phylogenetic Tree and Duplication Analysis
2.3. GRXs Gene Expression Analysis in Pepper
2.4. Submergence and Hypoxia Treatment and Quantitative RT-PCR
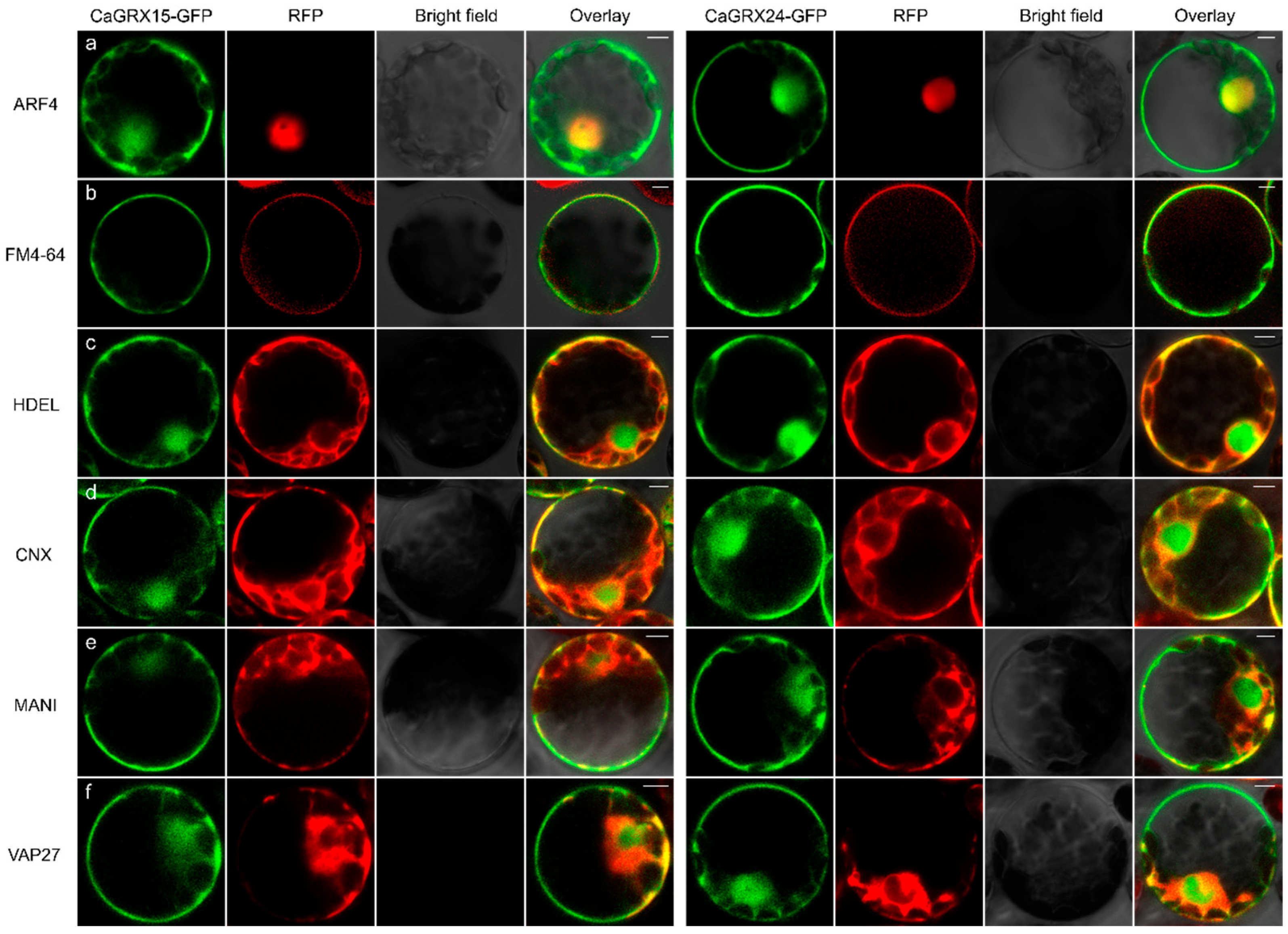
2.5. CaGRX Subcellular Localization Analysis
3. Results
3.1. Identification of GRX Members in Solanaceae Species Genomes
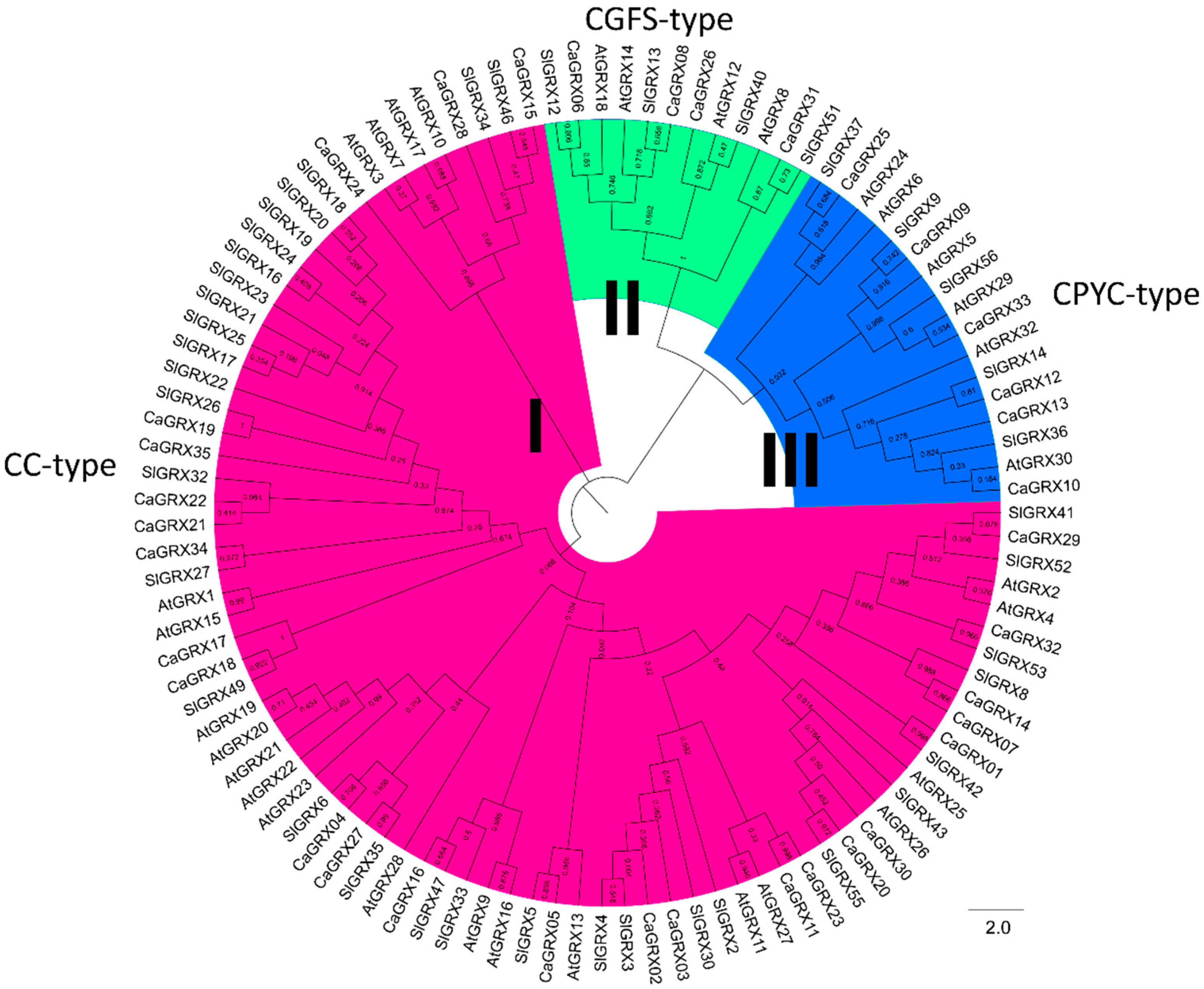
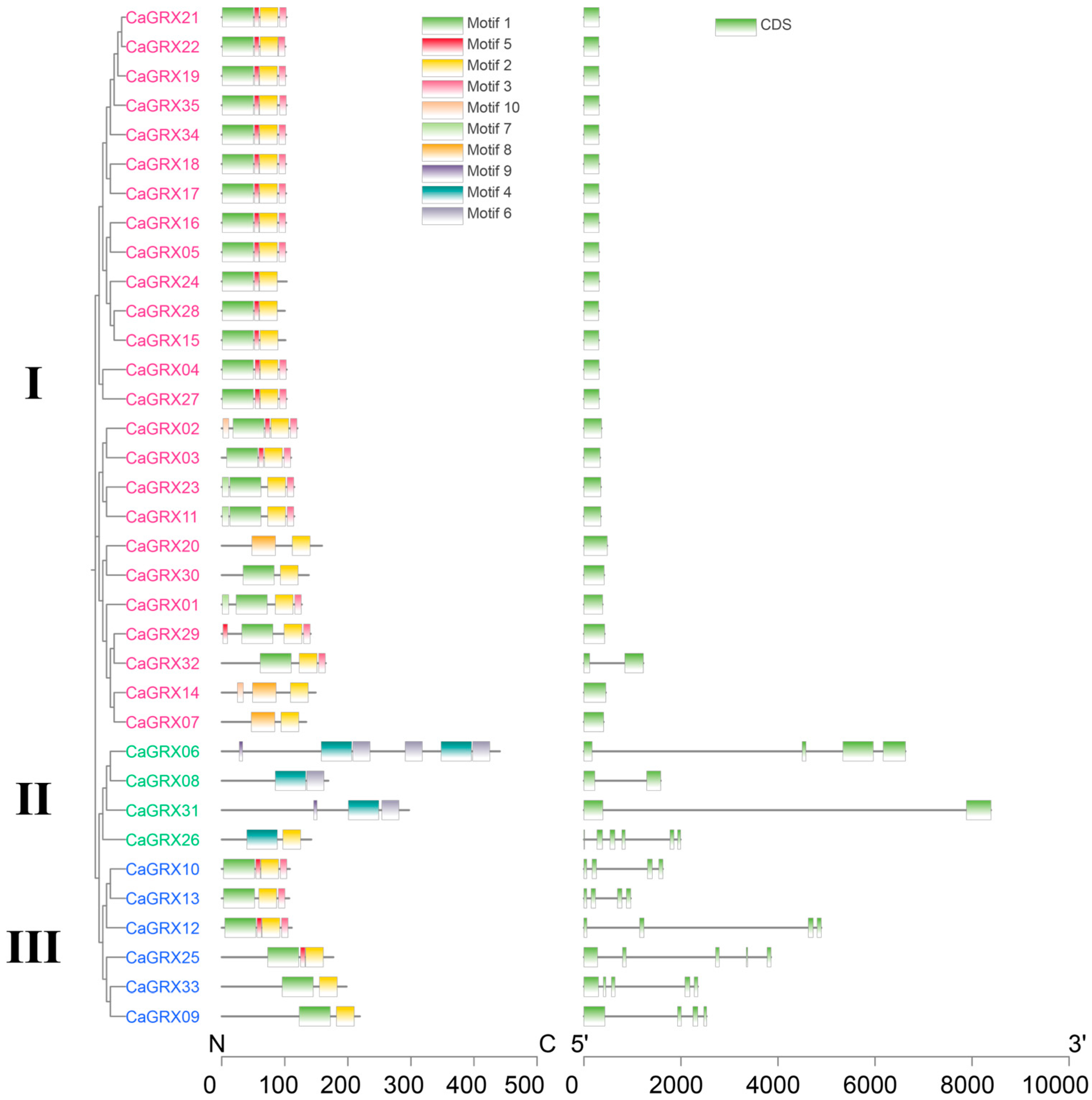
3.2. Phylogenetic Relationship and Structure Analysis
3.3. Pepper GRX Genes Location and Duplication
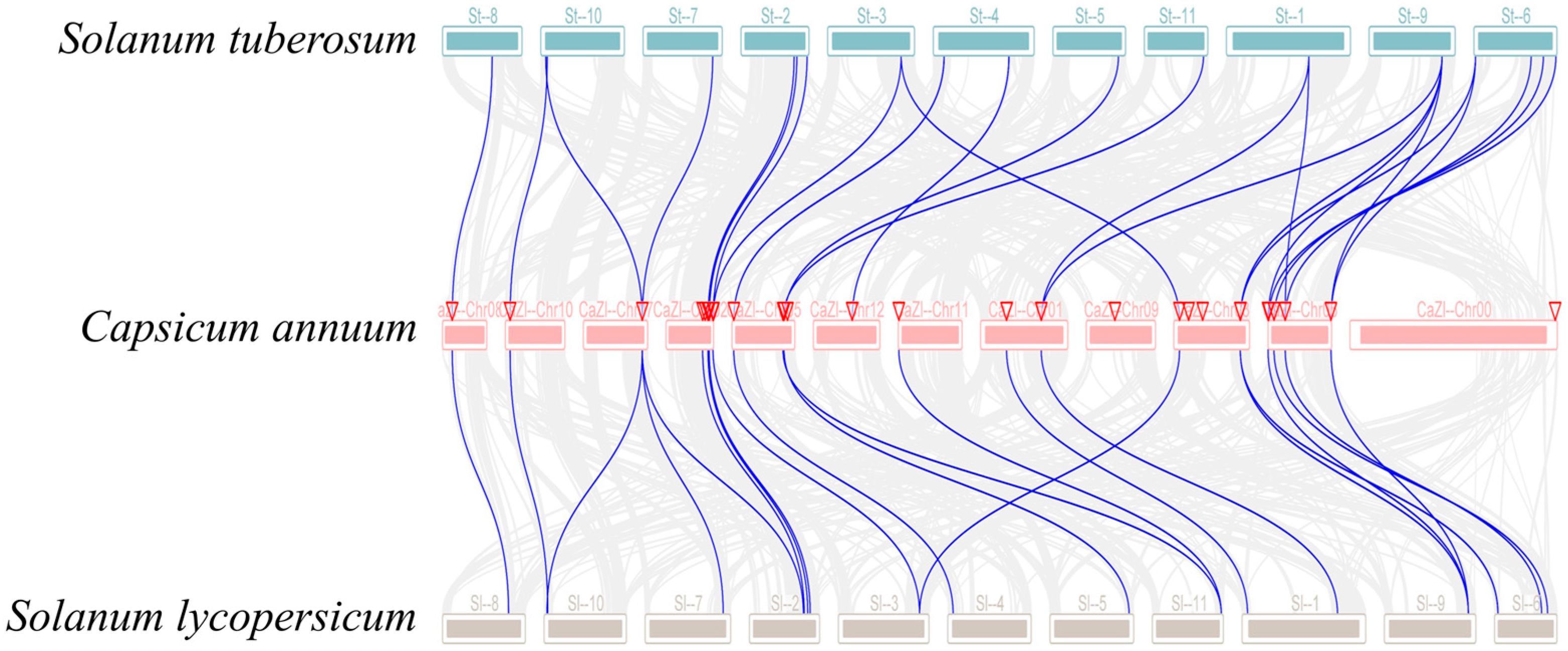
3.4. Collinearity Analysis of Solanaceae GRX Genes
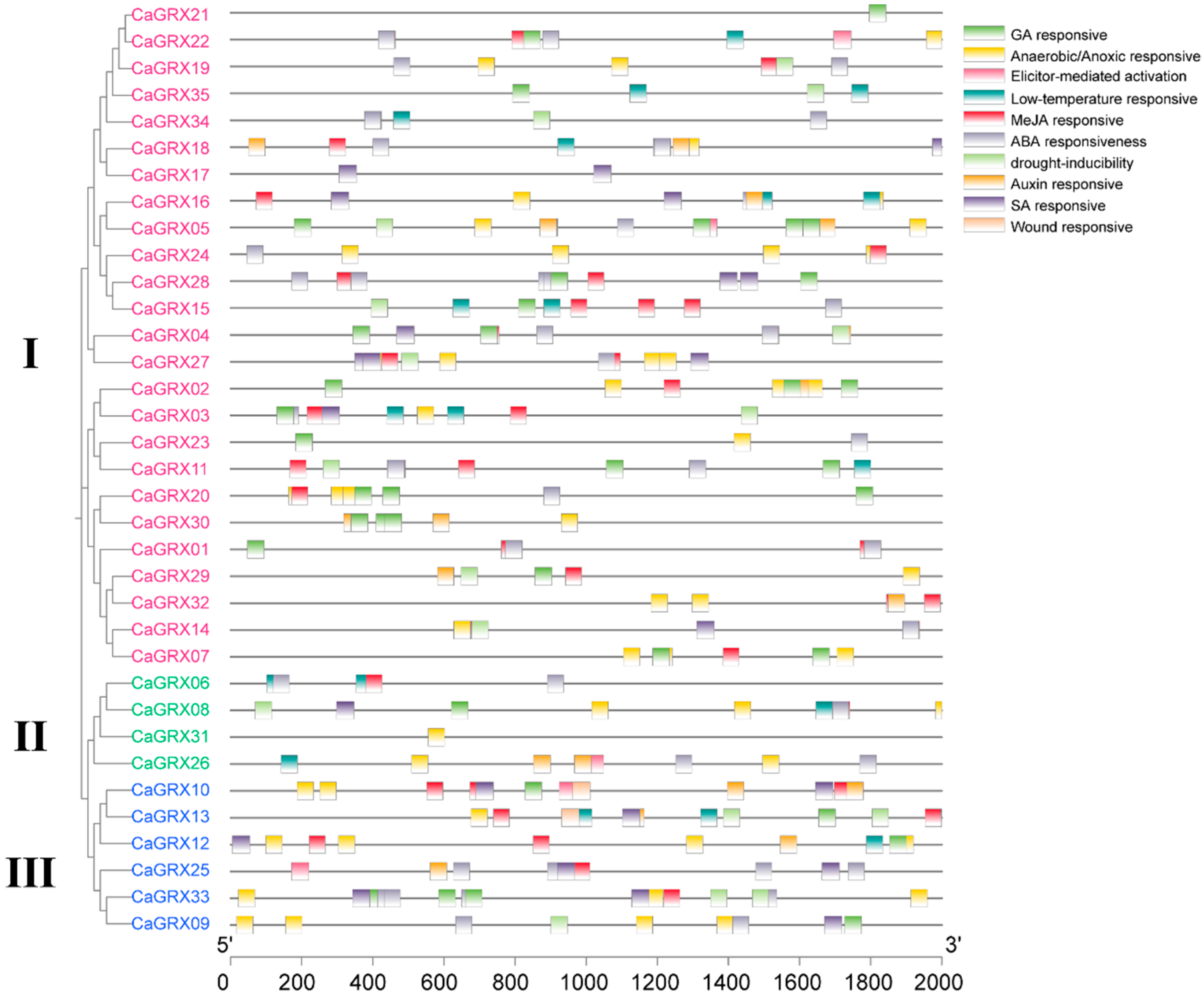
3.5. CRE Analysis of the CaGRX Gene Promoters
3.6. Expression Profile Analysis of CaGRX Genes
3.7. Expression Profiles of CaGRX Genes Under Phytohormone and Stress
3.8. Pepper GRX Genes in Response to Hypoxia and Submergence
3.9. Subcellular Localization of Selected CaGRX
4. Discussion
4.1. The CaGRX Genes Expansion Characterization
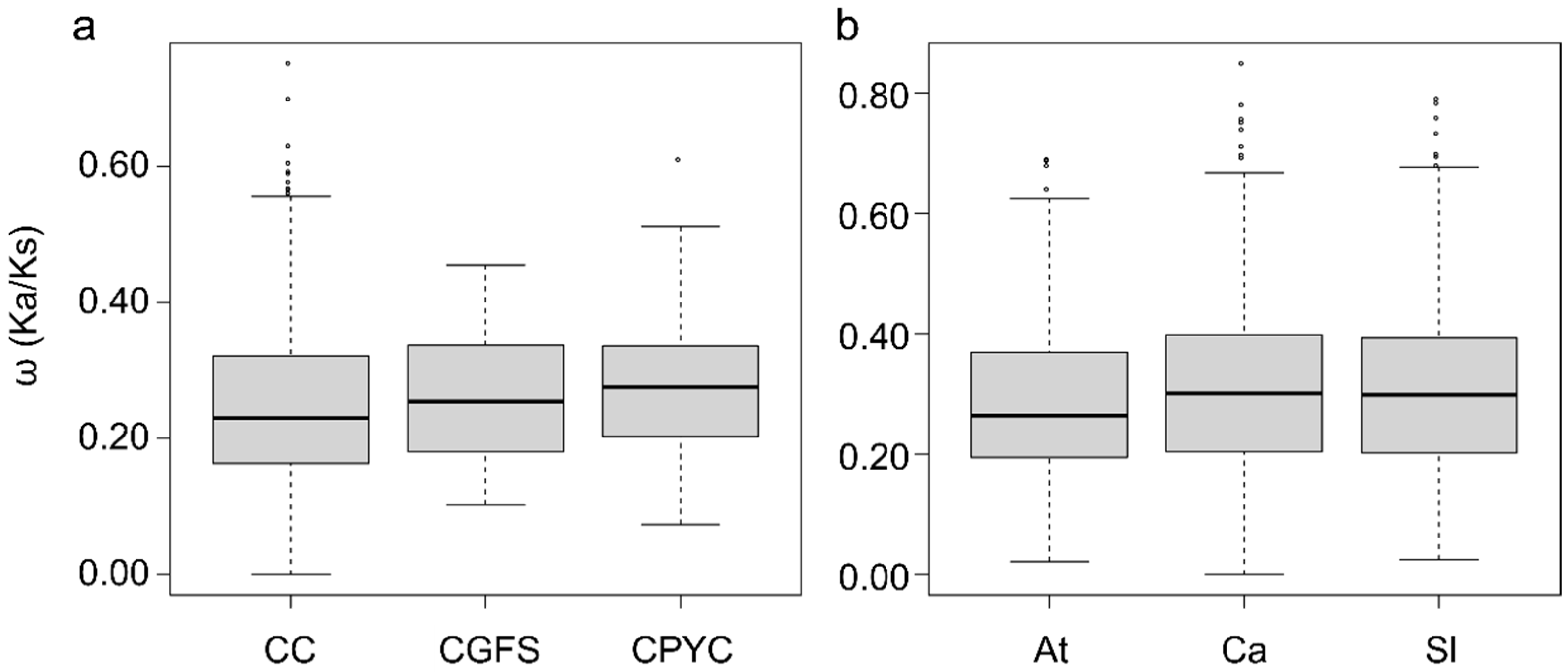
4.2. Solanaceae GRX Duplications and Collinearity Analysis
4.3. CaGRX Genes Roles in Plant Response to Stress
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- Mittler, R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef] [PubMed]
- Zang, J.; Klemm, S.; Pain, C.; Duckney, P.; Bao, Z.; Stamm, G.; Kriechbaumer, V.; Bürstenbinder, K.; Hussey, P.J.; Wang, P. A novel plant actin-microtubule bridging complex regulates cytoskeletal and ER structure at ER-PM contact sites. Curr. Biol. 2021, 31, 1251–1260.e1254. [Google Scholar] [CrossRef]
- Meyer, Y.; Belin, C.; Delorme-Hinoux, V.; Reichheld, J.P.; Riondet, C. Thioredoxin and glutaredoxin systems in plants: Molecular mechanisms, crosstalks, and functional significance. Antioxid. Redox Signal. 2012, 17, 1124–1160. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, A.; López-Martínez, R.; Martí, M.C.; Cano-Yelo, D.; Sevilla, F. The integration of TRX/GRX systems and phytohormonal signalling pathways in plant stress and development. Plant Physiol. Biochem. 2024, 207, 108298. [Google Scholar] [CrossRef]
- Meyer, Y.; Vignols, F.; Reichheld, J.P. Classification of plant thioredoxins by sequence similarity and intron position. Methods Enzymol. 2002, 347, 394–402. [Google Scholar]
- Rouhier, N.; Couturier, J.; Jacquot, J.P. Genome-wide analysis of plant glutaredoxin systems. J. Exp. Bot. 2006, 57, 1685–1696. [Google Scholar] [CrossRef]
- Esposito, S.; Cappetta, E.; Tranchida-Lombardo, V.; Batelli, G.; Ruggiero, A.; Ruocco, M.; Sportelli, G.; Cillo, F.; De Palma, M. Genome-wide survey of glutaredoxin gene family in four Solanaceae species and exploitation of duplicated CC-type following different environmental stimuli in tomato (Solanum lycopersicum). Sci. Hortic. 2023, 319, 112188. [Google Scholar] [CrossRef]
- Sha, S.; Minakuchi, K.; Higaki, N.; Sato, K.; Ohtsuki, K.; Kurata, A.; Yoshikawa, H.; Kotaru, M.; Masumura, T.; Ichihara, K.; et al. Purification and characterization of glutaredoxin (thioltransferase) from rice (Oryza sativa L.). J. Biochem. 1997, 121, 842–848. [Google Scholar] [CrossRef]
- Lemaire, S.D. The glutaredoxin family in oxygenic photosynthetic organisms. Photosynth. Res. 2004, 79, 305–318. [Google Scholar] [CrossRef]
- Rouhier, N.; Gelhaye, E.; Jacquot, J.P. Plant glutaredoxins: Still mysterious reducing systems. Cell. Mol. Life Sci. 2004, 61, 1266–1277. [Google Scholar] [CrossRef]
- Garg, R.; Jhanwar, S.; Tyagi, A.K.; Jain, M. Genome-wide survey and expression analysis suggest diverse roles of glutaredoxin gene family members during development and response to various stimuli in rice. DNA Res. Int. J. Rapid Publ. Rep. Genes Genomes 2010, 17, 353–367. [Google Scholar] [CrossRef]
- Laporte, D.; Olate, E.; Salinas, P.; Salazar, M.; Jordana, X.; Holuigue, L. Glutaredoxin GRXS13 plays a key role in protection against photooxidative stress in Arabidopsis. J. Biol. Chem. 2012, 63, 503–515. [Google Scholar] [CrossRef]
- Malik, W.A.; Wang, X.; Wang, X.; Shu, N.; Cui, R.; Chen, X.; Wang, D.; Lu, X.; Yin, Z.; Wang, J.; et al. Genome-wide expression analysis suggests glutaredoxin genes response to various stresses in cotton. Int. J. Biol. Macromol. 2020, 153, 470–491. [Google Scholar] [CrossRef]
- Couturier, J.; Jacquot, J.-P.; Rouhier, N. Evolution and diversity of glutaredoxins in photosynthetic organisms. Cell. Mol. Life Sci. 2009, 66, 2539–2557. [Google Scholar] [CrossRef] [PubMed]
- Boubakri, H.; Najjar, E.; Jihnaoui, N.; Chihaoui, S.-A.; Barhoumi, F.; Jebara, M. Genome-wide identification, characterization and expression analysis of glutaredoxin gene family (Grxs) in Phaseolus vulgaris. Gene 2022, 833, 146591. [Google Scholar] [CrossRef] [PubMed]
- Ehrary, A.; Rosas, M.; Carpinelli, S.; Davalos, O.; Cowling, C.; Fernandez, F.; Escobar, M. Glutaredoxin AtGRXS8 represses transcriptional and developmental responses to nitrate in Arabidopsis thaliana roots. Plant Direct 2020, 4, e00227. [Google Scholar] [CrossRef]
- Cheng, N.-H.; Liu, J.-Z.; Liu, X.; Wu, Q.; Thompson, S.M.; Lin, J.; Chang, J.; Whitham, S.A.; Park, S.; Cohen, J.D.; et al. Arabidopsis Monothiol Glutaredoxin, AtGRXS17, Is Critical for Temperature-dependent Postembryonic Growth and Development via Modulating Auxin Response. J. Biol. Chem. 2011, 286, 20398–20406. [Google Scholar] [CrossRef]
- Delorme-Hinoux, V.; Bangash, S.A.K.; Meyer, A.J.; Reichheld, J.-P. Nuclear thiol redox systems in plants. Plant Sci. 2016, 243, 84–95. [Google Scholar] [CrossRef]
- Xing, S.; Rosso, M.G.; Zachgo, S. ROXY1, a member of the plant glutaredoxin family, is required for petal development in Arabidopsis thaliana. Development 2005, 132, 1555–1565. [Google Scholar] [CrossRef] [PubMed]
- Xing, S.; Zachgo, S. ROXY1 and ROXY2, two Arabidopsis glutaredoxin genes, are required for anther development. Plant J. 2007, 53, 790–801. [Google Scholar] [CrossRef]
- Ndamukong, I.; Abdallat, A.A.; Thurow, C.; Fode, B.; Zander, M.; Weigel, R.; Gatz, C. SA-inducible Arabidopsis glutaredoxin interacts with TGA factors and suppresses JA-responsive PDF1.2 transcription. Plant J. 2007, 50, 128–139. [Google Scholar] [CrossRef]
- Kumar, A.; Dubey, A.K.; Kumar, V.; Ansari, M.A.; Narayan, S.; Meenakshi Kumar, S.; Pandey, V.; Shirke, P.A.; Pande, V.; Sanyal, I. Overexpression of rice glutaredoxin genes LOC_Os02g40500 and LOC_Os01g27140 regulate plant responses to drought stress. Ecotox. Environ. Safe 2020, 200, 110721. [Google Scholar] [CrossRef] [PubMed]
- Verma, P.K.; Verma, S.; Pande, V.; Mallick, S.; Deo Tripathi, R.; Dhankher, O.P.; Chakrabarty, D. Overexpression of Rice Glutaredoxin OsGrx_C7 and OsGrx_C2.1 Reduces Intracellular Arsenic Accumulation and Increases Tolerance in Arabidopsis thaliana. Front. Plant Sci. 2016, 7, 740. [Google Scholar]
- Liu, S.; Fu, H.; Jiang, J.; Chen, Z.; Gao, J.; Shu, H.; Zhang, S.; Yang, C.; Liu, J. Overexpression of a CPYC-Type Glutaredoxin, OsGrxC2.2, Causes Abnormal Embryos and an Increased Grain Weight in Rice. Front. Plant Sci. 2019, 10, 848. [Google Scholar] [CrossRef]
- Ning, X.; Sun, Y.; Wang, C.; Zhang, W.; Sun, M.; Hu, H.; Liu, J.; Yang, L. A Rice CPYC-Type Glutaredoxin OsGRX20 in Protection against Bacterial Blight, Methyl Viologen and Salt Stresses. Front. Plant Sci. 2018, 9, 111. [Google Scholar] [CrossRef]
- Son, S.; Kim, H.; Lee, K.S.; Kim, S.; Park, S.R. Rice glutaredoxin GRXS15 confers broad-spectrum resistance to Xanthomonas oryzae pv. oryzae and Fusarium fujikuroi. Biochem. Biophys. Res. Commun. 2020, 533, 1385–1392. [Google Scholar] [CrossRef]
- Sharma, R.; Priya, P.; Jain, M. Modified expression of an auxin-responsive rice CC-type glutaredoxin gene affects multiple abiotic stress responses. Planta 2013, 238, 871–884. [Google Scholar] [CrossRef]
- Yang, R.S.; Xu, F.; Wang, Y.M.; Zhong, W.S.; Dong, L.; Shi, Y.N.; Tang, T.J.; Sheng, H.J.; Jackson, D.; Yang, F. Glutaredoxins regulate maize inflorescence meristem development via redox control of TGA transcriptional activity. Nat. Plants 2021, 7, 1589–1601. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Huang, C.; Xie, Y.; Song, F.; Zhou, X. A tomato glutaredoxin gene SlGRX1 regulates plant responses to oxidative, drought and salt stresses. Planta 2010, 232, 1499–1509. [Google Scholar] [CrossRef]
- Kumar, A.; Dubey, A.K.; Kumar, V.; Ansari, M.A.; Narayan, S.; Meenakshi Kumar, S.; Pandey, V.; Shirke, P.A.; Pande, V.; Sanyal, I. Over-expression of chickpea glutaredoxin (CaGrx) provides tolerance to heavy metals by reducing metal accumulation and improved physiological and antioxidant defence system. Ecotox. Environ. Safe 2020, 192, 110252. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, X.; Zhang, L.; Chen, S. CC-type glutaredoxin gene CsGRX4 in cucumber responds to Botrytis cinerea via JA signaling pathway. Sci. Hortic. 2022, 306, 111440. [Google Scholar] [CrossRef]
- Hou, J.; Zhang, Q.; Zhou, Y.; Ahammed, G.J.; Zhou, Y.; Yu, J.; Fang, H.; Xia, X. Glutaredoxin GRXS16 mediates brassinosteroid-induced apoplastic H2O2 production to promote pesticide metabolism in tomato. Environ. Pollut. 2018, 240, 227–234. [Google Scholar] [CrossRef]
- Kumar, R.M.S.; Ramesh, S.V.; Sun, Z.; Thankappan, S.; Nulu, N.P.C.; Binodh, A.K.; Kalaipandian, S.; Srinivasan, R. Capsicum chinense Jacq.-derived glutaredoxin (CcGRXS12) alters redox status of the cells to confer resistance against pepper mild mottle virus (PMMoV-I). Plant Cell Rep. 2024, 43, 108. [Google Scholar] [CrossRef]
- Xu, H.; Li, Z.; Jiang, P.F.; Zhao, L.; Qu, C.; Van de Peer, Y.; Liu, Y.J.; Zeng, Q.Y. Divergence of active site motifs among different classes of Populus glutaredoxins results in substrate switches. Plant J. 2022, 110, 129–146. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.; He, F.; Tang, W.; Du, H.; Wang, H. Identification of Maize CC-Type Glutaredoxins That Are Associated with Response to Drought Stress. Genes 2019, 10, 610. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Li, M.; Jiang, Y.; Duan, X. Genome-wide identification, characterization and expression profile of glutaredoxin gene family in relation to fruit ripening and response to abiotic and biotic stresses in banana (Musa acuminata). Int. J. Biol. Macromol. 2021, 170, 636–651. [Google Scholar] [CrossRef] [PubMed]
- Ruan, M.B.; Yang, Y.L.; Li, K.M.; Guo, X.; Wang, B.; Yu, X.L.; Peng, M. Identification and characterization of drought-responsive CC-type glutaredoxins from cassava cultivars reveals their involvement in ABA signalling. BMC Plant Biol. 2018, 18, 329. [Google Scholar] [CrossRef]
- Bal, S.; Sharangi, A.B.; Upadhyay, T.K.; Khan, F.; Pandey, P.; Siddiqui, S.; Saeed, M.; Lee, H.J.; Yadav, D.K. Biomedical and Antioxidant Potentialities in Chilli: Perspectives and Way Forward. Molecules 2022, 27, 6380. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-H.; Siddique, M.I.; Jang, S.; Kim, G.-W.; Choi, G.J.; Kwon, J.-K.; Kang, B.-C. Identification of QTLs associated with resistance to bacterial wilt in pepper (Capsicum annuum L.) through bi-parental QTL mapping and genome-wide association analysis. Sci. Hortic. 2024, 329, 112987. [Google Scholar] [CrossRef]
- Pang, X.; Chen, J.; Li, L.Z.; Huang, W.J.; Liu, J. Deciphering Drought Resilience in Solanaceae Crops: Unraveling Molecular and Genetic Mechanisms. Biology 2024, 13, 1076. [Google Scholar] [CrossRef]
- Kim, S.; Park, M.; Yeom, S.I.; Kim, Y.M.; Lee, J.M.; Lee, H.A.; Seo, E.; Choi, J.; Cheong, K.; Kim, K.T.; et al. Genome sequence of the hot pepper provides insights into the evolution of pungency in species. Nat. Genet. 2014, 46, 270–278. [Google Scholar] [CrossRef]
- Liu, F.; Yu, H.; Deng, Y.; Zheng, J.; Liu, M.; Ou, L.; Yang, B.; Dai, X.; Ma, Y.; Feng, S.; et al. PepperHub, an Informatics Hub for the Chili Pepper Research Community. Mol. Plant 2017, 10, 1129–1132. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Ma, S.; Guo, Y.; Zhang, X.; Liu, D.; Gao, Y.; Zhai, C.; Chen, Q.; Xiao, S.; Zhang, Z.; et al. Evolution and Expression of the Meprin and TRAF Homology Domain-Containing Gene Family in Solanaceae. Int. J. Mol. Sci. 2023, 24, 8782. [Google Scholar] [CrossRef]
- Finn, R.D.; Mistry, J.; Schuster-Bockler, B.; Griffiths-Jones, S.; Hollich, V.; Lassmann, T.; Moxon, S.; Marshall, M.; Khanna, A.; Durbin, R.; et al. Pfam: Clans, web tools and services. Nucleic Acids Res. 2006, 34, D247–D251. [Google Scholar] [CrossRef]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013, 41, e121. [Google Scholar] [CrossRef]
- Kersey, P.J.; Allen, J.E.; Christensen, M.; Davis, P.; Falin, L.J.; Grabmueller, C.; Hughes, D.S.T.; Humphrey, J.; Kerhornou, A.; Khobova, J.; et al. Ensembl Genomes 2013: Scaling up access to genome-wide data. Nucleic Acids Res. 2014, 42, D546–D552. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.J.; Liu, D.; Chen, S.Y.; Dai, Y.S.; Guo, W.X.; Zhang, X.; Wang, L.N.; Ma, S.R.; Xiao, M.; Qi, H.; et al. Evolution and Expression of the Membrane Attack Complex and Perforin Gene Family in the Poaceae. Int. J. Mol. Sci. 2020, 21, 5736. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.R.; Guo, Y.X.; Zhang, T.Y.; Liu, D.; Wang, L.N.; Hu, R.W.; Zhou, D.M.; Zhou, Y.; Chen, Q.F.; Yu, L.J. Comprehensive Identification and Expression Analysis of the Multidrug and Toxic Compound Extrusion (MATE) Gene Family in Brachypodium distachyon. Plants 2024, 13, 2586. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Ji, W.; Mandal, S.; Rezenom, Y.H.; McKnight, T.D. Specialized metabolism by trichome-enriched Rubisco and fatty acid synthase components. Plant Physiol. 2023, 191, 1199–1213. [Google Scholar] [CrossRef]
- Mandal, S.; Rezenom, Y.H.; McKnight, T.D. Role of LEAFLESS, an AP2/ERF family transcription factor, in the regulation of trichome specialized metabolism. New Phytol. 2025, 247, 774–790. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Dai, Y.S.; Liu, D.; Guo, W.X.; Liu, Z.X.; Zhang, X.; Shi, L.L.; Zhou, D.M.; Wang, L.N.; Kang, K.; Wang, F.Z.; et al. Poaceae-specific β-1,3;1,4-D-glucans link jasmonate signalling to OsLecRK1-mediated defence response during rice-brown planthopper interactions. Plant Biotechnol. J. 2023, 21, 1286–1300. [Google Scholar] [CrossRef]
- Yu, L.J.; Chen, F.; Peng, Y.J.; Xie, L.J.; Liu, D.; Han, M.Q.; Chen, F.; Xiao, S.; Huang, J.C.; Li, J. Arabidopsis thaliana Plants Engineered To Produce Astaxanthin Show Enhanced Oxidative Stress Tolerance and Bacterial Pathogen Resistance. J. Agric. Food Chem. 2019, 67, 12590–12598. [Google Scholar] [CrossRef]
- Tang, B.Q.; Yang, H.P.; Zhang, X.H.; Du, J.; Xie, L.L.; Dai, X.Z.; Zou, X.X.; Liu, F. A global view of transcriptome dynamics during flower development in Capsicum annuum L. Hortic. Plant J. 2023, 9, 999–1012. [Google Scholar]
- Ma, S.; Guo, Y.; Liu, D.; Zhang, X.; Guo, J.; Zhang, T.; Lai, L.; Li, Y.; Chen, Q.; Yu, L. Genome-Wide Analysis of the Membrane Attack Complex and Perforin Genes and Their Expression Pattern under Stress in the Solanaceae. Int. J. Mol. Sci. 2023, 24, 13193. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, X.; Qi, T.C.; Wang, Z.H.; Wang, Y.; Wang, L.N.; Zeng, Y.L.; He, H.J.; Jiang, L.W.; Xie, D.X.; et al. Arabidopsis MACP2 contributes to autophagy induction by modulating starvation-induced reactive oxygen species homeostasis. Adv. Biotechnol. 2025, 3, 25. [Google Scholar] [CrossRef]
- Huang, L.; Yu, L.-J.; Zhang, X.; Fan, B.; Wang, F.-Z.; Dai, Y.-S.; Qi, H.; Zhou, Y.; Xie, L.-J.; Xiao, S. Autophagy regulates glucose-mediated root meristem activity by modulating ROS production in Arabidopsis. Autophagy 2018, 15, 407–422. [Google Scholar] [CrossRef] [PubMed]
- Fan, B.; Liao, K.; Wang, L.N.; Shi, L.L.; Zhang, Y.; Xu, L.J.; Zhou, Y.; Li, J.F.; Chen, Y.Q.; Chen, Q.F.; et al. Calcium-dependent activation of CPK12 facilitates its cytoplasm-to-nucleus translocation to potentiate plant hypoxia sensing by phosphorylating ERF-VII transcription factors. Mol. Plant 2023, 16, 979–998. [Google Scholar] [CrossRef]
- Yu, W.W.; Chen, Q.F.; Liao, K.; Zhou, D.M.; Yang, Y.C.; He, M.; Yu, L.J.; Guo, D.Y.; Xiao, S.; Xie, R.H.; et al. The calcium-dependent protein kinase CPK16 regulates hypoxia-induced ROS production by phosphorylating the NADPH oxidase RBOHD in Arabidopsis. Plant Cell 2024, 36, 3451–3466. [Google Scholar] [CrossRef]
- Akhunov, E.D.; Sehgal, S.; Liang, H.; Wang, S.; Akhunova, A.R.; Kaur, G.; Li, W.; Forrest, K.L.; See, D.; Simkova, H.; et al. Comparative analysis of syntenic genes in grass genomes reveals accelerated rates of gene structure and coding sequence evolution in polyploid wheat. Plant Physiol. 2013, 161, 252–265. [Google Scholar] [CrossRef]
- Hurst, L.D. The Ka/Ks ratio: Diagnosing the form of sequence evolution. Trends Genet. 2002, 18, 486. [Google Scholar] [CrossRef] [PubMed]
- Nei, M. The new mutation theory of phenotypic evolution. Proc. Natl. Acad. Sci. USA 2007, 104, 12235–12242. [Google Scholar] [CrossRef]
- Zhang, X.; Dai, Y.S.; Wang, Y.X.; Su, Z.Z.; Yu, L.J.; Zhang, Z.F.; Xiao, S.; Chen, Q.F. Overexpression of the Arabidopsis MACPF Protein AtMACP2 Promotes Pathogen Resistance by Activating SA Signaling. Int. J. Mol. Sci. 2022, 23, 8784. [Google Scholar] [CrossRef] [PubMed]
- Gomord, V.; Denmat, L.A.; Fitchette-Laine, A.C.; Satiat-Jeunemaitre, B.; Hawes, C.; Faye, L. The C-terminal HDEL sequence is sufficient for retention of secretory proteins in the endoplasmic reticulum (ER) but promotes vacuolar targeting of proteins that escape the ER. Plant J. 1997, 11, 313–325. [Google Scholar] [CrossRef]
- Ellgaard, L.; Molinari, M.; Helenius, A. Setting the standards: Quality control in the secretory pathway. Science 1999, 286, 1882–1888. [Google Scholar] [CrossRef]
- Liu, F.; Zhao, J.; Sun, H.; Xiong, C.; Sun, X.; Wang, X.; Wang, Z.; Jarret, R.; Wang, J.; Tang, B.; et al. Genomes of cultivated and wild Capsicum species provide insights into pepper domestication and population differentiation. Nat. Commun. 2023, 14, 5487. [Google Scholar] [CrossRef]
- Qin, C.; Yu, C.S.; Shen, Y.O.; Fang, X.D.; Chen, L.; Min, J.M.; Cheng, J.W.; Zhao, S.C.; Xu, M.; Luo, Y.; et al. Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc. Natl. Acad. Sci. USA 2014, 111, 5135–5140. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Tabata, S.; Hirakawa, H.; Asamizu, E.; Shirasawa, K.; Isobe, S.; Kaneko, T.; Nakamura, Y.; Shibata, D.; Aoki, K.; et al. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 2012, 485, 635–641. [Google Scholar] [CrossRef]
- Xu, X.; Pan, S.; Cheng, S.; Zhang, B.; Mu, D.; Ni, P.; Zhang, G.; Yang, S.; Li, R.; Wang, J.; et al. Genome sequence and analysis of the tuber crop potato. Nature 2011, 475, 189–195. [Google Scholar] [CrossRef]
- Wang, Z.; Xing, S.; Birkenbihl, R.P.; Zachgo, S. Conserved functions of Arabidopsis and rice CC-type glutaredoxins in flower development and pathogen response. Mol. Plant 2009, 2, 323–335. [Google Scholar] [CrossRef]
- La Camera, S.; L’Haridon, F.; Astier, J.; Zander, M.; Abou-Mansour, E.; Page, G.; Thurow, C.; Wendehenne, D.; Gatz, C.; Métraux, J.P.; et al. The glutaredoxin ATGRXS13 is required to facilitate Botrytis cinerea infection of Arabidopsis thaliana plants. Plant J. 2011, 68, 507–519. [Google Scholar] [CrossRef]
- Bisaro, D.M.; Zhao, W.; Zhou, Y.; Zhou, X.; Wang, X.; Ji, Y. Host GRXC6 restricts Tomato yellow leaf curl virus infection by inhibiting the nuclear export of the V2 protein. PLoS Pathog. 2021, 17, e1009844. [Google Scholar]
- Cui, J.; Luan, Y.; Jiang, N.; Bao, H.; Meng, J. Comparative transcriptome analysis between resistant and susceptible tomato allows the identification of lncRNA16397 conferring resistance to Phytophthora infestans by co-expressing glutaredoxin. Plant J. 2017, 89, 577–589. [Google Scholar] [CrossRef] [PubMed]
- Matsushima, R.; Hayashi, Y.; Kondo, M.; Shimada, T.; Nishimura, M.; Hara-Nishimura, I. An endoplasmic reticulum-derived structure that is induced under stress conditions in Arabidopsis. Plant Physiol. 2002, 130, 1807–1814. [Google Scholar] [CrossRef]
- Yang, J.; Zhai, N.; Chen, Y.; Wang, L.; Chen, R.; Pan, H. A signal peptide peptidase is required for ER-symbiosome proximal association and protein secretion. Nat. Commun. 2023, 14, 4355. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Richardson, C.; Hawkins, T.J.; Sparkes, I.; Hawes, C.; Hussey, P.J. Plant VAP27 proteins: Domain characterization, intracellular localization and role in plant development. New Phytol. 2016, 210, 1311–1326. [Google Scholar] [CrossRef]
- Zhang, T.; Li, Y.; Li, C.; Zang, J.; Gao, E.; Kroon, J.T.; Qu, X.; Hussey, P.J.; Wang, P. Exo84c interacts with VAP27 to regulate exocytotic compartment degradation and stigma senescence. Nat. Commun. 2023, 14, 4888. [Google Scholar] [CrossRef]
- Schlösser, M.; Moseler, A.; Bodnar, Y.; Homagk, M.; Wagner, S.; Pedroletti, L.; Gellert, M.; Ugalde, J.M.; Lillig, C.H.; Meyer, A.J. Localization of four class I glutaredoxins in the cytosol and the secretory pathway and characterization of their biochemical diversification. Plant J. 2024, 118, 1455–1474. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Suen, P.K.; Wang, X.; Lin, Y.; Lo, S.W.; Rojo, E.; Jiang, L. An in vivo expression system for the identification of cargo proteins of vacuolar sorting receptors in Arabidopsis culture cells. Plant J. 2013, 75, 1003–1017. [Google Scholar] [CrossRef]
- Wang, P.; Hawkins Timothy, J.; Richardson, C.; Cummins, I.; Deeks Michael, J.; Sparkes, I.; Hawes, C.; Hussey Patrick, J. The Plant Cytoskeleton, NET3C, and VAP27 Mediate the Link between the Plasma Membrane and Endoplasmic Reticulum. Curr. Biol. 2014, 24, 1397–1405. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, Y.; Ma, S.; Li, Z.; Yu, Y.; Liu, D.; Zhang, T.; Hu, R.; Zhou, D.; Zhou, Y.; Xiao, S.; et al. Systematic Survey and Expression Analysis of the Glutaredoxin Gene Family in Capsicum annuum Under Hypoxia Stress. Biology 2025, 14, 1106. https://doi.org/10.3390/biology14091106
Guo Y, Ma S, Li Z, Yu Y, Liu D, Zhang T, Hu R, Zhou D, Zhou Y, Xiao S, et al. Systematic Survey and Expression Analysis of the Glutaredoxin Gene Family in Capsicum annuum Under Hypoxia Stress. Biology. 2025; 14(9):1106. https://doi.org/10.3390/biology14091106
Chicago/Turabian StyleGuo, Yixian, Sirui Ma, Ziying Li, Yang Yu, Di Liu, Tianyi Zhang, Ruiwen Hu, Demian Zhou, Ying Zhou, Shi Xiao, and et al. 2025. "Systematic Survey and Expression Analysis of the Glutaredoxin Gene Family in Capsicum annuum Under Hypoxia Stress" Biology 14, no. 9: 1106. https://doi.org/10.3390/biology14091106
APA StyleGuo, Y., Ma, S., Li, Z., Yu, Y., Liu, D., Zhang, T., Hu, R., Zhou, D., Zhou, Y., Xiao, S., Chen, Q., & Yu, L. (2025). Systematic Survey and Expression Analysis of the Glutaredoxin Gene Family in Capsicum annuum Under Hypoxia Stress. Biology, 14(9), 1106. https://doi.org/10.3390/biology14091106




