A Genome-Wide Association Study of Rib Number and Thoracolumbar Vertebra Number in a Landrace × Yorkshire Crossbred Pig Population
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Phenotypic and Genotypic Data Preparation
2.2. Genetic Variance Estimation
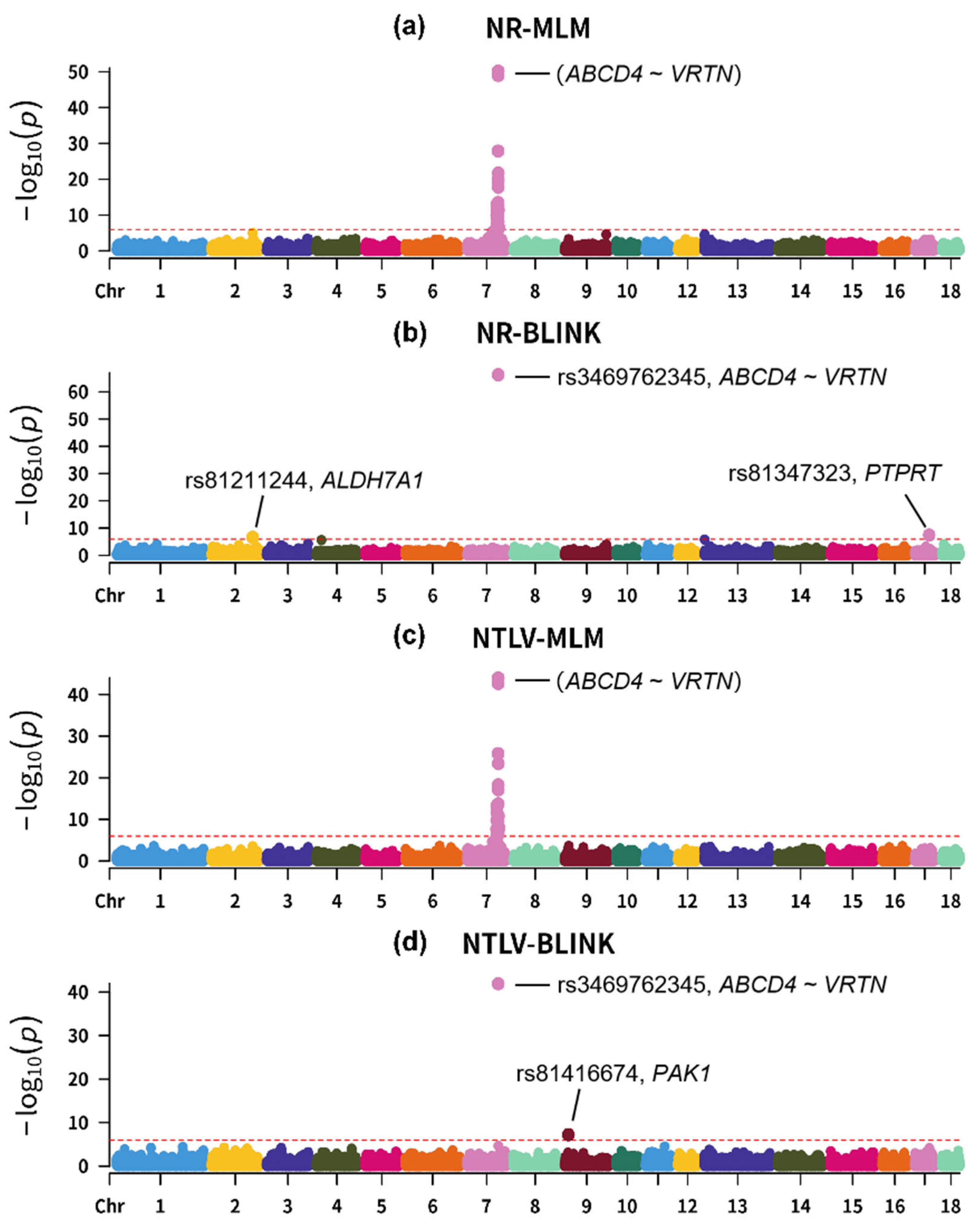
2.3. Genome-Wide Association Studies
3. Results and Discussion
3.1. Descriptive Statistics and Genetic Parameters for the Phenotype
3.2. Results of Genome-Wide Association Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, L.C.; Yue, J.W.; Pu, L.; Wang, L.G.; Liu, X.; Liang, J.; Yan, H.; Zhao, K.B.; Li, N.; Shi, H.B.; et al. Genome-wide study refines the quantitative trait locus for number of ribs in a Large White × Minzhu intercross pig population and reveals a new candidate gene. Mol. Genet. Genom. 2016, 291, 1885–1890. [Google Scholar] [CrossRef] [PubMed]
- Rohrer, G.A.; Nonneman, D.J.; Wiedmann, R.T.; Schneider, J.F. A study of vertebra number in pigs confirms the association of vertnin and reveals additional QTL. BMC Genet. 2015, 16, 129. [Google Scholar] [CrossRef]
- van Son, M.; Lopes, M.S.; Martell, H.J.; Derks, M.F.L.; Gangsei, L.E.; Kongsro, J.; Wass, M.N.; Grindflek, E.H.; Harlizius, B. A QTL for Number of Teats Shows Breed Specific Effects on Number of Vertebrae in Pigs: Bridging the Gap Between Molecular and Quantitative Genetics. Front. Genet. 2019, 10, 272. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.L.; Park, C.A.; Reecy, J.M. Bringing the Animal QTLdb and CorrDB into the future: Meeting new challenges and providing updated services. Nucleic Acids Res. 2022, 50, D956–D961. [Google Scholar] [CrossRef]
- Liu, K.; Hou, L.; Yin, Y.; Wang, B.; Liu, C.; Zhou, W.; Niu, P.; Li, Q.; Huang, R.; Li, P. Genome-wide association study reveals new QTL and functional candidate genes for the number of ribs and carcass length in pigs. Anim. Genet. 2023, 54, 435–445. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, M.; Yuan, J.; Zhou, X.; Xu, S.; Liu, B. Association of polymorphisms in NR6A1, PLAG1 and VRTN with the number of vertebrae in Chinese Tongcheng × Large White crossbred pigs. Anim. Genet. 2018, 49, 353–354. [Google Scholar] [CrossRef]
- Niu, N.; Liu, Q.; Hou, X.; Liu, X.; Wang, L.; Zhao, F.; Gao, H.; Shi, L.; Wang, L.; Zhang, L. Genome-wide association study revealed ABCD4 on SSC7 and GREB1L and MIB1 on SSC6 as crucial candidate genes for rib number in Beijing Black pigs. Anim. Genet. 2022, 53, 690–695. [Google Scholar] [CrossRef]
- Casiró, S.; Velez-Irizarry, D.; Ernst, C.W.; Raney, N.E.; Bates, R.O.; Charles, M.G.; Steibel, J.P. Genome-wide association study in an F2 Duroc x Pietrain resource population for economically important meat quality and carcass traits. J. Anim. Sci. 2017, 95, 545–558. [Google Scholar] [CrossRef]
- Jiang, N.; Liu, C.; Lan, T.; Zhang, Q.; Cao, Y.; Pu, G.; Niu, P.; Zhang, Z.; Li, Q.; Zhou, J.; et al. Polymorphism of VRTN Gene g.20311_20312ins291 Was Associated with the Number of Ribs, Carcass Diagonal Length and Cannon Bone Circumference in Suhuai Pigs. Animals 2020, 10, 484. [Google Scholar] [CrossRef]
- Ren, D.R.; Ren, J.; Ruan, G.F.; Guo, Y.M.; Wu, L.H.; Yang, G.C.; Zhou, L.H.; Li, L.; Zhang, Z.Y.; Huang, L.S. Mapping and fine mapping of quantitative trait loci for the number of vertebrae in a White Duroc × Chinese Erhualian intercross resource population. Anim. Genet. 2012, 43, 545–551. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Huang, L.; Yang, M.; Fan, Y.; Li, L.; Fang, S.; Deng, W.; Cui, L.; Zhang, Z.; Ai, H.; et al. Possible introgression of the VRTN mutation increasing vertebral number, carcass length and teat number from Chinese pigs into European pigs. Sci. Rep. 2016, 6, 19240. [Google Scholar] [CrossRef] [PubMed]
- Browning, B.L.; Zhou, Y.; Browning, S.R. A One-Penny Imputed Genome from Next-Generation Reference Panels. Am. J. Hum. Genet. 2018, 103, 338–348. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Zhang, H.; Tang, Z.; Yin, D.; Fu, Y.; Yuan, X.; Li, X.; Liu, X.; Zhao, S. HIBLUP: An integration of statistical models on the BLUP framework for efficient genetic evaluation using big genomic data. Nucleic Acids Res. 2023, 51, 3501–3512. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, Z. GAPIT Version 3: Boosting Power and Accuracy for Genomic Association and Prediction. Genom. Proteom. Bioinform. 2021, 19, 629–640. [Google Scholar] [CrossRef]
- Dyer, S.C.; Austine-Orimoloye, O.; Azov, A.G.; Barba, M.; Barnes, I.; Barrera-Enriquez, V.P.; Becker, A.; Bennett, R.; Beracochea, M.; Berry, A.; et al. Ensembl 2025. Nucleic Acids Res. 2025, 53, D948–D957. [Google Scholar] [CrossRef]
- Yang, L.; Li, X.; Zhuang, Z.; Zhou, S.; Wu, J.; Xu, C.; Ruan, D.; Qiu, Y.; Zhao, H.; Zheng, E.; et al. Genome-Wide Association Study Identifies the Crucial Candidate Genes for Teat Number in Crossbred Commercial Pigs. Animals 2023, 13, 1880. [Google Scholar] [CrossRef]
- Deng, S.; Qiu, Y.; Zhuang, Z.; Wu, J.; Li, X.; Ruan, D.; Xu, C.; Zheng, E.; Yang, M.; Cai, G.; et al. Genome-Wide Association Study of Body Conformation Traits in a Three-Way Crossbred Commercial Pig Population. Animals 2023, 13, 2414. [Google Scholar] [CrossRef]
- Sato, S.; Uemoto, Y.; Kikuchi, T.; Egawa, S.; Kohira, K.; Saito, T.; Sakuma, H.; Miyashita, S.; Arata, S.; Kojima, T.; et al. SNP- and haplotype-based genome-wide association studies for growth, carcass, and meat quality traits in a Duroc multigenerational population. BMC Genet. 2016, 17, 60. [Google Scholar] [CrossRef]
- Liu, Q.; Yue, J.; Niu, N.; Liu, X.; Yan, H.; Zhao, F.; Hou, X.; Gao, H.; Shi, L.; Wang, L.; et al. Genome-Wide Association Analysis Identified BMPR1A as a Novel Candidate Gene Affecting the Number of Thoracic Vertebrae in a Large White × Minzhu Intercross Pig Population. Animals 2020, 10, 2186. [Google Scholar] [CrossRef]
- Xue, Y.; Liu, S.; Li, W.; Mao, R.; Zhuo, Y.; Xing, W.; Liu, J.; Wang, C.; Zhou, L.; Lei, M.; et al. Genome-Wide Association Study Reveals Additive and Non-Additive Effects on Growth Traits in Duroc Pigs. Genes 2022, 13, 1454. [Google Scholar] [CrossRef]
- Huang, M.; Liu, X.; Zhou, Y.; Summers, R.M.; Zhang, Z. BLINK: A package for the next level of genome-wide association studies with both individuals and markers in the millions. Gigascience 2019, 8, giy154. [Google Scholar] [CrossRef]
- Horodyska, J.; Hamill, R.M.; Varley, P.F.; Reyer, H.; Wimmers, K. Genome-wide association analysis and functional annotation of positional candidate genes for feed conversion efficiency and growth rate in pigs. PLoS ONE 2017, 12, e0173482. [Google Scholar] [CrossRef]
- Guo, Y.; Tan, L.J.; Lei, S.F.; Yang, T.L.; Chen, X.D.; Zhang, F.; Chen, Y.; Pan, F.; Yan, H.; Liu, X.; et al. Genome-wide association study identifies ALDH7A1 as a novel susceptibility gene for osteoporosis. PLoS Genet. 2010, 6, e1000806. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.F.; Spector, T.D.; Richards, J.B. Insights into the genetics of osteoporosis from recent genome-wide association studies. Expert. Rev. Mol. Med. 2011, 13, e28. [Google Scholar] [CrossRef] [PubMed]
- Debaenst, S.; Jarayseh, T.; De Saffel, H.; Bek, J.W.; Boone, M.; Josipovic, I.; Kibleur, P.; Kwon, R.Y.; Coucke, P.J.; Willaert, A. Crispant analysis in zebrafish as a tool for rapid functional screening of disease-causing genes for bone fragility. Elife 2025, 13, RP100060. [Google Scholar] [CrossRef]
- Hendriks, W.J.; Elson, A.; Harroch, S.; Pulido, R.; Stoker, A.; den Hertog, J. Protein tyrosine phosphatases in health and disease. FEBS J. 2013, 280, 708–730. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Lin, R.; Hou, J.; Zhu, C.; Liu, G.; Lin, Y.; Su, J.; Yang, M.; Yang, B.; Ma, Y.; et al. Targeting endothelial PDGFR-β facilitates angiogenesis-associated bone formation through the PAK1/NICD axis. J. Cell Physiol. 2024, 239, e31291. [Google Scholar] [CrossRef]

| Trait | Mean | SD | Range | Genomic Variation () | Heritability () |
|---|---|---|---|---|---|
| NTLV | 21.3 | 0.581 | 20~23 | 0.223 | 0.700 |
| NR | 15.3 | 0.582 | 14~16 | 0.253 | 0.752 |
| NLV | 6.0 | 0.274 | 5~7 | 0.001 | 0.017 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bai, C.; Fei, J.; Zhang, X.; Liu, W.; Ke, J.; Chen, C.; He, Y.; Liang, S.; Sun, B.; Sun, H. A Genome-Wide Association Study of Rib Number and Thoracolumbar Vertebra Number in a Landrace × Yorkshire Crossbred Pig Population. Biology 2025, 14, 1068. https://doi.org/10.3390/biology14081068
Bai C, Fei J, Zhang X, Liu W, Ke J, Chen C, He Y, Liang S, Sun B, Sun H. A Genome-Wide Association Study of Rib Number and Thoracolumbar Vertebra Number in a Landrace × Yorkshire Crossbred Pig Population. Biology. 2025; 14(8):1068. https://doi.org/10.3390/biology14081068
Chicago/Turabian StyleBai, Chunyan, Junwen Fei, Xiaoran Zhang, Wuyang Liu, Juan Ke, Changyi Chen, Yu He, Shuang Liang, Boxing Sun, and Hao Sun. 2025. "A Genome-Wide Association Study of Rib Number and Thoracolumbar Vertebra Number in a Landrace × Yorkshire Crossbred Pig Population" Biology 14, no. 8: 1068. https://doi.org/10.3390/biology14081068
APA StyleBai, C., Fei, J., Zhang, X., Liu, W., Ke, J., Chen, C., He, Y., Liang, S., Sun, B., & Sun, H. (2025). A Genome-Wide Association Study of Rib Number and Thoracolumbar Vertebra Number in a Landrace × Yorkshire Crossbred Pig Population. Biology, 14(8), 1068. https://doi.org/10.3390/biology14081068






