Molecular Mechanisms Underlying Differences in Athletic Ability in Racehorses Based on Whole Transcriptome Sequencing
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Animals and Sample Collection
2.2. RNA Extraction and Quality Control, Library Construction, and High-Throughput Sequencing
2.3. Differential Expression Gene Analysis
2.4. Functional Enrichment Analysis
2.5. Protein Interaction Network Analysis
3. Results
3.1. Transcriptome Quality Control Analysis
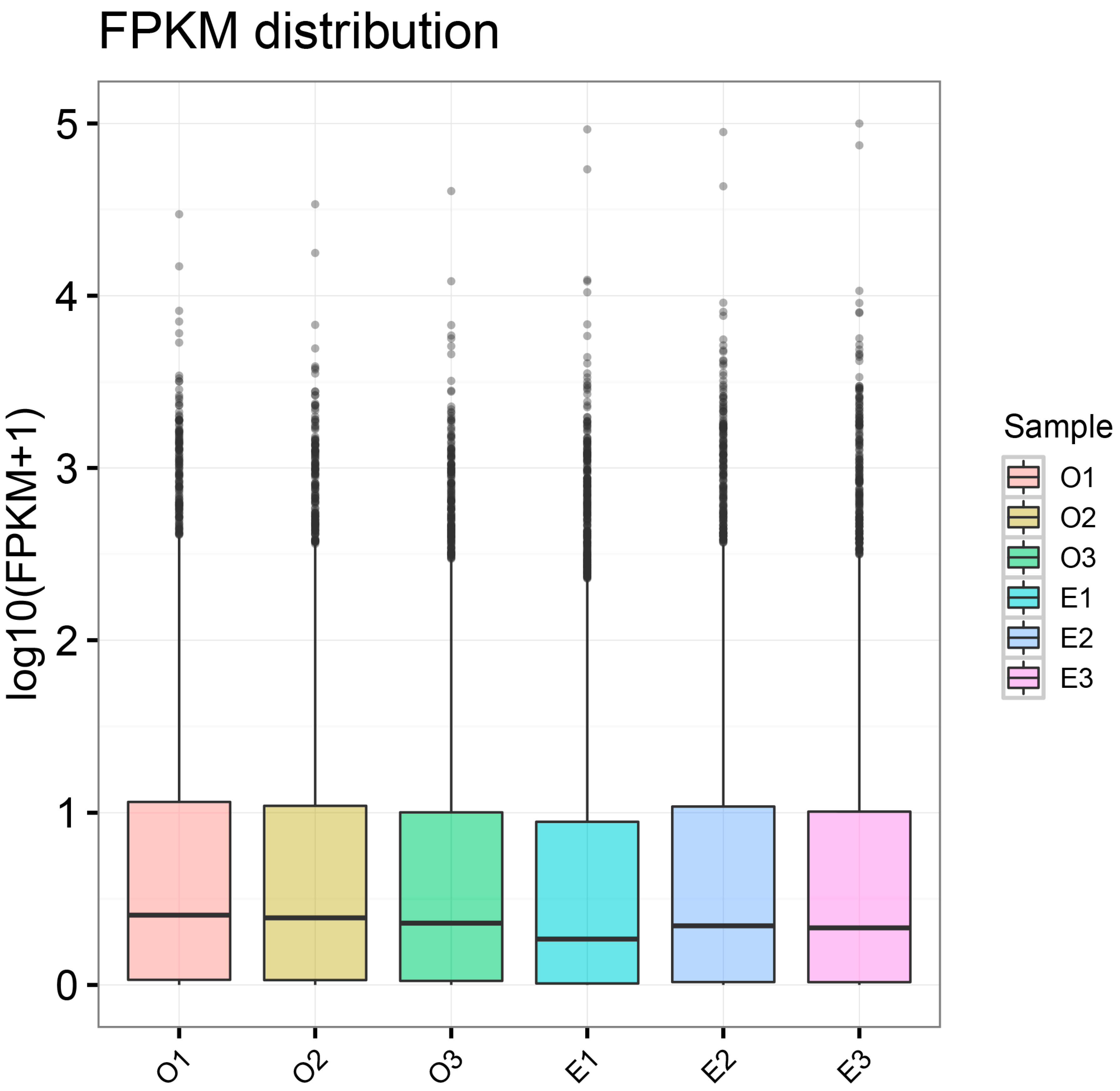
3.2. Gene Expression Level Analysis
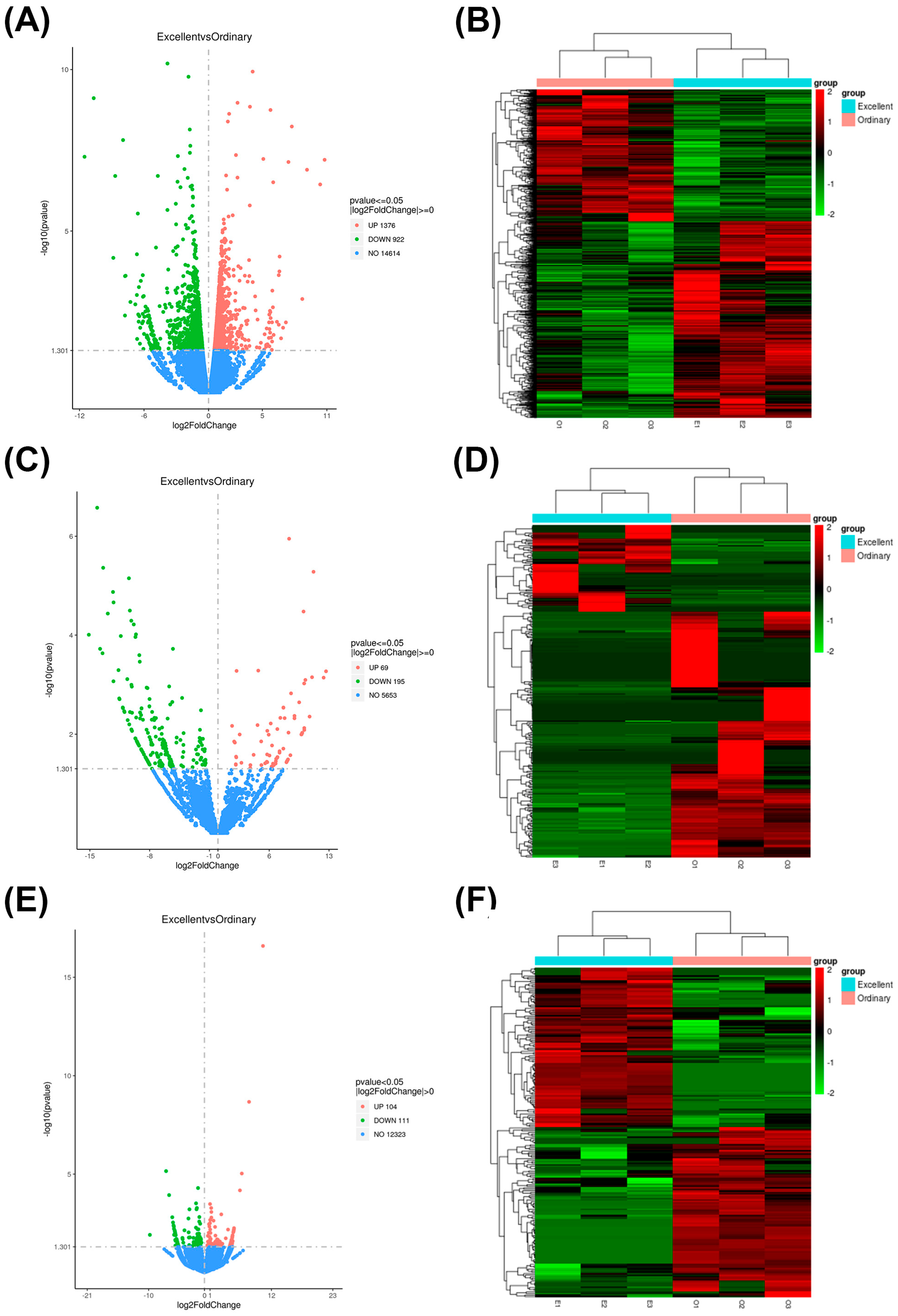
3.3. Identification of Differentially Protein-Coding Genes and Statistical Identification of Differentially Non-Coding Genes
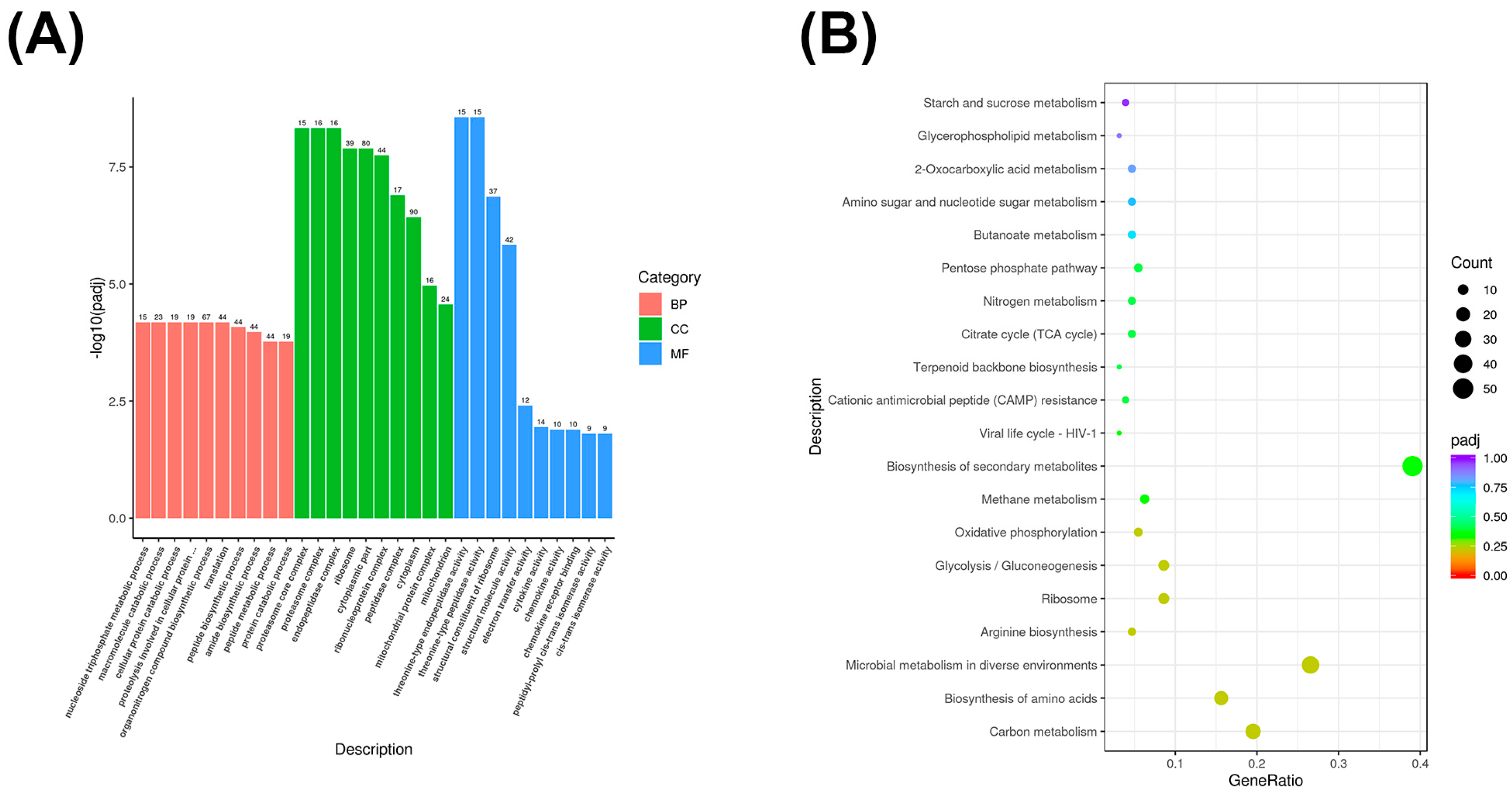
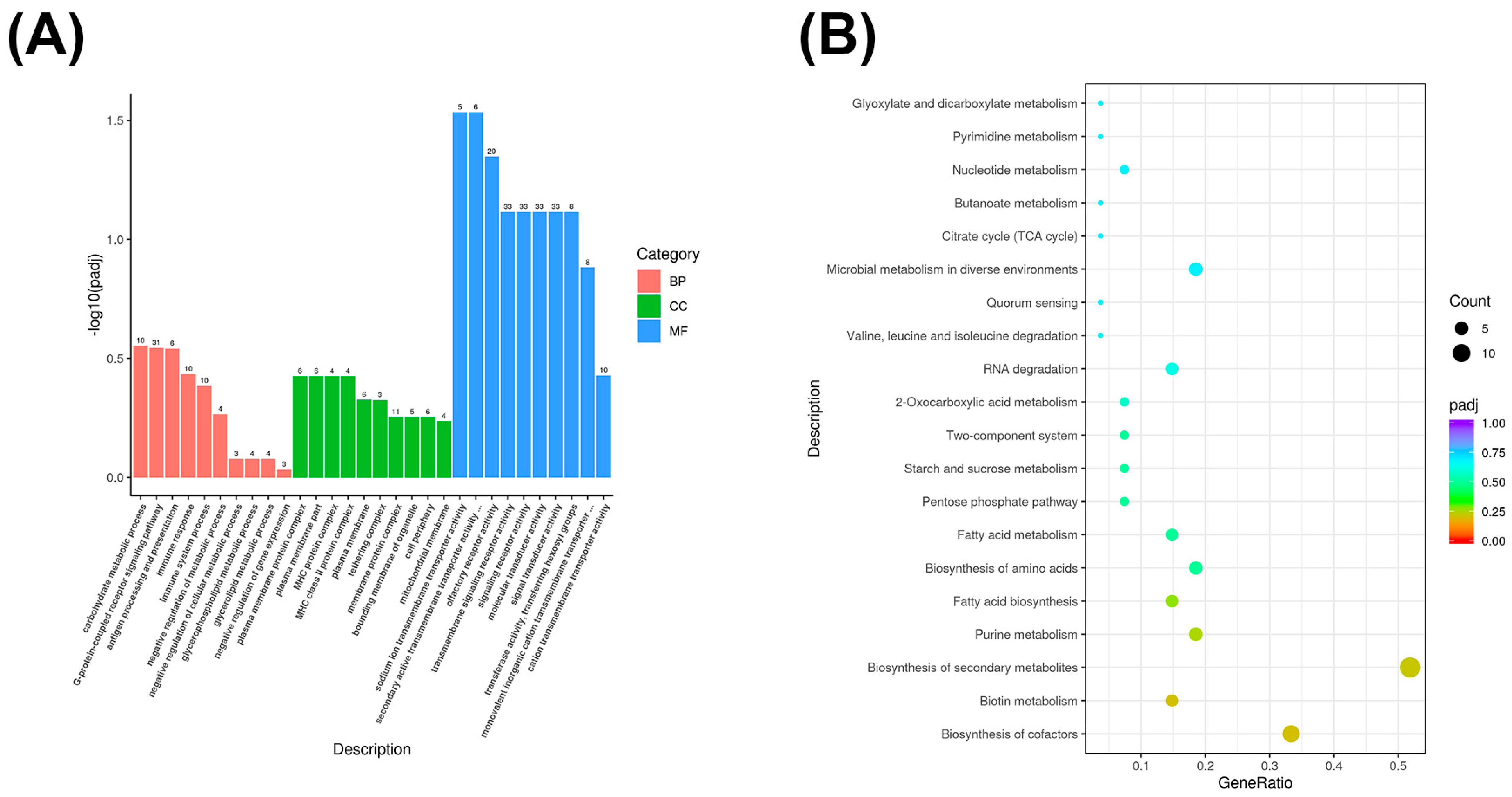
3.4. GO and KEGG Enrichment Analysis of Differentially Expressed mRNA Genes
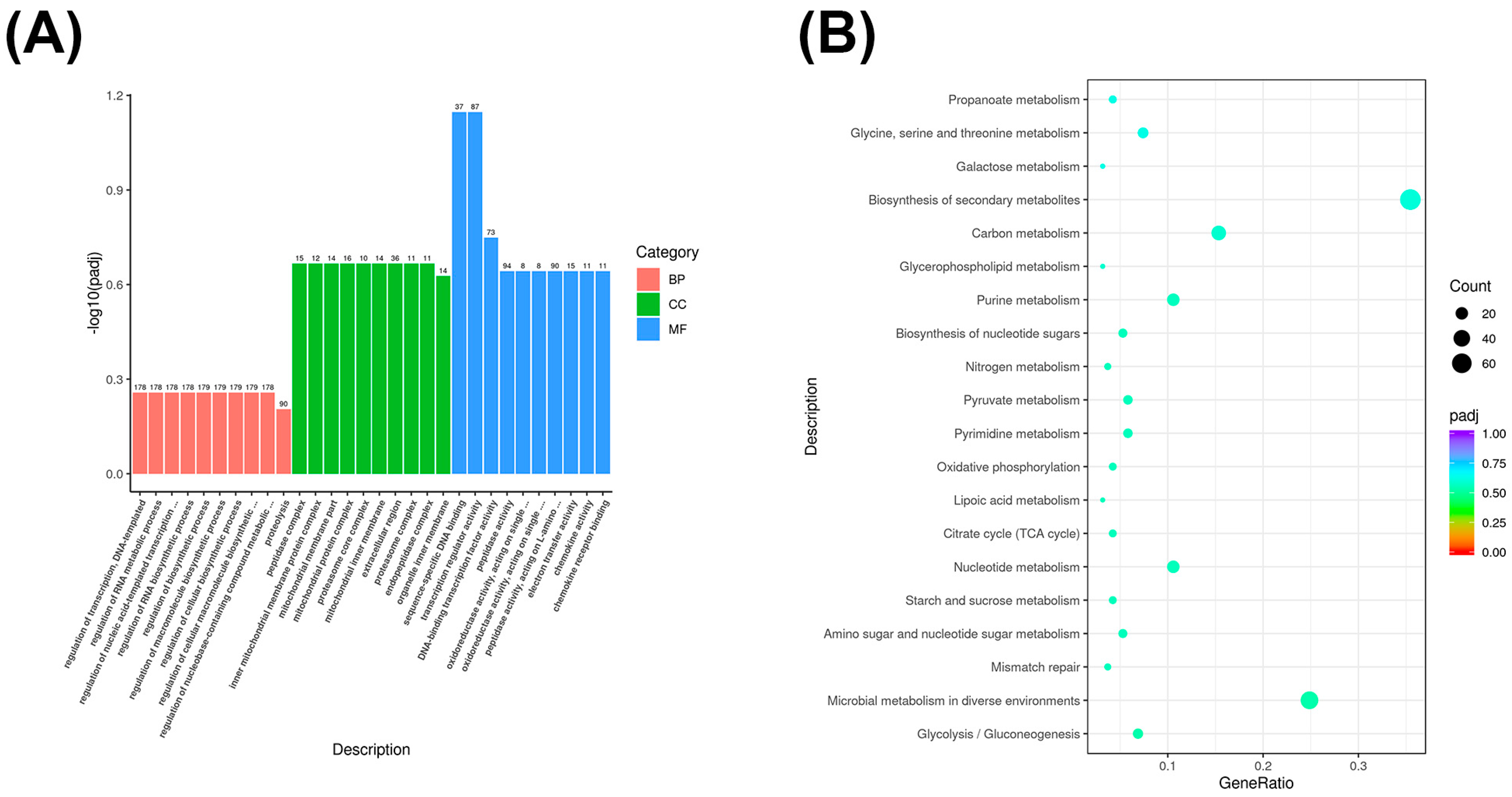
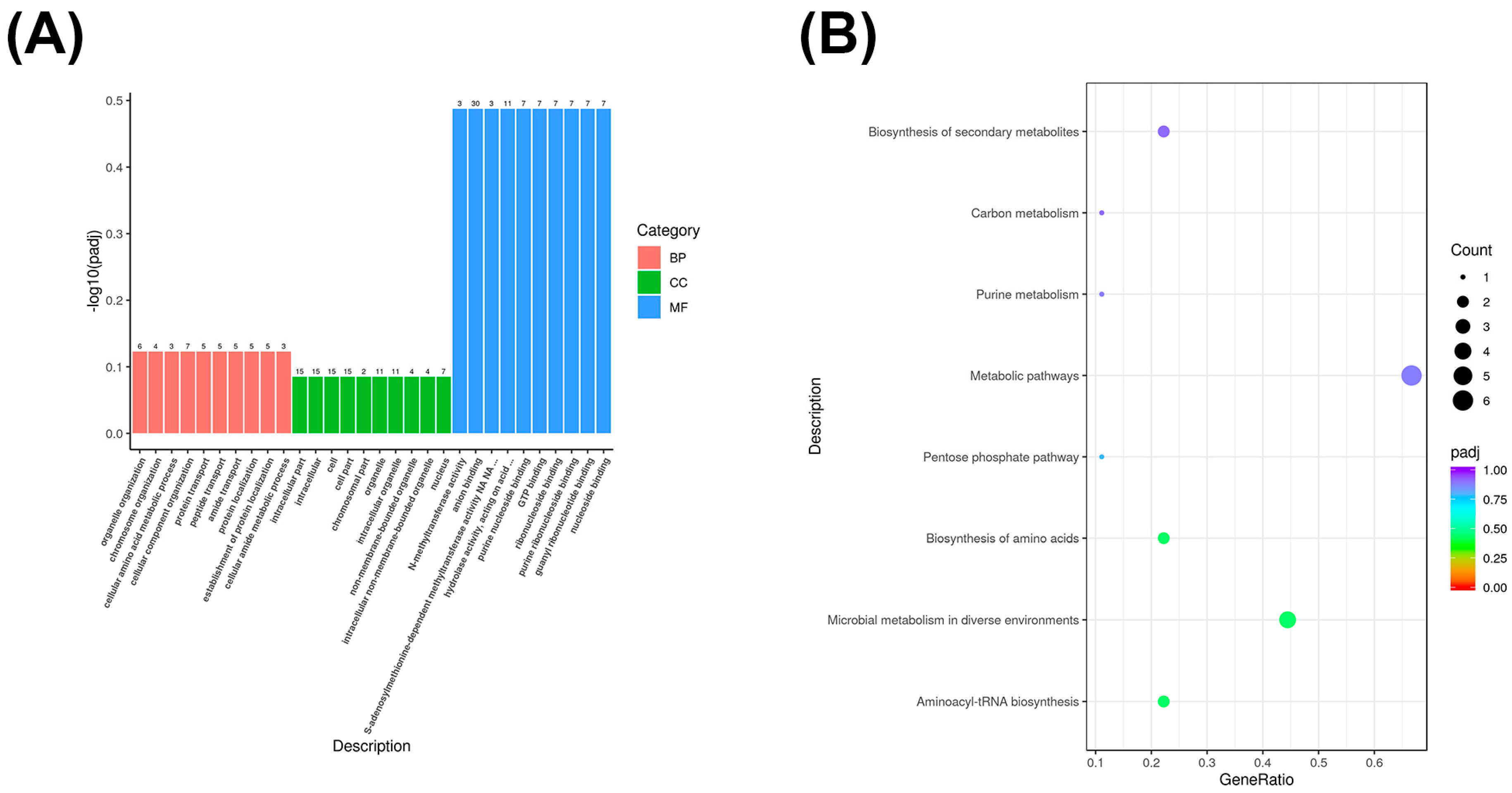
3.5. Co-Expressed Long Non-Coding RNA (lncRNA) Target Gene GO and KEGG Enrichment Analysis
3.6. Co-Localized Long Non-Coding RNA (lncRNA) Target Gene GO and KEGG Enrichment Analysis
3.7. GO and KEGG Enrichment Analysis of Differentially Expressed circRNA Genes
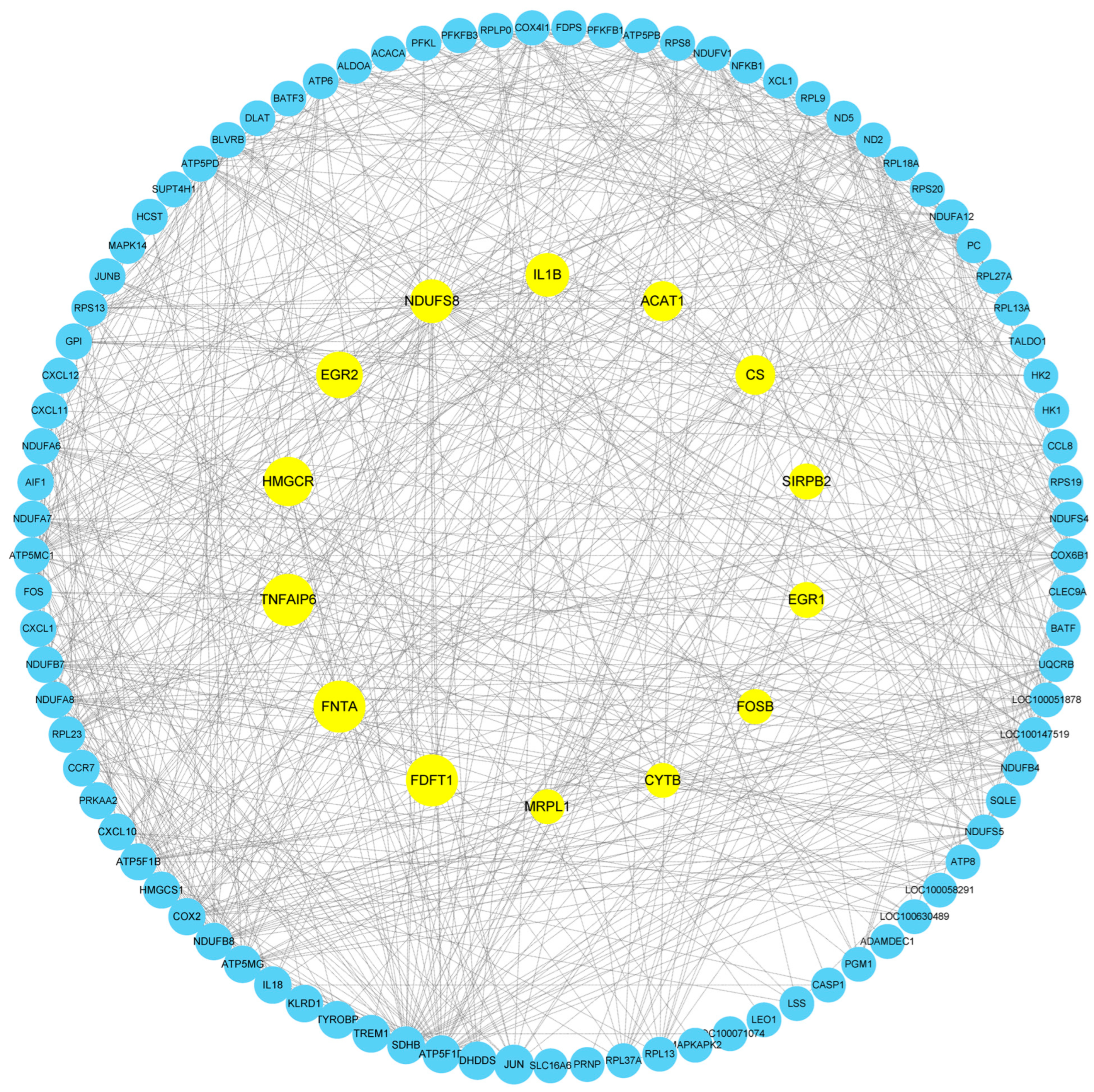
3.8. Protein Network Interactions Results Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kirsch, K.; Düe, M.; Holzhausen, H.; Sandersen, C. Correlation of competition performance with heart rate and blood lactate response during interval training sessions in eventing horses. Comp. Exerc. Physiol. 2019, 15, 187–198. [Google Scholar] [CrossRef]
- Zuluaga Cabrera, A.M.; Casas Soto, M.J.; Martínez Aranzales, J.R.; Correa Valencia, N.M.; Arias Gutiérrez, M.P. Blood lactate concentrations and heart rates of Colombian Paso horses during a field exercise test. Veter-Anim. Sci. 2021, 13, 100185. [Google Scholar] [CrossRef] [PubMed]
- McGivney, B.A.; Browne, J.A.; Fonseca, R.G.; Katz, L.M.; MacHugh, D.E.; Whiston, R.; Hill, E.W. MSTN genotypes in T horoughbred horses influence skeletal muscle gene expression and racetrack performance. Anim. Genet. 2012, 43, 810–812. [Google Scholar] [CrossRef]
- Pira, E.; Vacca, G.M.; Dettori, M.L.; Piras, G.; Moro, M.; Paschino, P.; Pazzola, M. Polymorphisms at myostatin gene (MSTN) and the associations with sport performances in Anglo-Arabian racehorses. Animals 2021, 11, 964. [Google Scholar] [CrossRef] [PubMed]
- Eivers, S.; McGivney, B.; Gu, J.; MacHugh, D.; Katz, L.; Hill, E. PGC-1α encoded by the PPARGC1A gene regulates oxidative energy metabolism in equine skeletal muscle during exercise. Anim. Genet. 2012, 43, 153–162. [Google Scholar] [CrossRef]
- Pereira, G.L.; de Matteis, R.; Meira, C.T.; Regitano, L.C.; Silva, J.A.I.V.; Chardulo, L.A.L.; Curi, R.A. Comparison of sequence variants in the PDK4 and COX4I2 genes between racing and cutting lines of quarter horses and associations with the speed index. J. Equine Vet. Sci. 2016, 39, 1–6. [Google Scholar] [CrossRef]
- Yang, L.; Li, P.; Huang, X.; Wang, C.; Zeng, Y.; Wang, J.; Yao, X.; Meng, J. Effects of combined transcriptome and metabolome analysis training on athletic performance of 2-year-old trot-type Yili horses. Genes 2025, 16, 197. [Google Scholar] [CrossRef]
- McGee, S.L.; Hargreaves, M. Epigenetics and exercise. Trends Endocrinol. Metab. 2019, 30, 636–645. [Google Scholar] [CrossRef]
- Bou, T.; Ding, W.; Ren, X.; Liu, H.; Gong, W.; Jia, Z.; Zhang, X.; Dugarjaviin, M.; Bai, D. Muscle fibre transition and transcriptional changes of horse skeletal muscles during traditional Mongolian endurance training. Equine Vet. J. 2024, 56, 178–192. [Google Scholar] [CrossRef]
- Wang, J.; Ren, W.; Li, Z.; Li, L.; Wang, R.; Ma, S.; Zeng, Y.; Meng, J.; Yao, X. Regulatory Mechanisms of Yili Horses During an 80 km Race Based on Transcriptomics and Metabolomics Analyses. Int. J. Mol. Sci. 2025, 26, 2426. [Google Scholar] [CrossRef]
- Ma, B.; Wang, S.; Wu, W.; Shan, P.; Chen, Y.; Meng, J.; Xing, L.; Yun, J.; Hao, L.; Wang, X. Mechanisms of circRNA/lncRNA-miRNA interactions and applications in disease and drug research. Biomed. Pharmacother. 2023, 162, 114672. [Google Scholar] [CrossRef]
- Li, R.; Li, B.; Jiang, A.; Cao, Y.; Hou, L.; Zhang, Z.; Zhang, X.; Liu, H.; Kim, K.-H.; Wu, W. Exploring the lncRNAs related to skeletal muscle fiber types and meat quality traits in pigs. Genes 2020, 11, 883. [Google Scholar] [CrossRef]
- Mao, C.; You, W.; Yang, Y.; Cheng, H.; Hu, X.; Lan, X.; Song, E. Comprehensive characterization of lncRNA N6-methyladenosine modification dynamics throughout bovine skeletal muscle development. J. Anim. Sci. Biotechnol. 2025, 16, 36. [Google Scholar] [CrossRef]
- Ma, M.; Chen, M.; Wu, X.; Sooranna, S.R.; Liu, Q.; Shi, D.; Wang, J.; Li, H. A newly identified lncRNA lnc000100 regulates proliferation and differentiation of cattle skeletal muscle cells. Epigenetics 2023, 18, 2270864. [Google Scholar] [CrossRef] [PubMed]
- Stefaniuk-Szmukier, M.; Szmatoła, T.; Ropka-Molik, K. Molecular Signatures of Exercise Adaptation in Arabian Racing Horses: Transcriptomic Insights from Blood and Muscle. Genes 2025, 16, 431. [Google Scholar] [CrossRef]
- Xu, B.; Yang, G.; Jiao, B.; Zhu, H. Analysis of ancient and modern horse genomes reveals the critical impact of lncRNA-mediated epigenetic regulation on horse domestication. Front. Genet. 2022, 13, 944933. [Google Scholar] [CrossRef]
- Su, Y.; Ren, W.; Ma, S.; Meng, J.; Yao, X.; Zeng, Y.; Li, Z.; Li, L.; Wang, R.; Wang, J. Comparative analysis of blood whole transcriptome profiles in Yili horses pre-and post-5000-meter racing. Front. Genet. 2025, 16, 1651628. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Wang, J.; Zhao, F. CIRI: An efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015, 16, 4. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Zhou, L.; Chen, J.; Li, Z.; Li, X.; Hu, X.; Huang, Y.; Zhao, X.; Liang, C.; Wang, Y.; Sun, L. Integrated profiling of microRNAs and mRNAs: microRNAs located on Xq27. 3 associate with clear cell renal cell carcinoma. PLoS ONE 2010, 5, e15224. [Google Scholar] [CrossRef]
- Han, H.; McGivney, B.A.; Allen, L.; Bai, D.; Corduff, L.R.; Davaakhuu, G.; Davaasambuu, J.; Dorjgotov, D.; Hall, T.J.; Hemmings, A.J. Common protein-coding variants influence the racing phenotype in galloping racehorse breeds. Commun. Biol. 2022, 5, 1320. [Google Scholar] [CrossRef] [PubMed]
- Ropka-Molik, K.; Stefaniuk-Szmukier, M.; Musiał, A.; Velie, B. The genetics of racing performance in Arabian horses. Int. J. Genom. 2019, 2019, 9013239. [Google Scholar] [CrossRef]
- Allen, J.; Ramsden, M.; Nisar, S. Skeletal muscle structure, function and pathology. Orthop. Trauma 2024, 38, 137–144. [Google Scholar] [CrossRef]
- Yang, Y.; Fan, X.; Yan, J.; Chen, M.; Zhu, M.; Tang, Y.; Liu, S.; Tang, Z. A comprehensive epigenome atlas reveals DNA methylation regulating skeletal muscle development. Nucleic Acids Res. 2021, 49, 1313–1329. [Google Scholar] [CrossRef]
- Smith, J.A.; Murach, K.A.; Dyar, K.A.; Zierath, J.R. Exercise metabolism and adaptation in skeletal muscle. Nat. Rev. Mol. Cell Biol. 2023, 24, 607–632. [Google Scholar] [CrossRef]
- Ji, F.; Lee, H.S.; Kim, J.-H. Resistance exercise and skeletal muscle: Protein synthesis, degradation, and controversies. Eur. J. Appl. Physiol. 2025, 125, 2353–2382. [Google Scholar] [CrossRef]
- Pardo, P.S.; Mohamed, J.S.; Lopez, M.A.; Boriek, A.M. Induction of Sirt1 by mechanical stretch of skeletal muscle through the early response factor EGR1 triggers an antioxidative response. J. Biol. Chem. 2011, 286, 2559–2566. [Google Scholar] [CrossRef] [PubMed]
- Pardo, P.S.; Boriek, A.M. An autoregulatory loop reverts the mechanosensitive Sirt1 induction by EGR1 in skeletal muscle cells. Aging 2012, 4, 456. [Google Scholar] [CrossRef] [PubMed]
- Haasper, C.; Jagodzinski, M.; Drescher, M.; Meller, R.; Wehmeier, M.; Krettek, C.; Hesse, E. Cyclic strain induces FosB and initiates osteogenic differentiation of mesenchymal cells. Exp. Toxicol. Pathol. 2008, 59, 355–363. [Google Scholar] [CrossRef] [PubMed]
- Inoue, D.; Kido, S.; Matsumoto, T. Transcriptional induction of FosB/ΔFosB gene by mechanical stress in osteoblasts. J. Biol. Chem. 2004, 279, 49795–49803. [Google Scholar] [CrossRef]
- Wei, X.; Li, H.; Zhao, G.; Yang, J.; Li, L.; Huang, Y.; Lan, X.; Ma, Y.; Hu, L.; Zheng, H. ΔFosB regulates rosiglitazone-induced milk fat synthesis and cell survival. J. Cell. Physiol. 2018, 233, 9284–9298. [Google Scholar] [CrossRef]
- Xiong, G.; Xie, Y.; Tan, Y.; Ye, Y.; Tan, X.; Jiang, L.; Qin, E.; Wei, X.; Li, J.; Liang, T. HMGB1-Mediated Pyroptosis Promotes Inflammation and Contributes to Skeletal Muscle Atrophy induced by Cigarette Smoke. Am. J. Physiol.-Cell Physiol. 2025, 329, C325–C340. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Lu, G.; Xiao, L.; Tan, B.; Yang, Y.; Hong, L.; Li, Z.; Cai, G.; Gu, T. Effect of m6A Recognition Protein YTHDC1 on Skeletal Muscle Growth. Animals 2025, 15, 1978. [Google Scholar] [CrossRef]
- Zhou, L.; Mudianto, T.; Ma, X.; Riley, R.; Uppaluri, R. Targeting EZH2 enhances antigen presentation, antitumor immunity, and circumvents anti–PD-1 resistance in head and neck cancer. Clin. Cancer Res. 2020, 26, 290–300. [Google Scholar] [CrossRef]
- Galmiche, L.; Serre, V.; Beinat, M.; Assouline, Z.; Lebre, A.S.; Chretien, D.; Nietschke, P.; Benes, V.; Boddaert, N.; Sidi, D. Exome sequencing identifies MRPL3 mutation in mitochondrial cardiomyopathy. Hum. Mutat. 2011, 32, 1225–1231. [Google Scholar] [CrossRef]
- Chatterjee, S.D.; Zhou, J.; Dasgupta, R.; Cramer-Blok, A.; Timmer, M.; van der Stelt, M.; Ubbink, M. Protein dynamics influence the enzymatic activity of phospholipase A/Acyltransferases 3 and 4. Biochemistry 2021, 60, 1178–1190. [Google Scholar] [CrossRef]
- Agarwal, A.K.; Garg, A. Phospholipid biosynthetic pathways and lipodystrophies: A novel syndrome due to PLAAT3 deficiency. Nat. Rev. Endocrinol. 2024, 20, 128–129. [Google Scholar] [CrossRef]
- Uyama, T.; Tsuboi, K.; Ueda, N. An involvement of phospholipase A/acyltransferase family proteins in peroxisome regulation and plasmalogen metabolism. FEBS Lett. 2017, 591, 2745–2760. [Google Scholar] [CrossRef] [PubMed]
- Koeberle, A.; Shindou, H.; Harayama, T.; Yuki, K.; Shimizu, T. Polyunsaturated fatty acids are incorporated into maturating male mouse germ cells by lysophosphatidic acid acyltransferase 3. FASEB J. 2012, 26, 169–180. [Google Scholar] [CrossRef] [PubMed]
- Blaauw, R.; Calder, P.C.; Martindale, R.G.; Berger, M.M. Combining proteins with n-3 PUFAs (EPA+ DHA) and their inflammation pro-resolution mediators for preservation of skeletal muscle mass. Crit. Care 2024, 28, 38. [Google Scholar] [CrossRef]
- Valentine, W.J.; Tokuoka, S.M.; Hishikawa, D.; Kita, Y.; Shindou, H.; Shimizu, T. LPAAT3 incorporates docosahexaenoic acid into skeletal muscle cell membranes and is upregulated by PPARδ activation. J. Lipid Res. 2018, 59, 184–194. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.A.; Viggars, M.R.; Esser, K.A. Metabolism and exercise: The skeletal muscle clock takes centre stage. Nat. Rev. Endocrinol. 2023, 19, 272–284. [Google Scholar] [CrossRef]
- Visser, N.; Nelemans, L.C.; He, Y.; Lourens, H.J.; Corrales, M.G.; Huls, G.; Wiersma, V.R.; Schuringa, J.J.; Bremer, E. Signal regulatory protein beta 2 is a novel positive regulator of innate anticancer immunity. Front. Immunol. 2023, 14, 1287256. [Google Scholar] [CrossRef]
- Nakatsu, Y.; Matsunaga, Y.; Nakanishi, M.; Yamamotoya, T.; Sano, T.; Kanematsu, T.; Asano, T. Prolyl isomerase Pin1 in skeletal muscles contributes to systemic energy metabolism and exercise capacity through regulating SERCA activity. Biochem. Biophys. Res. Commun. 2024, 715, 150001. [Google Scholar] [CrossRef]
- Barclay, A.N.; Brown, M.H. The SIRP family of receptors and immune regulation. Nat. Rev. Immunol. 2006, 6, 457–464. [Google Scholar] [CrossRef]
- Londino, J.D.; Gulick, D.; Isenberg, J.S.; Mallampalli, R.K. Cleavage of signal regulatory protein α (SIRPα) enhances inflammatory signaling. J. Biol. Chem. 2015, 290, 31113–31125. [Google Scholar] [CrossRef]
- Wu, J.-I.; Lin, Y.-P.; Tseng, C.-W.; Chen, H.-J.; Wang, L.-H. Crabp2 promotes metastasis of lung cancer cells via HuR and integrin β1/FAK/ERK signaling. Sci. Rep. 2019, 9, 845. [Google Scholar] [CrossRef] [PubMed]
- Du, C.; Chen, B.; Huang, Y.; Liang, W.; Su, Y.H.; Liu, X. Roads diverged in a wood: Proteins and RNAs encoded by cytochrome b (CYTB) gene in health and disease. Clin. Transl. Discov. 2025, 5, e70026. [Google Scholar] [CrossRef]
- Massie, R.; Wong, L.J.C.; Milone, M. Exercise intolerance due to cytochrome b mutation. Muscle Nerve 2010, 42, 136–140. [Google Scholar] [CrossRef]
- Islam, R.; Rahman, M.; Fatema, K. A case report of methylmalonic acidemia associated with CYBT gene mutation. Paediatr. Nephrol. J. Bangladesh 2022, 7, 81–83. [Google Scholar] [CrossRef]
- Miyamoto-Mikami, E.; Fuku, N. Variation of Mitochondrial DNA and elite athletic performance. In Sports, Exercise, and Nutritional Genomics; Elsevier: Amsterdam, The Netherlands, 2019; pp. 129–145. [Google Scholar]
- Soci, U.P.R.; Melo, S.F.S.; Gomes, J.L.P.; Silveira, A.C.; Nóbrega, C.; de Oliveira, E.M. Exercise training and epigenetic regulation: Multilevel modification and regulation of gene expression. In Exercise for Cardiovascular Disease Prevention and Treatment: From Molecular to Clinical, Part 2; Springer: Singapore, 2017; pp. 281–322. [Google Scholar]
- Furrer, R.; Handschin, C. Molecular aspects of the exercise response and training adaptation in skeletal muscle. Free Radic. Biol. Med. 2024, 223, 53–68. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhang, X.; Baker, J.S.; Davison, G.W.; Yan, X. Redox signaling and skeletal muscle adaptation during aerobic exercise. Iscience 2024, 27, 109643. [Google Scholar] [CrossRef] [PubMed]
- McGivney, B.; Herdan, C.; Gough, K.; Katz, L.; Hill, E. Effect of Training on PPARGC1A and FNDC5 Gene Expression in T horoughbred Horses. Equine Vet. J. 2014, 46, 35. [Google Scholar] [CrossRef]
- Ropka-Molik, K.; Stefaniuk-Szmukier, M.; Ukowski, K.Z.; Piórkowska, K.; Bugno-Poniewierska, M. Exercise-induced modification of the skeletal muscle transcriptome in Arabian horses. Physiol. Genom. 2017, 49, 318–326. [Google Scholar] [CrossRef]
- Godet, A.-C.; Roussel, E.; Laugero, N.; Morfoisse, F.; Lacazette, E.; Garmy-Susini, B.; Prats, A.-C. Translational control by long non-coding RNAs. Biochimie 2024, 217, 42–53. [Google Scholar] [CrossRef]
- Lissek, T. Enhancement of physiology via adaptive transcription. Pflügers Arch.-Eur. J. Physiol. 2025, 477, 187–199. [Google Scholar] [CrossRef]
- Jaiswal, A.; Kaushik, N.; Choi, E.H.; Kaushik, N.K. Functional impact of non-coding RNAs in high-grade breast carcinoma: Moving from resistance to clinical applications: A comprehensive review. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2023, 1878, 188915. [Google Scholar] [CrossRef]
- Liu, D.; Dong, Z.; Wang, J.; Tao, Y.; Sun, X.; Yao, X. The existence and function of mitochondrial component in extracellular vesicles. Mitochondrion 2020, 54, 122–127. [Google Scholar] [CrossRef]
- Koshinaka, K.; Honda, A.; Masuda, H.; Sato, A. Effect of quercetin treatment on mitochondrial biogenesis and exercise-induced AMP-activated protein kinase activation in rat skeletal muscle. Nutrients 2020, 12, 729. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Zhang, Y.; Xu, L.; Zhou, D.; Jin, Z.; Zhou, H.; Lin, S.; Cao, J.; Huang, L. CircRNA expression pattern and ceRNA and miRNA–mRNA networks involved in anther development in the CMS line of Brassica campestris. Int. J. Mol. Sci. 2019, 20, 4808. [Google Scholar] [CrossRef]
- Mach, N.; Plancade, S.; Pacholewska, A.; Lecardonnel, J.; Rivière, J.; Moroldo, M.; Vaiman, A.; Morgenthaler, C.; Beinat, M.; Nevot, A. Integrated mRNA and miRNA expression profiling in blood reveals candidate biomarkers associated with endurance exercise in the horse. Sci. Rep. 2016, 6, 22932. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Tan, Z.; Zhou, J.; Wu, Y.; Hu, Q.; Ling, Q.; Ling, J.; Liu, M.; Ma, J.; Zhang, D. The regulation of circRNA and lncRNAprotein binding in cardiovascular diseases: Emerging therapeutic targets. Biomed. Pharmacother. 2023, 165, 115067. [Google Scholar] [CrossRef] [PubMed]
| Raw_Reads | Clean_Reads | Error_Rate | Q20% | Q30% | GC_pct |
|---|---|---|---|---|---|
| 91,493,206 | 89,858,044 | 0.01 | 99.12 | 96.24 | 46.09 |
| 103,900,482 | 100,689,662 | 0.01 | 98.97 | 96 | 46.48 |
| 94,957,100 | 90,466,348 | 0.01 | 98.82 | 95.63 | 47.21 |
| 78,769,258 | 76,519,920 | 0.01 | 98.54 | 96.11 | 49.91 |
| 86,281,582 | 84,191,164 | 0.01 | 98.1 | 94.88 | 49.61 |
| 78,833,206 | 77,067,014 | 0.01 | 98.67 | 96.42 | 49.35 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Q.; Ren, W.; Shan, D.; Su, Y.; Li, Z.; Li, L.; Wang, R.; Ma, S.; Wang, J. Molecular Mechanisms Underlying Differences in Athletic Ability in Racehorses Based on Whole Transcriptome Sequencing. Biology 2025, 14, 1364. https://doi.org/10.3390/biology14101364
Huang Q, Ren W, Shan D, Su Y, Li Z, Li L, Wang R, Ma S, Wang J. Molecular Mechanisms Underlying Differences in Athletic Ability in Racehorses Based on Whole Transcriptome Sequencing. Biology. 2025; 14(10):1364. https://doi.org/10.3390/biology14101364
Chicago/Turabian StyleHuang, Qiuping, Wanlu Ren, Dehaxi Shan, Yi Su, Zexu Li, Luling Li, Ran Wang, Shikun Ma, and Jianwen Wang. 2025. "Molecular Mechanisms Underlying Differences in Athletic Ability in Racehorses Based on Whole Transcriptome Sequencing" Biology 14, no. 10: 1364. https://doi.org/10.3390/biology14101364
APA StyleHuang, Q., Ren, W., Shan, D., Su, Y., Li, Z., Li, L., Wang, R., Ma, S., & Wang, J. (2025). Molecular Mechanisms Underlying Differences in Athletic Ability in Racehorses Based on Whole Transcriptome Sequencing. Biology, 14(10), 1364. https://doi.org/10.3390/biology14101364






