Deciphering the Gene Expression and Alternative Splicing Basis of Muscle Development Through Interpretable Machine Learning Models
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
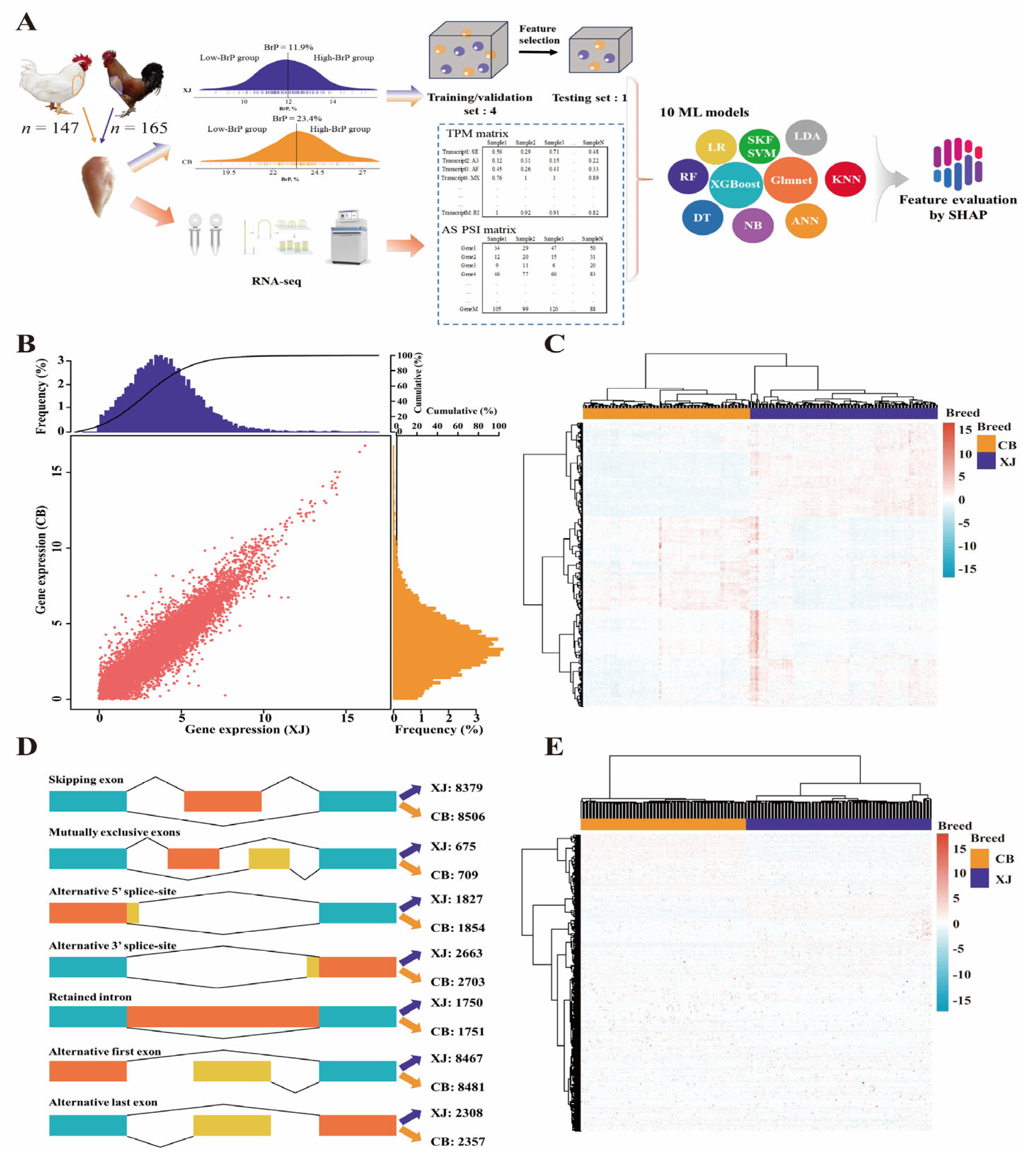
2.1. Animals and Trait
2.2. RNA Extraction, Library Preparation, and Sequencing
2.3. Construction of the Gene Expression Matrix
2.4. Identification of Differentially Expressed Genes (DEGs)
2.5. Identification of Alternative Splicing Events
2.6. Machine Learning Modeling
2.7. Interpretation of ML Models by Shapley Values
2.8. Annotation of Feature DEGs and DSTs
2.9. Statistics
3. Results
3.1. Descriptive Summary of the Sequencing Results
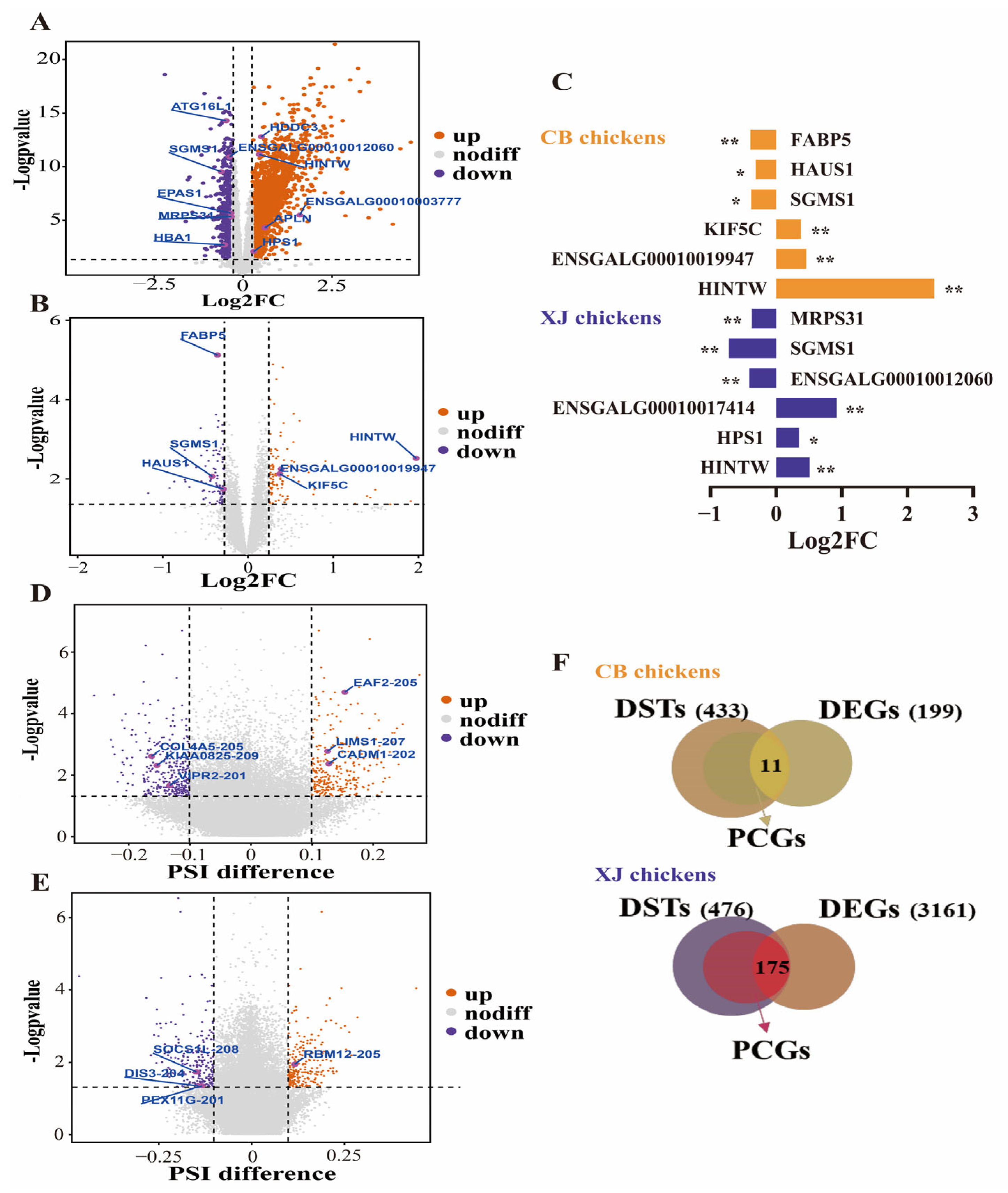
3.2. Detection of DEGs and DSGs in Each Population
3.3. ML Models for BrP
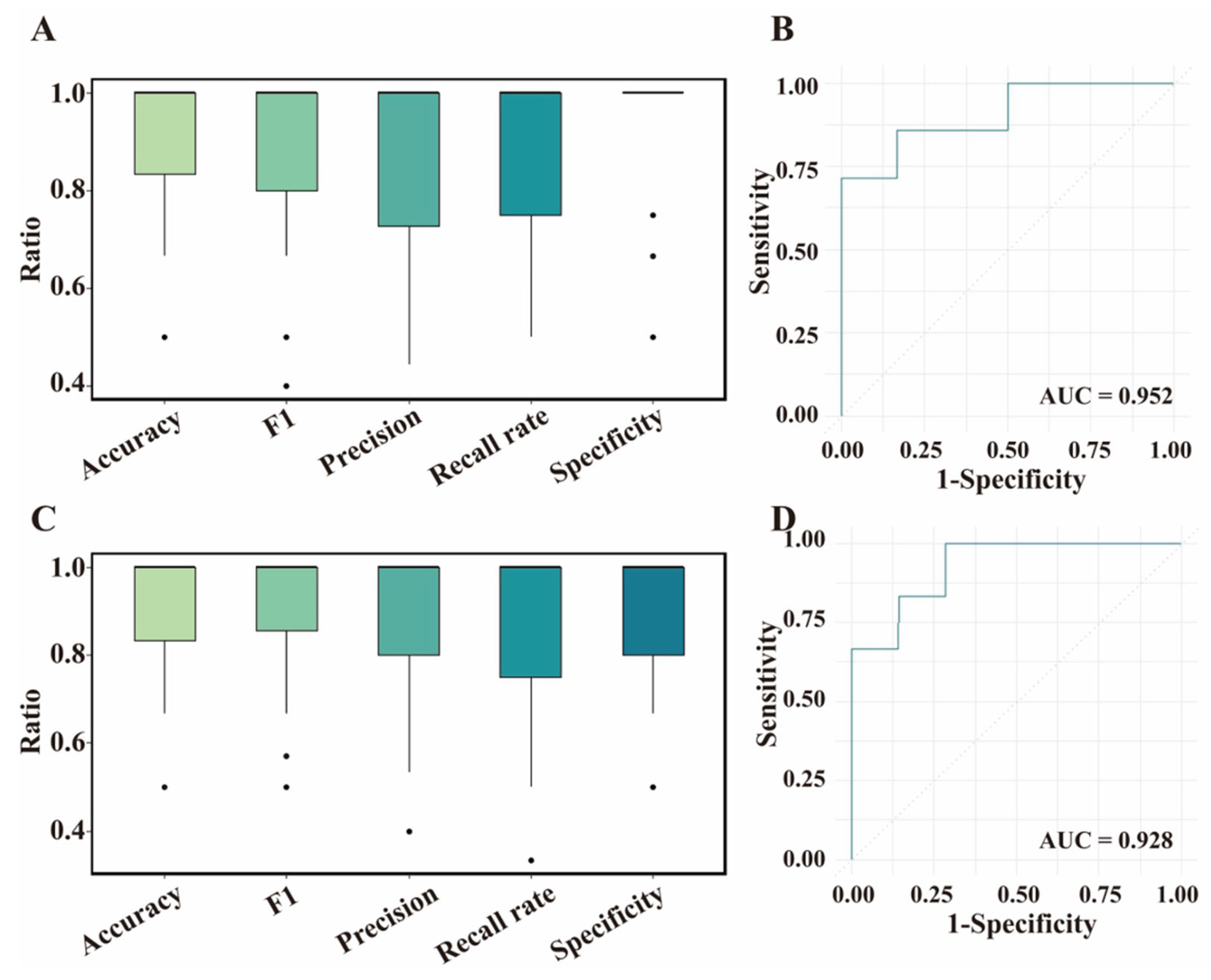
3.4. Validation for ML Models Based on Test Dataset
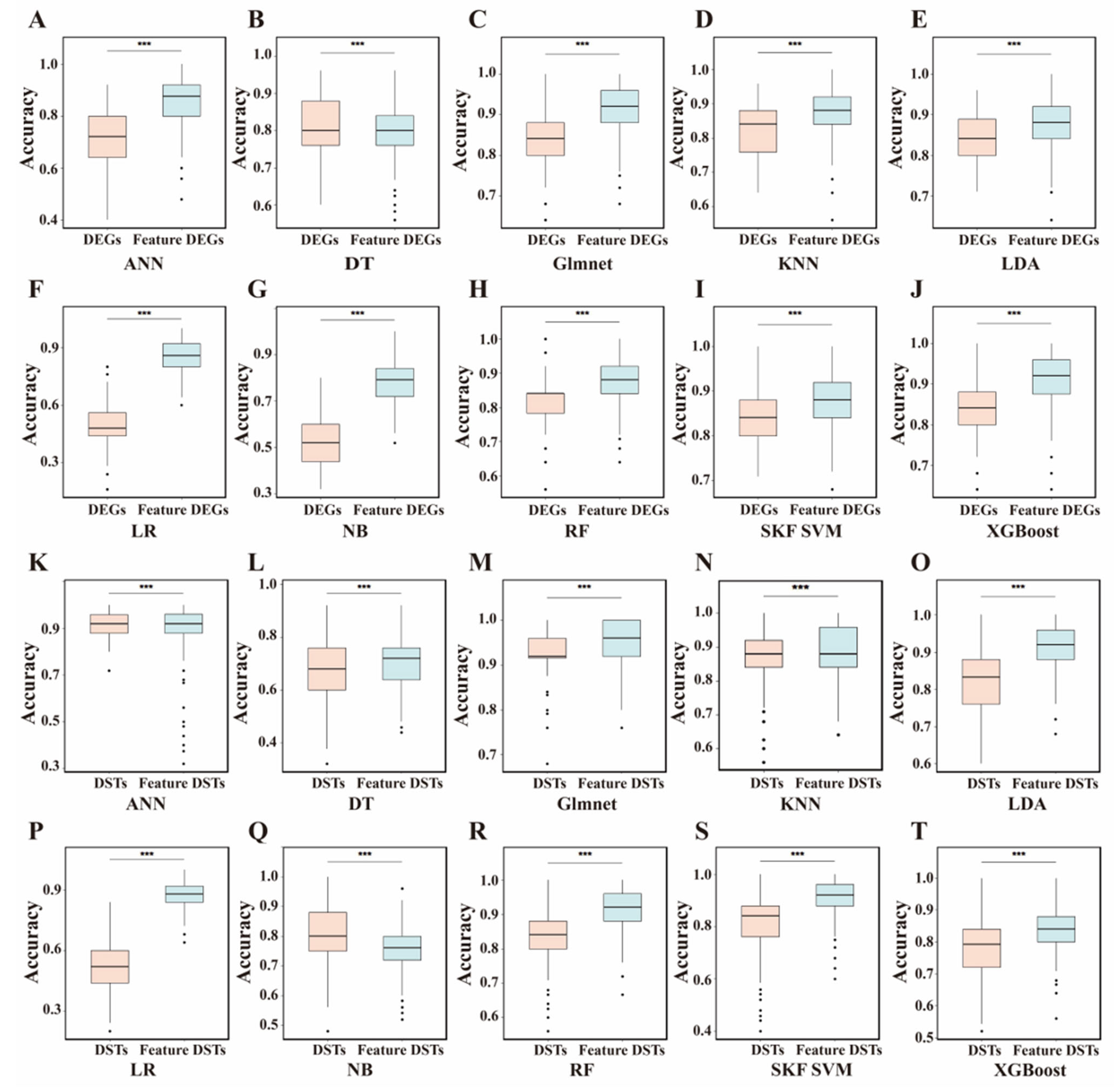
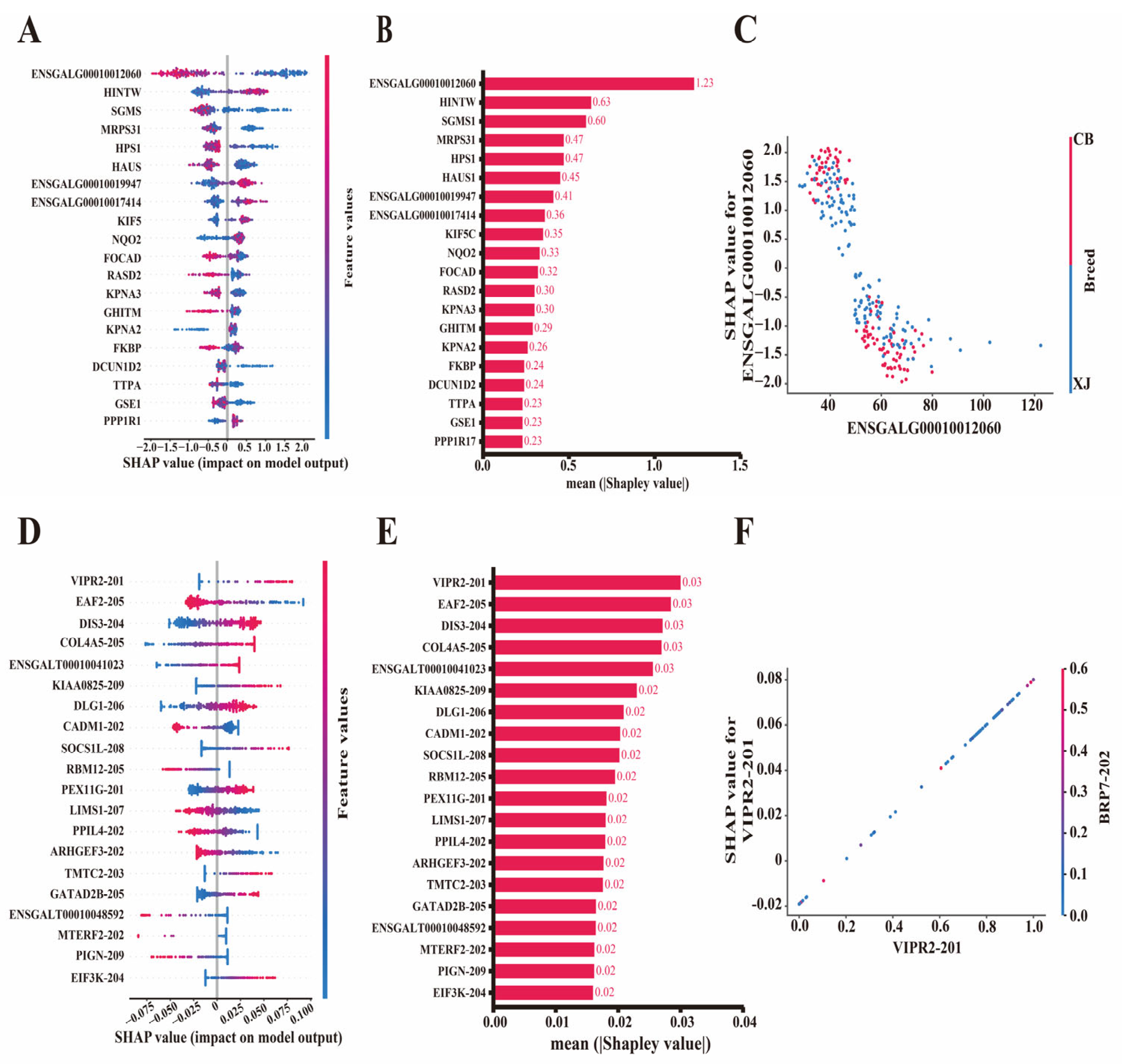
3.5. Evaluation of Feature Contributions by Shapley Values
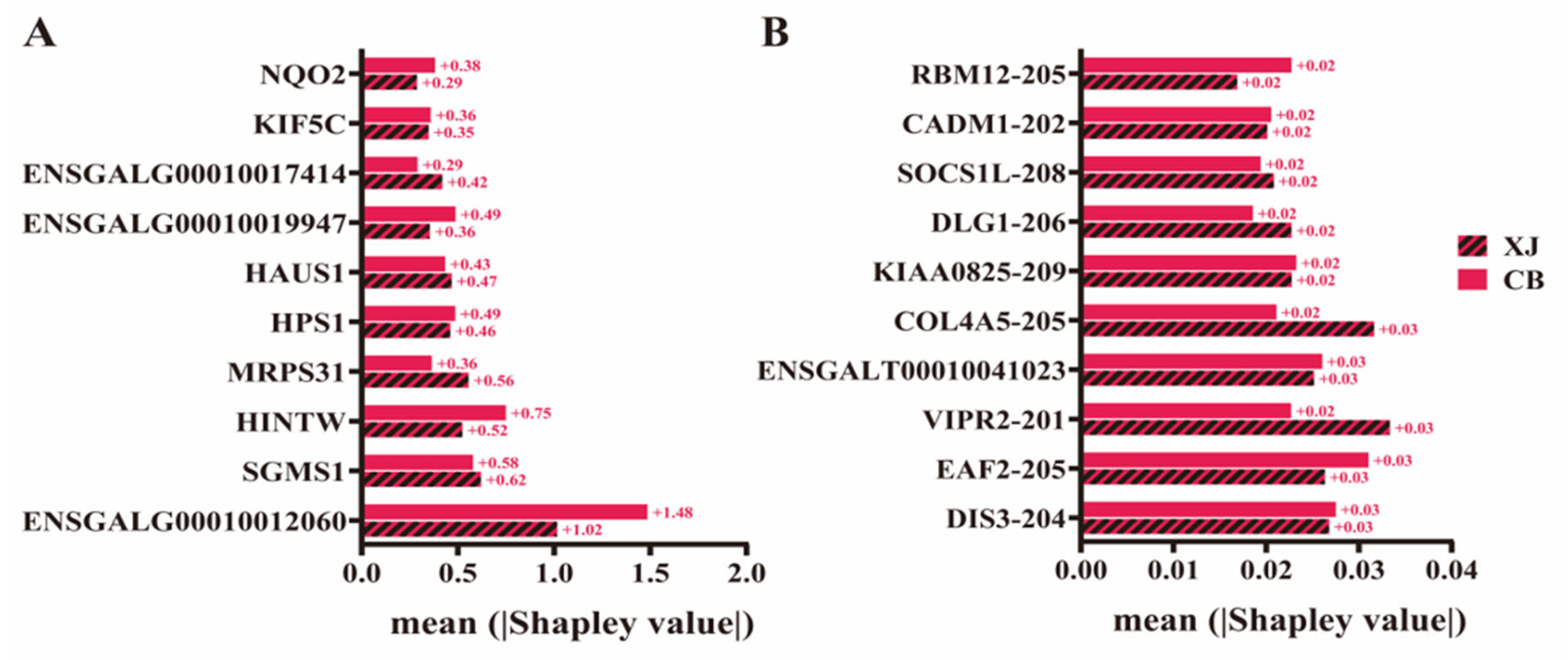
3.6. Evaluation of Breed Effect on the Prediction Results
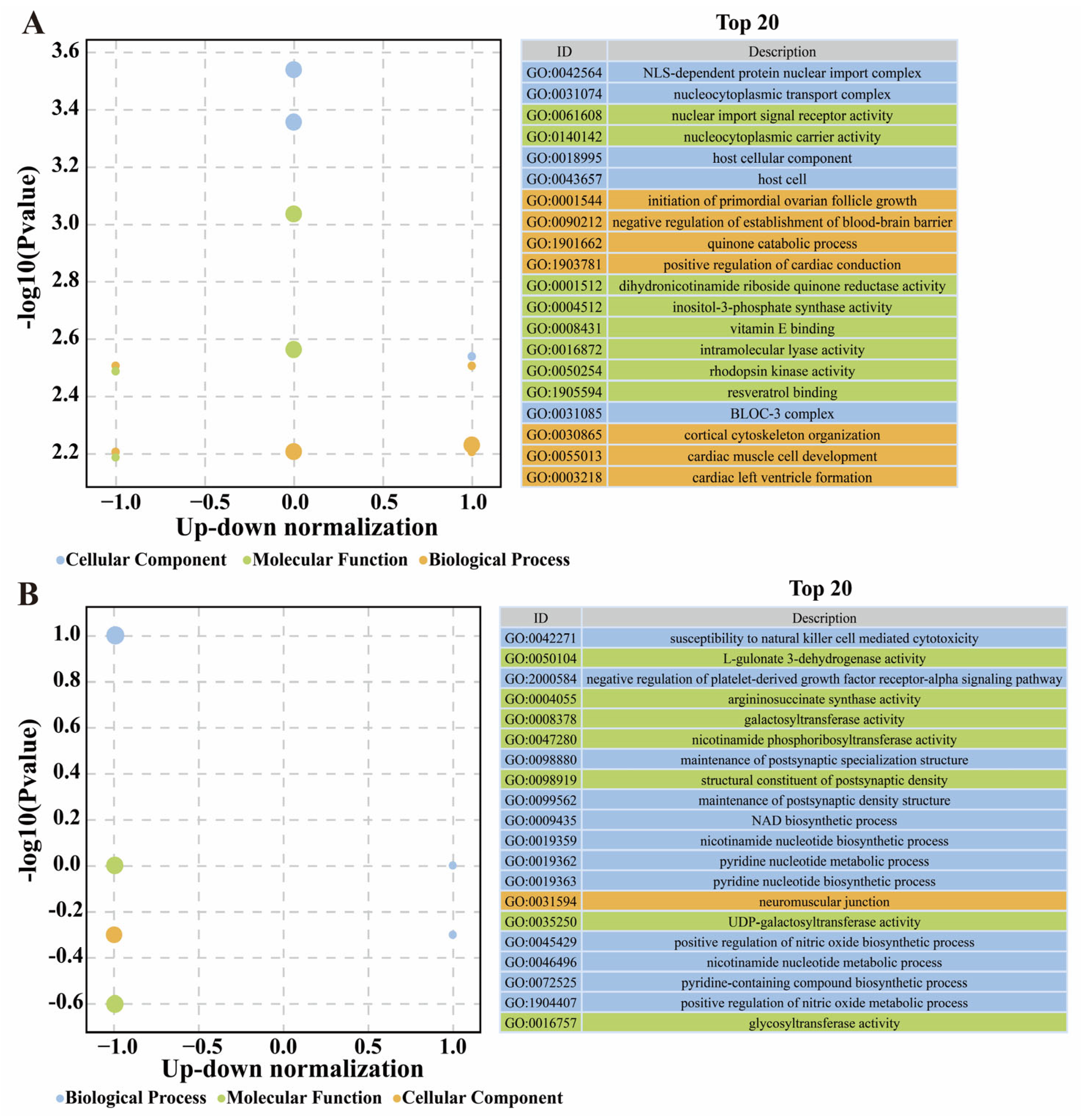
3.7. Annotation of Feature DEGs and DSTs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ANN | Artificial Neural Network |
| AS | Alternative Splicing |
| BrP | Percentage of breast muscle weight |
| DEG | Differentially expressed genes |
| DST | Differentially spliced transcripts |
| DT | Decision Tree |
| Glmnet | Generalized Linear Model Network |
| KNN | K-nearest Neighbor |
| LDA | Linear Discriminant Analysis |
| LR | Logistic Regression |
| ML | Machine learning |
| NB | Naïve Bayes |
| RF | Random Forest |
| SHAP | Shapley Additive exPlanations |
| SKF SVM | Sigmoid Kernel Function Support Vector Machine |
| XGBoost | eXtreme Gradient Boosting |
References
- FAOSTAT Statistical Database; FA. Available online: https://www.fao.org/faostat/en/#search/2019 (accessed on 30 May 2025).
- Poore, J.; Nemecek, T. Reducing food’s environmental impacts through producers and consumers. Science 2018, 360, 987–992. [Google Scholar] [CrossRef] [PubMed]
- Mottet, A.; Tempio, G. Global poultry production: Current state and future outlook and challenges. World’s Poult. Sci. J. 2017, 73, 245–256. [Google Scholar] [CrossRef]
- Zuidhof, M.J.; Schneider, B.L.; Carney, V.L.; Korver, D.R.; Robinson, F.E. Growth, efficiency, and yield of commercial broilers from 1957, 1978, and 2005. Poult. Sci. 2014, 93, 2970–2982. [Google Scholar] [CrossRef]
- Tan, X.; Liu, R.; Zhao, D.; He, Z.; Li, W.; Zheng, M.; Li, Q.; Wang, Q.; Liu, D.; Feng, F.; et al. Large-scale genomic and transcriptomic analyses elucidate the genetic basis of high meat yield in chickens. J. Adv. Res. 2024, 55, 1–16. [Google Scholar] [CrossRef]
- Zambonelli, P.; Zappaterra, M.; Soglia, F.; Petracci, M.; Sirri, F.; Cavani, C.; Davoli, R. Detection of differentially expressed genes in broiler pectoralis major muscle affected by White Striping—Wooden Breast myopathies. Poult. Sci. 2016, 95, 2771–2785. [Google Scholar] [CrossRef]
- Takeda, J.; Suzuki, Y.; Nakao, M.; Barrero, R.A.; Koyanagi, K.O.; Jin, L.; Motono, C.; Hata, H.; Isogai, T.; Nagai, K.; et al. Large-scale identification and characterization of alternative splicing variants of human gene transcripts using 56,419 completely sequenced and manually annotated full-length cDNAs. Nucleic Acids Res. 2006, 34, 3917–3928. [Google Scholar] [CrossRef]
- Liu, J.; Tan, S.; Huang, S.; Huang, W. ASlive: A database for alternative splicing atlas in livestock animals. BMC Genom. 2020, 21, 97. [Google Scholar] [CrossRef]
- Kim, E.; Magen, A.; Ast, G. Different levels of alternative splicing among eukaryotes. Nucleic Acids Res. 2007, 35, 125–131. [Google Scholar] [CrossRef]
- Bland, C.S.; Wang, E.T.; Vu, A.; David, M.P.; Castle, J.C.; Johnson, J.M.; Cooper, T.A.; Burge, C.B. Global regulation of alternative splicing during myogenic differentiation. Nucleic Acids Res. 2010, 38, 7651–7664. [Google Scholar] [CrossRef]
- Shin, S.; Song, Y.; Ahn, J.; Kim, E.; Chen, P.; Yang, S.; Suh, Y.; Lee, K. A novel mechanism of myostatin regulation by its alternative splicing variant during myogenesis in avian species. Am. J. Physiol. Cell Physiol. 2015, 309, C650–C659. [Google Scholar] [CrossRef]
- Chen, G.; Chen, J.; Qi, L.; Yin, Y.; Lin, Z.; Wen, H.; Zhang, S.; Xiao, C.; Bello, S.F.; Zhang, X.; et al. Bulk and single-cell alternative splicing analyses reveal roles of TRA2B in myogenic differentiation. Cell Prolif. 2024, 57, e13545. [Google Scholar] [CrossRef]
- Reel, P.S.; Reel, S.; Pearson, E.; Trucco, E.; Jefferson, E. Using machine learning approaches for multi-omics data analysis: A review. Biotechnol. Adv. 2021, 49, 107739. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016. [Google Scholar]
- Pirooznia, M.; Deng, Y. SVM Classifier—A comprehensive java interface for support vector machine classification of microarray data. BMC Bioinform. 2006, 7, S25. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Li, X.; Zong, M.; Zhu, X.; Wang, R. Efficient kNN classification with different numbers of nearest neighbors. IEEE Trans. Neural Netw. Learn. Syst. 2017, 29, 1774–1785. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Tran, H. An XGBoost-Based Fitted Q Iteration for Finding the Optimal STI Strategies for HIV Patients. IEEE Trans. Neural Netw. Learn. Syst. 2022, 35, 648–656. [Google Scholar] [CrossRef]
- Nazari, M.; Shiri, I.; Zaidi, H. Radiomics-based machine learning model to predict risk of death within 5-years in clear cell renal cell carcinoma patients. Comput. Biol. Med. 2021, 129, 104135. [Google Scholar] [CrossRef]
- Friedman, J.; Hastie, T.; Tibshirani, R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J. Stat. Softw. 2010, 33, 1–22. [Google Scholar] [CrossRef]
- Kou, J.; Wang, Y.; Chen, Z.; Shi, Y.; Guo, Q. Gait planning and multimodal human-exoskeleton cooperative control based on central pattern generator. IEEE/ASME Trans. Mechatron. 2024. [Google Scholar] [CrossRef]
- Chen, Y.; Yu, W.; Benali, A.; Lu, D.; Kok, S.Y.; Wang, R. Towards Human-like Walking with Biomechanical and Neuromuscular Control Features: Personalized Attachment Point Optimization Method of Cable-Driven Exoskeleton. Front. Aging Neurosci. 2024, 16, 1327397. [Google Scholar] [CrossRef]
- Zhang, G.; Song, C.; Yin, M.; Liu, L.; Zhang, Y.; Li, Y.; Zhang, J.; Guo, M.; Li, C. TRAPT: A multi-stage fused deep learning framework for predicting transcriptional regulators based on large-scale epigenomic data. Nat. Commun. 2025, 16, 3611. [Google Scholar] [CrossRef]
- Li, Z.; Zheng, J.; An, B.; Ma, X.; Ying, F.; Kong, F.; Wen, J.; Zhao, G. Several models combined with ultrasound techniques to predict breast muscle weight in broilers. Poult. Sci. 2023, 102, 102911. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.T.; He, P.G.; Jiang, J.S.; Yang, Y.F.; Wang, S.Y.; Pan, C.H.; Zeng, L.; He, Y.-F.; Chen, Z.-H.; Lin, H.-J.; et al. In vivo prediction of abdominal fat and breast muscle in broiler chicken using live body measurements based on machine learning. Poult. Sci. 2023, 102, 102239. [Google Scholar] [CrossRef] [PubMed]
- Cho, E.; Cho, S.; Kim, M.; Ediriweera, T.K.; Seo, D.; Lee, S.S.; Cha, J.; Jin, D.; Kim, Y.-K.; Lee, J.H. Single nucleotide polymorphism marker combinations for classifying Yeonsan Ogye chicken using a machine learning approach. J. Anim. Sci. Technol. 2022, 64, 830–841. [Google Scholar] [CrossRef] [PubMed]
- Fu, W.; Zhang, R.; Ding, H.; Wang, W.; Liu, H.; Zang, S.; Zhou, R. Identification of Taihang-chicken-specific genetic markers using genome-wide SNPs and machine learning: BREED-SPECIFIC SNPS OF TAIHANG CHICKEN. Poult. Sci. 2025, 104, 104585. [Google Scholar]
- Liu, H.; Xing, K.; Jiang, Y.; Liu, Y.; Wang, C.; Ding, X. Using Machine Learning to Identify Biomarkers Affecting Fat Deposition in Pigs by Integrating Multisource Transcriptome Information. J. Agric. Food Chem. 2022, 70, 10359–10370. [Google Scholar] [CrossRef]
- Chen, S. Ultrafast one-pass FASTQ data preprocessing, quality control, and deduplication using fastp. Imeta 2023, 2, e107. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Kovaka, S.; Zimin, A.V.; Pertea, G.M.; Razaghi, R.; Salzberg, S.L.; Pertea, M. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 2019, 20, 278. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Trincado, J.L.; Entizne, J.C.; Hysenaj, G.; Singh, B.; Skalic, M.; Elliott, D.J.; Eyras, E. SUPPA2: Fast, accurate, and uncertainty-aware differential splicing analysis across multiple conditions. Genome Biol. 2018, 19, 40. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Lang, M.; Binder, M.; Richter, J.; Schratz, P.; Pfisterer, F.; Coors, S.; Au, Q.; Casalicchio, G.; Kotthoff, L.; Bischl, B. mlr3: A modern object-oriented machine learning framework in R. J. Open Source Softw. 2019, 4, 1903. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Lee, S.-I. A unified approach to interpreting model predictions. Adv. Neural Inf. Process. Syst. 2017, 30, 4765–4774. [Google Scholar]
- Bu, D.; Luo, H.; Huo, P.; Wang, Z.; Zhang, S.; He, Z.; Wu, Y.; Zhao, L.; Liu, J.; Guo, J.; et al. KOBAS-i: Intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 2021, 49, W317–W325. [Google Scholar] [CrossRef] [PubMed]
- Villanueva, R.A.M.; Chen, Z.J. ggplot2: Elegant Graphics for Data Analysis; Taylor & Francis: Abingdon, UK, 2019. [Google Scholar]
- Mukherjee, K.; LaConte, L.E.W.; Srivastava, S. The Non-Linear Path from Gene Dysfunction to Genetic Disease: Lessons from the MICPCH Mouse Model. Cells 2022, 11, 1131. [Google Scholar] [CrossRef]
- Tan, X.; Liu, L.; Liu, X.; Cui, H.; Liu, R.; Zhao, G.; Wen, J. Large-Scale Whole Genome Sequencing Study Reveals Genetic Architecture and Key Variants for Breast Muscle Weight in Native Chickens. Genes 2021, 13, 3. [Google Scholar] [CrossRef]
- Xiang, T.; Li, T.; Li, J.; Li, X.; Wang, J. Using machine learning to realize genetic site screening and genomic prediction of productive traits in pigs. FASEB J. 2023, 37, e22961. [Google Scholar] [CrossRef]
- Zhu, R.; Li, J.; Yang, J.; Sun, R.; Yu, K. In Vivo Prediction of Breast Muscle Weight in Broiler Chickens Using X-ray Images Based on Deep Learning and Machine Learning. Animals 2024, 14, 628. [Google Scholar] [CrossRef]
- Mantica, F.; Irimia, M. The 3D-Evo Space: Evolution of Gene Expression and Alternative Splicing Regulation. Annu. Rev. Genet. 2022, 56, 315–337. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Yan, Y.; Wang, Y.; Liu, Y.; Yang, R. Splicing complexity as a pivotal feature of alternative exons in mammalian species. BMC Genom. 2023, 24, 198. [Google Scholar] [CrossRef] [PubMed]
- Ule, J.; Blencowe, B.J. Alternative Splicing Regulatory Networks: Functions, Mechanisms, and Evolution. Mol. Cell 2019, 76, 329–345. [Google Scholar] [CrossRef] [PubMed]
- Guidotti, R.; Monreale, A.; Ruggieri, S.; Turini, F.; Giannotti, F.; Pedreschi, D. A survey of methods for explaining black box models. ACM Comput. Surv. 2018, 51, 1–42. [Google Scholar] [CrossRef]
- Strumbelj, E.; Kononenko, I. An efficient explanation of individual classifications using game theory. J. Mach. Learn. Res. 2010, 11, 1–18. [Google Scholar]
- Ji, H.; Xu, Y.; Teng, G. Predicting egg production rate and egg weight of broiler breeders based on machine learning and Shapley additive explanations. Poult. Sci. 2025, 104, 104458. [Google Scholar] [CrossRef]
- Guo, Y.; Li, S.; Na, R.; Guo, L.; Huo, C.; Zhu, L.; Shi, C.; Na, R.; Gu, M.; Zhang, W. Comparative Transcriptome Analysis of Bovine, Porcine, and Sheep Muscle Using Interpretable Machine Learning Models. Animals 2024, 14, 2947. [Google Scholar] [CrossRef]
- Rengaraj, D.; Cha, D.G.; Park, K.J.; Lee, K.Y.; Woo, S.J.; Han, J.Y. Finer resolution analysis of transcriptional programming during the active migration of chicken primordial germ cells. Comput. Struct. Biotechnol. J. 2022, 20, 5911–5924. [Google Scholar] [CrossRef]
- Onteru, S.K.; Gorbach, D.M.; Young, J.M.; Garrick, D.J.; Dekkers, J.C.; Rothschild, M.F.; Liu, Z. Whole Genome Association Studies of Residual Feed Intake and Related Traits in the Pig. PLoS ONE 2013, 8, e61756. [Google Scholar] [CrossRef]
- Nikonova, E.; Kao, S.Y.; Spletter, M.L. Contributions of alternative splicing to muscle type development and function. Semin. Cell Dev. Biol. 2020, 104, 65–80. [Google Scholar] [CrossRef]
- Cuppens, T.; Moisse, M.; Depreeuw, J.; Annibali, D.; Colas, E.; Gil-Moreno, A.; Huvila, J.; Carpén, O.; Zikán, M.; Matias-Guiu, X.; et al. Integrated genome analysis of uterine leiomyosarcoma to identify novel driver genes and targetable pathways. Int. J. Cancer 2018, 142, 1230–1243. [Google Scholar] [CrossRef]
- Liu, X.; Chen, Z.; Ouyang, G.; Song, T.; Liang, H.; Liu, W.; Xiao, W. ELL Protein-associated Factor 2 (EAF2) Inhibits Transforming Growth Factor β Signaling through a Direct Interaction with Smad3. J. Biol. Chem. 2015, 290, 25933–25945. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, X.; Huang, M.; Jin, Y.; Li, J.; Dong, J.; Wang, D. Deciphering the Gene Expression and Alternative Splicing Basis of Muscle Development Through Interpretable Machine Learning Models. Biology 2025, 14, 1059. https://doi.org/10.3390/biology14081059
Tan X, Huang M, Jin Y, Li J, Dong J, Wang D. Deciphering the Gene Expression and Alternative Splicing Basis of Muscle Development Through Interpretable Machine Learning Models. Biology. 2025; 14(8):1059. https://doi.org/10.3390/biology14081059
Chicago/Turabian StyleTan, Xiaodong, Minjie Huang, Yuting Jin, Jiahua Li, Jie Dong, and Deqian Wang. 2025. "Deciphering the Gene Expression and Alternative Splicing Basis of Muscle Development Through Interpretable Machine Learning Models" Biology 14, no. 8: 1059. https://doi.org/10.3390/biology14081059
APA StyleTan, X., Huang, M., Jin, Y., Li, J., Dong, J., & Wang, D. (2025). Deciphering the Gene Expression and Alternative Splicing Basis of Muscle Development Through Interpretable Machine Learning Models. Biology, 14(8), 1059. https://doi.org/10.3390/biology14081059





