The Gradient Heterogeneity of Deserts Alters the Interaction Relationships Between Xerophytic Plants and Soils
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Areas and Experimental Design
2.2. Plant Collection
2.3. Soil Collection
2.4. DNA Extraction and Amplicon Sequencing
2.5. Statistical Analysis
3. Results
3.1. Plant Functional Traits
3.2. Soil Environmental Factors
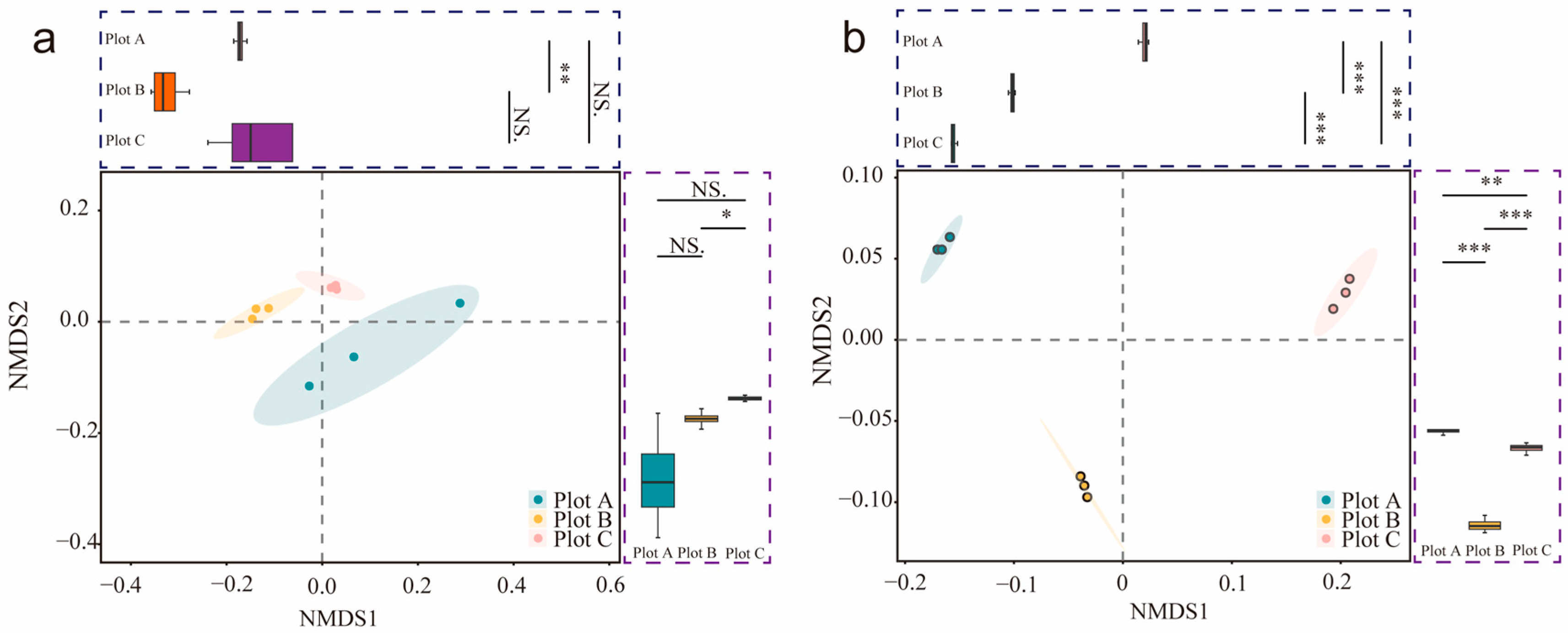
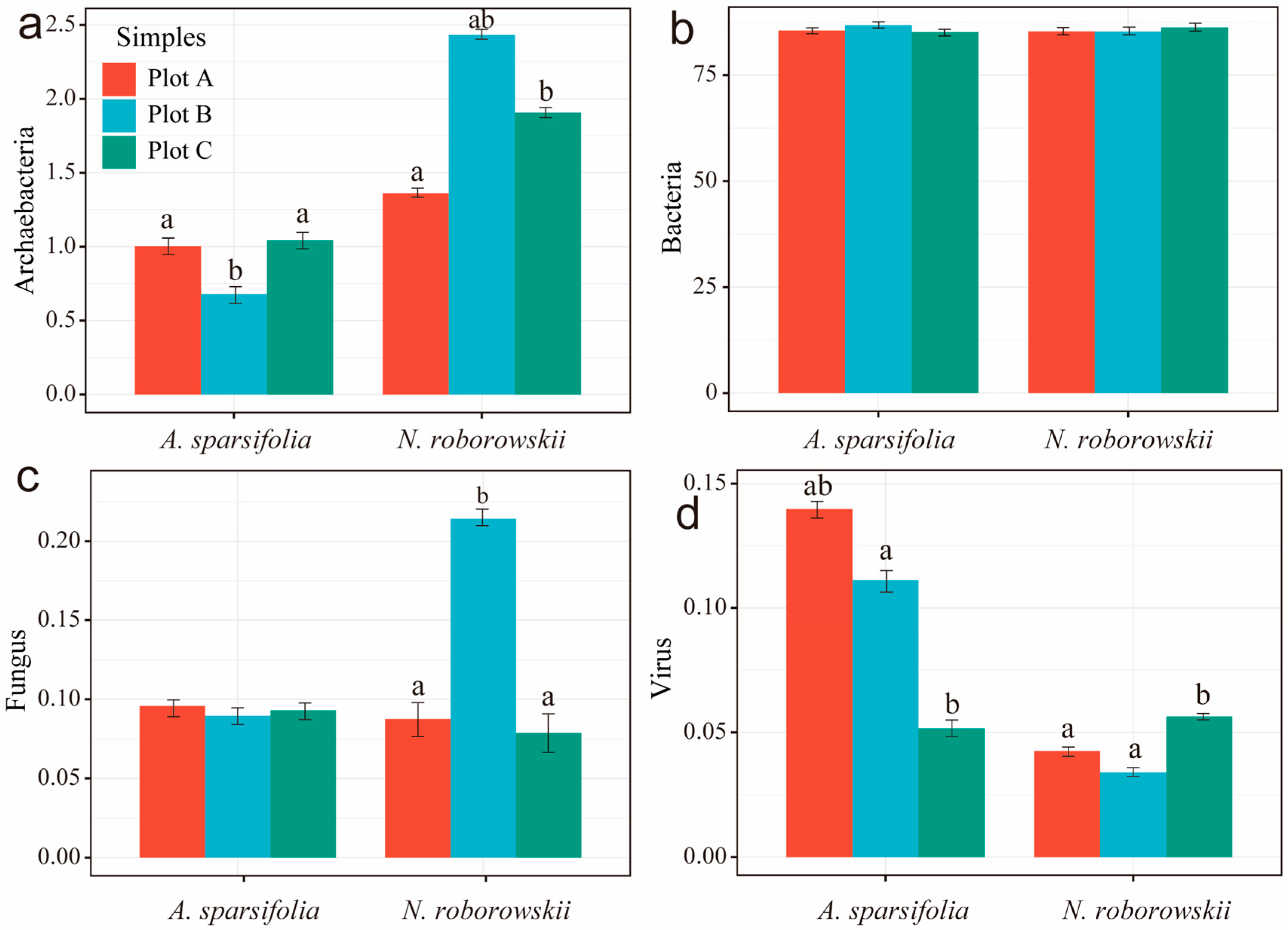
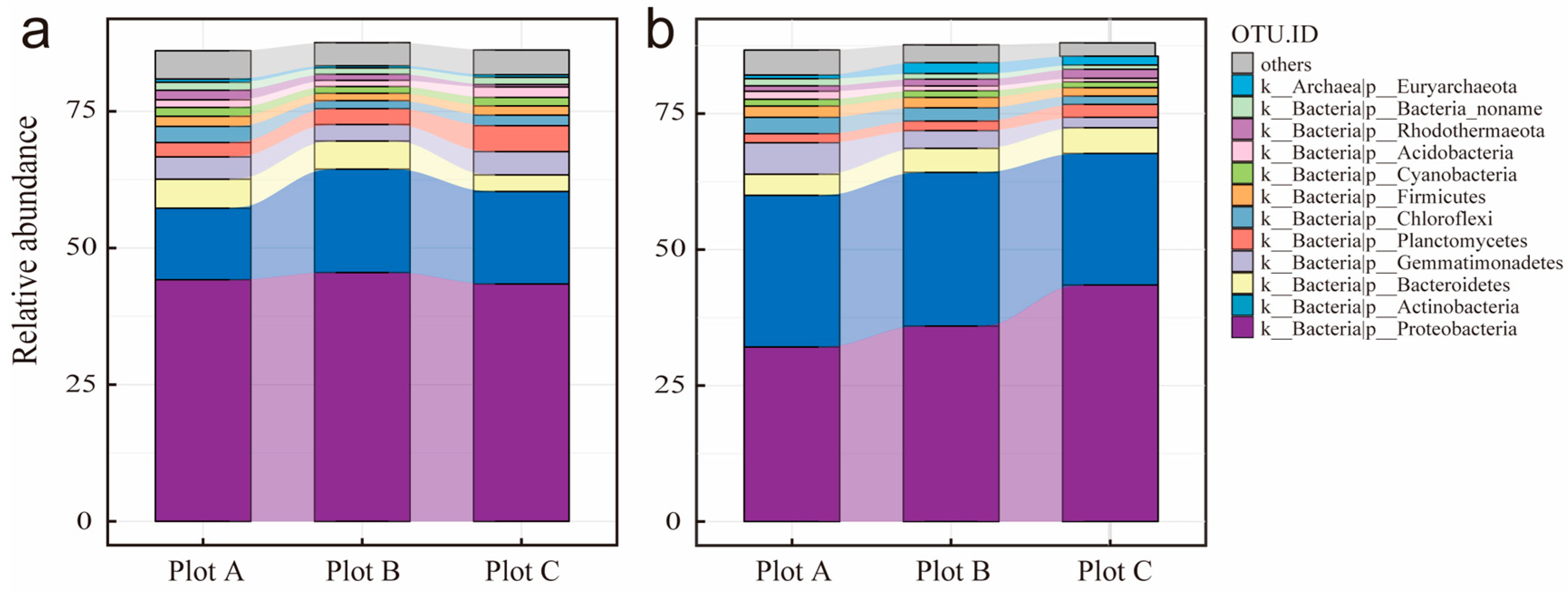
3.3. Rhizosphere Soil Microorganisms
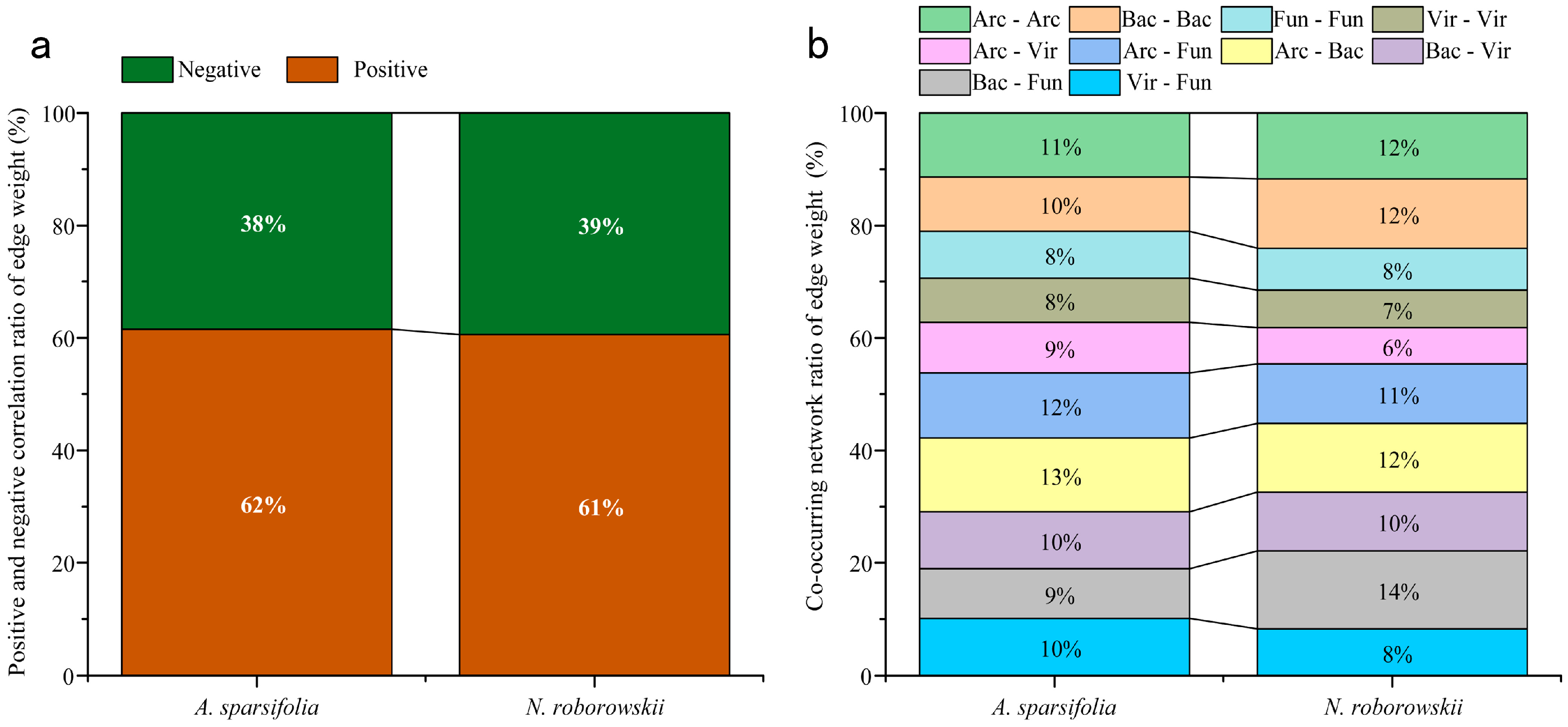
3.4. Relationships Among Plant Functional Traits, Rhizosphere Soil Physicochemical Properties, Enzyme Activities, and Microorganisms
4. Discussion
4.1. Gradient Heterogeneity of Functional Traits in Two Desert Halophytes
4.2. Gradient Heterogeneity of Soil Properties and Rhizosphere Microbial Community Structure in Two Desert Halophytes
4.3. Interactions Between Functional Traits, Soil Properties, and Rhizosphere Microbial Structure in Two Desert Halophytes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xiao, X.; Fan, M.; Wang, E.; Chen, W.; Wei, G. Interactions of Plant Growth-Promoting Rhizobacteria and Soil Factors in Two Leguminous Plants. Appl. Microbiol. Biotechnol. 2017, 101, 8485–8497. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, D.S.; Lebeis, S.L.; Paredes, S.H.; Yourstone, S.; Gehring, J.; Malfatti, S.; Tremblay, J.; Engelbrektson, A.; Kunin, V.; Rio, T.G.D.; et al. Defining the Core Arabidopsis Thaliana Root Microbiome. Nature 2012, 488, 86–90. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Lake, B.B.; Zhang, K. High-throughput sequencing of the transcriptome and chromatin accessibility in the same cell. Nat. Biotechnol. 2019, 37, 1452–1457. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Kuzyakov, Y. Rhizosphere engineering for soil carbon sequestration. Trends Plant Sci. 2024, 29, 447–468. [Google Scholar] [CrossRef]
- Xun, W.; Liu, Y.; Ma, A.; Yan, H.; Miao, Y.; Shao, J.; Zhang, N.; Xu, Z.; Shen, Q.; Zhang, R. Dissection of rhizosphere microbiome and exploiting strategies for sustainable agriculture. New Phytol. 2024, 242, 10. [Google Scholar] [CrossRef] [PubMed]
- Wankhade, A.; Wilkinson, E.; Britt, D.W.; Kaundal, A. A Review of Plant–Microbe Interactions in the Rhizosphere and the Role of Root Exudates in Microbiome Engineering. Appl. Sci. 2025, 15, 7127. [Google Scholar] [CrossRef]
- Gamalero, E.; Glick, B.R. Bacterial Modulation of Plant Ethylene Levels. Plant Physiol. 2015, 169, 13–22. [Google Scholar] [CrossRef]
- Hasan, A.; Tabassum, B.; Hashim, M.; Khan, N. Role of Plant Growth Promoting Rhizobacteria (PGPR) as a Plant Growth Enhancer for Sustainable Agriculture: A Review. Bacteria 2024, 3, 59–75. [Google Scholar] [CrossRef]
- Anderson, M.; Habiger, J. Characterization and Identification of Productivity-Associated Rhizobacteria in Wheat. Appl. Environ. Microbiol. 2012, 78, 4434–4446. [Google Scholar] [CrossRef] [PubMed]
- Vacheron, J.; Desbrosses, G.; Bouffaud, M.-L.; Touraine, B.; Moënne-Loccoz, Y.; Muller, D.; Legendre, L.; Wisniewski-Dyé, F.; Prigent-Combaret, C. Plant Growth-Promoting Rhizobacteria and Root System Functioning. Front. Plant Sci. 2013, 4, 356. [Google Scholar] [CrossRef]
- Nuccio, E.E.; Anderson-Furgeson, J.; Estera, K.Y.; Pett-Ridge, J.; de Valpine, P.; Brodie, E.L.; Firestone, M.K. Climate and Edaphic Controllers Influence Rhizosphere Community Assembly for a Wild Annual Grass. Ecology 2016, 97, 1307–1318. [Google Scholar] [CrossRef] [PubMed]
- Pii, Y.; Borruso, L.; Brusetti, L.; Crecchio, C.; Cesco, S.; Mimmo, T. The Interaction between Iron Nutrition, Plant Species and Soil Type Shapes the Rhizosphere Microbiome. Plant Physiol. Biochem. 2016, 99, 39–48. [Google Scholar] [CrossRef]
- Edwards, J.; Johnson, C.; Santos-Medellín, C.; Lurie, E.; Podishetty, N.K.; Bhatnagar, S.; Eisen, J.A.; Sundaresan, V. Structure, Variation, and Assembly of the Root-Associated Microbiomes of Rice. Proc. Natl. Acad. Sci. USA 2015, 112, E911–E920. [Google Scholar] [CrossRef]
- Zhao, J.; Ni, T.; Li, Y.; Xiong, W.; Ran, W.; Shen, B.; Shen, Q.; Zhang, R. Responses of Bacterial Communities in Arable Soils in a Rice-Wheat Cropping System to Different Fertilizer Regimes and Sampling Times. PLoS ONE 2014, 9, e85301. [Google Scholar] [CrossRef]
- Zhalnina, K.; Dias, R.; De Quadros, P.D.; Davis-Richardson, A.; Camargo, F.A.O.; Clark, I.M.; McGrath, S.P.; Hirsch, P.R.; Triplett, E.W. Soil pH Determines Microbial Diversity and Composition in the Park Grass Experiment. Microb. Ecol. 2015, 69, 395–406. [Google Scholar] [CrossRef]
- Chaparro, J.M.; Badri, D.V.; Vivanco, J.M. Rhizosphere Microbiome Assemblage Is Affected by Plant Development. ISME J. 2014, 8, 790–803. [Google Scholar] [CrossRef]
- Chai, Y.N.; Schachtman, D.P. Root Exudates Impact Plant Performance under Abiotic Stress. Trends Plant Sci. 2022, 27, 80–91. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.P.; Yu, J.M.; Wang, J.F.; Liu, D.Y.; Gong, J.; Jiang, L.Q.; Zhang, S.-T.; Yu, X.-W.; Li, S.-L.; Yang, L.; et al. Soil properties affect bacterial community assembly and co-occurrence network in tobacco rhizosphere. AMS 2023, 63, 1168–1184. [Google Scholar] [CrossRef]
- Colchado-López, J.; Rougon-Cardoso, A.; Vélez, P.; Rosas, U. Meta-Analysis of Community Composition Patterns of Halophyte and Xerophyte Rhizosphere Associated Bacteria. Rhizosphere 2022, 24, 100588. [Google Scholar] [CrossRef]
- Mathur, V.; Ulanova, D. Microbial Metabolites Beneficial to Plant Hosts Across Ecosystems. Microb. Ecol. 2023, 86, 25–48. [Google Scholar] [CrossRef]
- Mukhtar, S.; Mehnaz, S.; Malik, K.A. Microbial Diversity in the Rhizosphere of Plants Growing under Extreme Environments and Its Impact on Crop Improvement. Environ. Sustain. 2019, 2, 329–338. [Google Scholar] [CrossRef]
- Chowaniec, K.; Zubek, S.; Zalewska-Gałosz, J.; Stanek, M.; Skubała, K. Mosaic of Biological Soil Crusts and Vascular Plants Contributes to the Spatial Heterogeneity of Key Soil Properties at Different Successional Stages of Restored Inland Sand Dunes. Plant Soil 2025, 510, 603–626. [Google Scholar] [CrossRef]
- Wang, J.; Teng, D.; He, X.; Qin, L.; Yang, X.; Lv, G. Spatial Non-Stationarity Effects of Driving Factors on Soil Respiration in an Arid Desert Region. Catena 2021, 207, 105617. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, J.; Jiang, L.; Li, H.; Wang, H.; Lv, G.; Li, X. Prediction of Spatial Distribution Characteristics of Ecosystem Functions Based on a Minimum Data Set of Functional Traits of Desert Plants. Front. Plant Sci. 2023, 14, 1131778. [Google Scholar] [CrossRef]
- Li, X.; Chen, Y.; Lv, G.; Wang, J.; Jiang, L.; Wang, H.; Yang, X. Predicting Spatial Variability of Species Diversity with the Minimum Data Set of Soil Properties in an Arid Desert Riparian Forest. Front. Plant Sci. 2022, 13, 1014643. [Google Scholar] [CrossRef]
- Li, W.; Li, Z.; Li, Z.; Chen, Y.; Lv, G.; Tian, J.; Yang, G. Coupling of Leaf Economic and Hydraulic Traits of Desert Plants of Different Life Forms in the Ebinur Lake Basin. Environ. Exp. Bot. 2024, 219, 105644. [Google Scholar] [CrossRef]
- Cao, W.; Chen, Y.; Jiang, L.; Wu, Q.; Zhu, M.; Wang, Y.; Li, W.; Lv, G. Ecoenzymatic Stoichiometry Reveals That Increasing Altitude Exacerbates Soil Microbial Phosphorus Limitation in Alpine Grassland Ecosystems in Xinjiang. Environ. Res. 2025, 275, 121320. [Google Scholar] [CrossRef] [PubMed]
- Carvajal, D.E.; Loayza, A.P.; Squeo, F.A. Functional diversity and spatial association analyses at different spatial scales reveal no changes in community assembly processes along an aridity gradient in the Atacama Desert. Sci. Rep. 2023, 13, 19905. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Shi, C.; Wang, L.; Han, X.; Zhu, Y.; Liu, J.; Yang, X. Functional Trait Responses of Sophora Alopecuroides L. Seedlings to Diverse Environmental Stresses in the Desert Steppe of Ningxia, China. Plants 2023, 13, 69. [Google Scholar] [CrossRef]
- Yao, Z.; Pan, X.; Yang, X.; Shao, X.; Wang, B.; Deng, Y.; Zhang, Z.; Li, Q.; Liu, L. Canopy structural heterogeneity drives α and β species–genetic diversity correlations in a Chinese subtropical forest. Plant Divers. 2025, 47, 47–106. [Google Scholar] [CrossRef] [PubMed]
- García-Palacios, P.; Maestre, F.T.; Bardgett, R.D.; De Kroon, H. Plant Responses to Soil Heterogeneity and Global Environmental Change. J. Ecol. 2012, 100, 1303–1314. [Google Scholar] [CrossRef]
- Faucon, M.-P.; Houben, D.; Lambers, H. Plant Functional Traits: Soil and Ecosystem Services. Trends Plant Sci. 2017, 22, 385–394. [Google Scholar] [CrossRef]
- Fernández-Marín, B.; Gulías, J.; Figueroa, C.M.; Iñiguez, C.; Clemente-Moreno, M.J.; Nunes-Nesi, A.; Fernie, A.R.; Cavieres, L.A.; Bravo, L.A.; García-Plazaola, J.I.; et al. How Do Vascular Plants Perform Photosynthesis in Extreme Environments? An Integrative Ecophysiological and Biochemical Story. Plant J. 2020, 101, 979–1000. [Google Scholar] [CrossRef]
- Asefa, M.; Worthy, S.J.; Cao, M.; Song, X.; Lozano, Y.M.; Yang, J. Above- and below-Ground Plant Traits Are Not Consistent in Response to Drought and Competition Treatments. Ann. Bot. 2022, 130, 939–950. [Google Scholar] [CrossRef]
- Shryock, D.F.; DeFalco, L.A.; Esque, T.C. Life-History Traits Predict Perennial Species Response to Fire in a Desert Ecosystem. Ecol. Evol. 2014, 4, 3046–3059. [Google Scholar] [CrossRef]
- Schöb, C.; Armas, C.; Guler, M.; Prieto, I.; Pugnaire, F.I. Variability in Functional Traits Mediates Plant Interactions along Stress Gradients. J. Ecol. 2013, 101, 753–762. [Google Scholar] [CrossRef]
- Akman, M.; Carlson, J.E.; Latimer, A.M. Climate Explains Population Divergence in Drought-Induced Plasticity of Functional Traits and Gene Expression in a South African Protea. Mol. Ecol. 2021, 30, 255–273. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Xu, Y.; Tang, Z.; Jin, S.; Yang, S. The Impact of Alkaline Stress on Plant Growth and Its Alkaline Resistance Mechanisms. Int. J. Mol. Sci. 2024, 25, 13719. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, S.; Zhao, Z.; Zhang, K.; Tian, C.; Mai, W. Progress of Euhalophyte Adaptation to Arid Areas to Remediate Salinized Soil. Agriculture 2023, 13, 704. [Google Scholar] [CrossRef]
- Chang, D.; Lu, X.; Sun, Y.; Fan, H.; Wang, K. Responses of Rhizosphere Microbial Communities and Resource Competition to Soil Amendment in Saline and Alkaline Soils. Plant Soil 2025, 1–17. [Google Scholar] [CrossRef]
- Hodge, A.; Fitter, A.H. Microbial Mediation of Plant Competition and Community Structure. Funct. Ecol. 2013, 27, 865–875. [Google Scholar] [CrossRef]
- Wen, Y.; Wu, R.; Qi, D.; Xu, T.; Chang, W.; Li, K.; Fang, X.; Song, F. The Effect of AMF Combined with Biochar on Plant Growth and Soil Quality under Saline-Alkali Stress: Insights from Microbial Community Analysis. Ecotoxicol. Environ. Saf. 2024, 281, 116592. [Google Scholar] [CrossRef]
- Luo, J.; Zeng, H.; Zhou, Q.; Hu, X.; Qu, Q.; Ouyang, S.; Wang, Y. Anthropogenic Impacts on the Biodiversity and Anti-Interference Ability of Microbial Communities in Lakes. Sci. Total Environ. 2022, 820, 153264. [Google Scholar] [CrossRef]
- Ling, N.; Wang, T.; Kuzyakov, Y. Rhizosphere Bacteriome Structure and Functions. Nat. Commun. 2022, 13, 836. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Huang, Y.; An, S.; Sun, H.; Bhople, P.; Chen, Z. Soil Physicochemical and Microbial Characteristics of Contrasting Land-Use Types along Soil Depth Gradients. Catena 2018, 162, 345–353. [Google Scholar] [CrossRef]
- Li, H.; La, S.; Zhang, X.; Gao, L.; Tian, Y. Salt-Induced Recruitment of Specific Root-Associated Bacterial Consortium Capable of Enhancing Plant Adaptability to Salt Stress. ISME J. 2021, 15, 2865–2882. [Google Scholar] [CrossRef]
- Vives-Peris, V.; De Ollas, C.; Gómez-Cadenas, A.; Pérez-Clemente, R.M. Root Exudates: From Plant to Rhizosphere and Beyond. Plant Cell Rep. 2020, 39, 3–17. [Google Scholar] [CrossRef]
- Marcellus, M.; Goud, E.M.; Swartz, N.; Brown, E.; Soper, F.M. Evolutionary history and root trait coordination predict nutrient strategy in tropical legume trees. New Phytol. 2024, 243, 1711–1723. [Google Scholar] [CrossRef] [PubMed]
- Fang, S.; Hou, X.; Liang, X. Response Mechanisms of Plants Under Saline-Alkali Stress. Front. Plant Sci. 2021, 12, 667458. [Google Scholar] [CrossRef]
- Pérez-Ramos, I.M.; Matías, L.; Gómez-Aparicio, L.; Godoy, Ó. Functional traits and phenotypic plasticity modulate species coexistence across contrasting climatic conditions. Nat. Commun. 2019, 10, 2555. [Google Scholar] [CrossRef]
- Gross, N.; Le Bagousse-Pinguet, Y.; Liancourt, P.; Berdugo, M.; Gotelli, N.J.; Maestre, F.T. Functional trait diversity maximizes ecosystem multifunctionality. Nat. Ecol. Evol. 2017, 1, 132. [Google Scholar] [CrossRef]
- Burns, R.G.; DeForest, J.L.; Marxsen, J.; Sinsabaugh, R.L.; Stromberger, M.E.; Wallenstein, M.D.; Weintraub, M.N.; Zoppini, A. Soil Enzymes in a Changing Environment: Current Knowledge and Future Directions. Soil Biol. Biochem. 2013, 58, 216–234. [Google Scholar] [CrossRef]
- Cui, H.; Wang, S.; Wei, T.; Yang, X.; Li, X.; Fan, M.; Zhang, X.; Song, W.; Ma, J.; Sun, W. Soil phoD-harboring bacteria mediate the responses of phosphorus availability to N addition and mowing among soil aggregates. Geoderma 2025, 454, 117170. [Google Scholar] [CrossRef]
- Liang, M.; Wu, Y.; Jiang, Y.; Zhao, Z.; Yang, J.; Liu, G.; Xue, S. Microbial functional genes play crucial roles in enhancing soil nutrient availability of halophyte rhizospheres in salinized grasslands. Sci. Total Environ. 2024, 958, 178160. [Google Scholar] [CrossRef]
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; Van Der Putten, W.H. Going Back to the Roots: The Microbial Ecology of the Rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789–799. [Google Scholar] [CrossRef]
- Feng, H.; Qiao, Y.; Xia, L.; Yang, W.; Zhao, Y.; Jeelani, N.; An, S. The Responses of Soil Bacterial and Archaeal Communities to Coastal Embankments in Three Typical Salt Marshes of Eastern China. Plant Soil 2022, 477, 439–459. [Google Scholar] [CrossRef]
- Fierer, N. Embracing the Unknown: Disentangling the Complexities of the Soil Microbiome. Nat. Rev. Microbiol. 2017, 15, 579–590. [Google Scholar] [CrossRef]
- Zhou, T.; Liang, G.; Reich, P.B.; Delgado-Baquerizo, M.; Wang, C.; Zhou, Z. Promoting Effect of Plant Diversity on Soil Microbial Functionality Is Amplified over Time. One Earth 2024, 7, 2139–2148. [Google Scholar] [CrossRef]



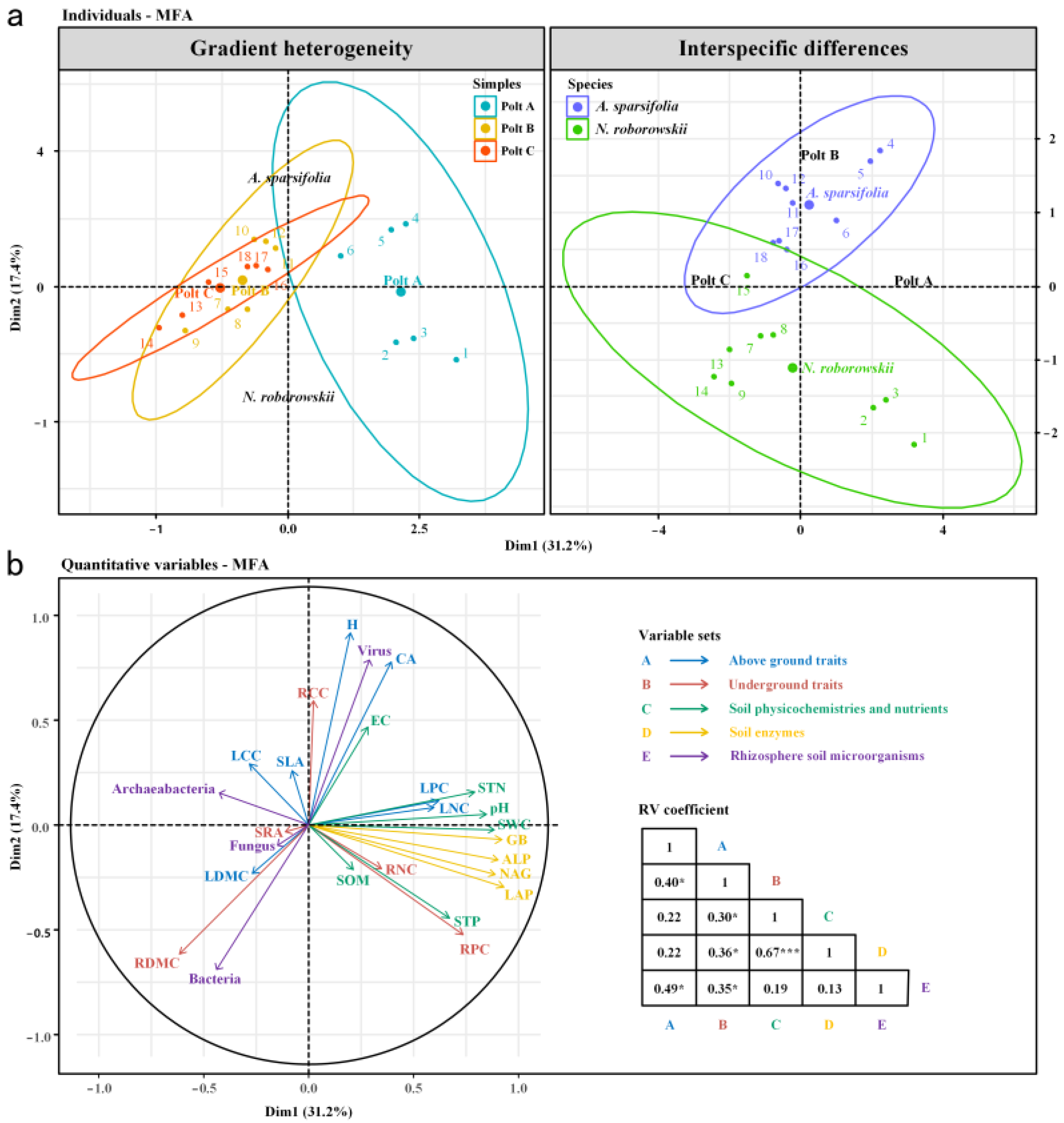
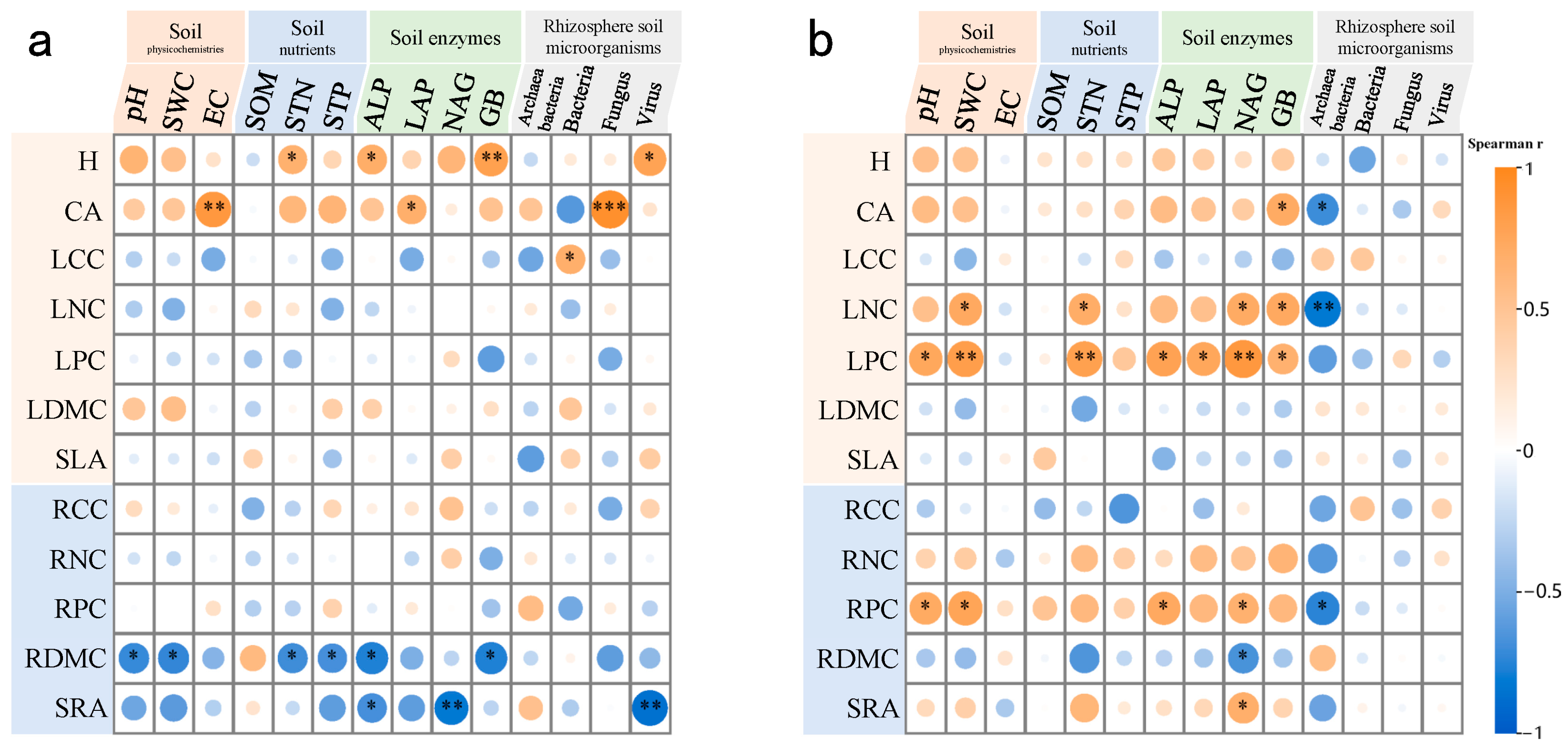
| Samples | Variable | A. sparsifolia | N. roborowskii | Variable | A. sparsifolia | N. roborowskii |
|---|---|---|---|---|---|---|
| Plot A | H (cm) | 74.33 ± 6.63 a | 0.50 ± 0.09 a | SLA (cm2 g−1) | 113.44 ± 30.83 a | 95.46 ± 43.24 a |
| Plot B | 69.33 ± 4.67 a | 0.38 ± 0.01 a | 141.69 ± 10.77 a | 106.48 ± 13.69 a | ||
| Plot C | 59.00 ± 2.65 a | 0.35 ± 0.02 a | 123.51 ± 6.24 a | 121.38 ± 9.93 a | ||
| Plot A | CA (cm2) | 4448.50 ± 683.64 a | 3.19 ± 1.90 a | RCC (g kg−1) | 256.15 ± 15.67 a | 238.36 ± 9.05 a |
| Plot B | 1271.04 ± 204.36 b | 0.24 ± 0.01 a | 264.46 ± 5.84 a | 245.73 ± 2.05 a | ||
| Plot C | 1975.54 ± 265.13 b | 0.56 ± 0.30 a | 239.24 ± 5.00 a | 16.97 ± 3.81 a | ||
| Plot A | LCC (g kg−1) | 380.86 ± 4.72 a | 374.93 ± 11.17 a | RNC (g kg−1) | 13.304 ± 2.39 a | 12.53 ± 0.86 a |
| Plot B | 428.50 ± 19.86 a | 391.34 ± 11.96 a | 14.09 ± 2.17 a | 14.42 ± 1.28 a | ||
| Plot C | 385.62 ± 25.52 a | 393.55 ± 16.56 a | 14.31 ± 0.26 a | 1.12 ± 1.57 a | ||
| Plot A | LNC (g kg−1) | 20.309 ± 1.11 a | 22.93 ± 0.88 a | RPC (g kg−1) | 0.59 ± 0.05 a | 0.46 ± 0.10 a |
| Plot B | 19.08 ± 0.97 a | 16.87 ± 0.74 b | 0.50 ± 0.04 a | 0.51 ± 0.07 b | ||
| Plot C | 22.30 ± 0.53 a | 17.66 ± 2.60 ab | 0.57 ± 0.01 a | 0.44 ± 0.13 b | ||
| Plot A | LPC (g kg−1) | 1.083 ± 0.08 a | 1.34 ± 0.13 a | RDMC (g g−1) | 0.28 ± 0.00 b | 0.69 ± 0.01 a |
| Plot B | 1.23 ± 0.07 a | 0.94 ± 0.08 b | 0.41 ± 0.01 a | 0.57 ± 0.05 a | ||
| Plot C | 1.13 ± 0.05 a | 0.84 ± 0.09 b | 0.43 ± 0.03 a | 6.43 ± 0.12 a | ||
| Plot A | LDMC (g g−1) | 0.30 ± 0.01 a | 0.32 ± 0.11 a | SRA (cm2 g−1) | 3.34 ± 1.22 a | 3.01 ± 0.47 a |
| Plot B | 0.31 ± 0.01 a | 0.32 ± 0.04 a | 2.43 ± 0.12 a | 13.13 ± 1.23 a | ||
| Plot C | 0.28 ± 0.01 a | 0.39 ± 0.12 a | 7.79 ± 2.65 a | 238.36 ± 11.20 a |
| Samples | Variable | A. sparsifolia | N. roborowskii | Variable | A. sparsifolia | N. roborowskii |
|---|---|---|---|---|---|---|
| Plot A | pH | 8.28 ± 0.08 a | 8.21 ± 0.10 a | STP (g kg−1) | 0.84 ± 0.02 a | 0.94 ± 0.11 a |
| Plot B | 7.87 ± 0.02 b | 7.89 ± 0.11 b | 0.62 ± 0.03 b | 0.78 ± 0.02 ab | ||
| Plot C | 7.60 ± 0.09 c | 7.40 ± 0.04 c | 0.58 ± 0.03 b | 0.65 ± 0.06 b | ||
| Plot A | SWC | 15.92 ± 0.88 a | 16.16 ± 0.20 a | ALP (nmol g−1 h−1) | 4.92 ± 0.58 a | 5.93 ± 0.54 a |
| Plot B | 9.15 ± 0.48 b | 8.05 ± 0.56 b | 1.80 ± 0.02 b | 2.33 ± 0.09 b | ||
| Plot C | 3.75 ± 0.54 c | 2.75 ± 0.34 c | 1.24 ± 0.24 b | 0.60 ± 0.05 c | ||
| Plot A | EC (μS cm−1) | 8.09 ± 0.49 a | 4.59 ± 0.44 a | LAP (nmol g−1 h−1) | 14.91 ± 0.02 a | 21.09 ± 0.58 a |
| Plot B | 5.35 ± 0.47 b | 6.06 ± 1.36 a | 4.97 ± 0.02 b | 5.84 ± 0.06 b | ||
| Plot C | 5.76 ± 1.12 ab | 4.49 ± 0.53 a | 4.79 ± 0.17 b | 3.58 ± 0.23 b | ||
| Plot A | SOM (g kg−1) | 15.24 ± 2.32 a | 18.37 ± 2.6 a | NAG (nmol g−1 h−1) | 15.77 ± 4.61 a | 26.32 ± 1.72 a |
| Plot B | 15.07 ± 2.81 a | 18.10 ± 4.25 a | 8.57 ± 0.29 a | 5.91 ± 1.37 b | ||
| Plot C | 17.41 ± 3.65 a | 12.56 ± 4.46 a | 6.85 ± 0.06 a | 5.94 ± 0.11 b | ||
| Plot A | STN (g kg−1) | 0.99 ± 0.10 a | 0.91 ± 0.05 a | GB (nmol g−1 h−1) | 28.51 ± 2.8 a | 30.53 ± 3.32 a |
| Plot B | 0.73 ± 0.07 ab | 0.76 ± 0.07 a | 8.77 ± 1.95 b | 5.39 ± 0.54 b | ||
| Plot C | 0.66 ± 0.08 b | 0.53 ± 0.07 b | 7.83 ± 0.44 b | 5.31 ± 0.31 b |
| Properties | A. sparsifolia | N. roborowskii |
|---|---|---|
| Edge number | 650 | 1176 |
| Edge density | 0.206 | 0.372 |
| Diameter | 6 | 4 |
| Average path length | 2.299 | 1.868 |
| Modularity | 2.497 | 1.865 |
| Modules | 7 | 3 |
| Average degree | 16.250 | 29.375 |
| Clustering coefficient | 0.646 | 0.786 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Chen, Y.; Li, X.; Cao, X.; Tang, H.; Lv, G. The Gradient Heterogeneity of Deserts Alters the Interaction Relationships Between Xerophytic Plants and Soils. Biology 2025, 14, 1048. https://doi.org/10.3390/biology14081048
Wang J, Chen Y, Li X, Cao X, Tang H, Lv G. The Gradient Heterogeneity of Deserts Alters the Interaction Relationships Between Xerophytic Plants and Soils. Biology. 2025; 14(8):1048. https://doi.org/10.3390/biology14081048
Chicago/Turabian StyleWang, Jinlong, Yudong Chen, Xiaotong Li, Xiaojuan Cao, Hongli Tang, and Guanghui Lv. 2025. "The Gradient Heterogeneity of Deserts Alters the Interaction Relationships Between Xerophytic Plants and Soils" Biology 14, no. 8: 1048. https://doi.org/10.3390/biology14081048
APA StyleWang, J., Chen, Y., Li, X., Cao, X., Tang, H., & Lv, G. (2025). The Gradient Heterogeneity of Deserts Alters the Interaction Relationships Between Xerophytic Plants and Soils. Biology, 14(8), 1048. https://doi.org/10.3390/biology14081048






