Simple Summary
SRS, a transcription factor unique to plants, plays important roles in regulating plant growth and development and in responding to stress. Although SRS genes have been studied in many plants, in cucurbit crops, they have only been identified in cucumber thus far. This study not only presents a comprehensive bioinformatics examination of all SRS genes in the genomes of seven Cucurbitaceae crops, but also investigates the expression levels of CmSRS genes in different tissues. In addition, the expression of CmSRS genes under abiotic (drought and salt) and biotic (wilt and powdery mildew) stresses, as well as their subcellular localization, are analysed. These results lay the foundation for studying the biological functions of SRS genes in Cucurbitaceae crops.
Abstract
Background: The short strand-related sequence (SRS) gene family is a class of plant-specific transcription factors related to a group of genes known as the short internode (SHI) or SRS/STY gene family, which plays important roles in regulating plant growth and development and stress responses. Although the SRS genes have been studied in many plants, in cucurbit crops, they have thus far only been identified in cucumber. Methods: In the Cucurbitaceae database from melon (Cucumis melo), cucumber (Cucumis sativus), watermelon (Citrullus lanatus), bottle gourd (Lagenaria siceraria), wax gourd (Benincasa hispida), moschata pumpkin (Cucurbita moschata), and pumpkin (Cucurbita maxima), a total of 60 SRS genes were identified in seven Cucurbitaceae crops, which were classified into three subfamilies. Results: The same subfamily showed conserved motifs and gene structures. The differences in the number of SRS genes in different Cucurbitaceae crops implied likely gene loss or duplication events during evolution. Analysis of promoter cis-regulatory elements indicated that these SRS genes may be involved in hormone response, growth and development, and biotic and abiotic stress responses in plants. Most of the CmSRS genes in melons were expressed in the roots, with a few expressed in the leaves and ovaries. In addition, CmSRS expression was induced by biotic (wilt and powdery mildew) and abiotic (drought and salt) stresses. Subcellular localization of CmSRS proteins showed predominant expression in the nucleus. Conclusions: A total of 60 Cucurbitaceae SRS genes are present in the genomes of seven Cucurbitaceae crops. These cucurbit SRS genes seem to have maintained similar characteristics and functions during the evolutionary process. These results lay the foundation for the study of biological functions of SRS genes in Cucurbitaceae crops.
1. Introduction
The SHI-related sequence (SRS) gene family is a family of genes related to plant-specific transcription factors, also known as the short internode (SHI) or SRS/STY family, consisting of two highly conserved structural domains, RING and IXGH [1]. The RING domain is located at the N-terminal end of a CH3CH3 motif-containing RING zinc finger structure (CX2CX7CX4CX2C2X6C) [2], which was initially identified as a DNA-binding motif in the African clawed frog [3]. In plant cells, the RING domain binds to RNA, proteins, and lipid substrates and is involved in a variety of physiological and biochemical processes [4]. The IXGH domain, located at the C-terminal end, contains acidic amino acids and is homodimerized [5], a trait which has not been found in other proteins and may be a unique feature of the SRS family [6].
The SRS protein family plays key roles in a variety of physiological and biochemical processes in plants, including hormone synthesis and signal transduction, abiotic stress response, and plant organ growth and development [7]. The SRS family is widely distributed in plants. In Arabidopsis thaliana, a total of 10 SRS genes were identified, including SHI, STY1, STY2, LPR1, and SRS3/4/5/6/7/8. Except for the SRS8 gene missing the IXGH structural domain, the remaining nine genes contain both the RING structural domain and the IXGH structural domain [7,8,9,10]. Functionally, STY1 (SRS1) plays an important role in the development of apical meristematic tissue auxin biosynthesis [11]. LPR1 is able to form protein complexes with SHI, STY1, SRS3, SRS6, and SRS7, which regulate lateral root development by modulating auxin signalling and chromatin modification [8,12,13]. However, LPR1 overexpression inhibits root development, which is mainly due to elevated auxin levels [14]. Overexpression of AtSHI in Arabidopsis thaliana, Longevity Flower, and Pongamia monnieri showed a dwarfing phenotype. These studies suggest that AtSHI, as a negative regulator in gibberellin (GA) signalling, affects plant stature by regulating stem elongation [5,15,16]. Meanwhile, in rice (Oryza sativa L.), OsSHI increases tiller number and reduces spike size by regulating the transcriptional activity of IPA1 (ideal plant architecture 1) [17].
SRS genes may function as key regulators in plant abiotic stress response. It has been shown that the 9 MaSRS genes of Melilotus albus exhibited significant up- and downregulated expression at different time points under salt, low temperature, salicylic acid (SA) and methyl jasmonate (MeJA) treatments [18]. In soybean, GmSRS18 has been identified as a key gene for negatively regulating drought and salt tolerance [1]. In cotton, GhSRS21 is also involved in salt stress response as a negative regulator [19]. However, in cucurbit crops, the identification of SRS genes and their expression patterns have been reported only in cucumber, despite the increasing research on the function of SRS genes in various plants.
Cucurbitaceae crops are widely distributed in subtropical and tropical regions and play critical roles in global economic development [20]. These crops are rich in important nutrients [21,22], exhibit medicinal value in promoting cardiovascular health [23,24], and are used in the treatment of many diseases [25,26]. Among them, melon (Cucumis melo L.) is one of the major cash crops in China [27]. According to statistics, the global annual production of melon is estimated to be more than 40 million tonnes [28]. However, its final yield is often affected by pests, diseases, and abiotic stresses [29]. In particular, abiotic factors such as high temperatures, high salinity, and drought can lead to an average yield reduction of about 50% in crops [30]. In order to gain a deeper understanding of the functions of SRS genes in Cucurbitaceae crops, the present study was carried out to systematically analyse seven cucurbit crops. A total of 60 SRS genes were identified, and their gene structures, conserved motifs, conserved structural domains, chromosomal localization, cis-acting elements, and phylogenies were comprehensively analysed. The covariance relationship between the SRS genes of melon and six other Cucurbitaceae crops was also explored. Finally, the expression pattern of the melon CmSRS gene under Fusarium acnes and Fusarium powdery mildew infestation was analysed using transcriptome data. The expression characteristics of the CmSRS gene in different tissues, as well as under drought stress, were analysed by qRT-PCR. This study aims to provide a theoretical basis for the functional identification of SRS genes in cucurbit crops, as well as an important reference for further studies related to SRS genes and melon growth, development, disease resistance, and abiotic stress tolerance.
2. Materials and Methods
2.1. Identification of SRS Genes in Cucurbitaceae
The HMM model file (PF05142) for the SRS family was downloaded from the Pfam database (http: //pfam.xfam.org/ (accessed on 15 September 2024)). Protein sequences, genomes, and annotation files for seven cucurbit crops were obtained from the Cucurbitaceae database (http://www.cucurbitgenomics.org/ (accessed on 15 September 2024)), while protein sequences, genomes, and annotation files for Arabidopsis thaliana and rice were downloaded from the Ensemble Plants database (https://plants.ensembl.org/ (accessed on 15 September 2024)). Protein sequences were extracted using Fasta Extract (Recommended) plug-in of Tbtools. SRS gene family members were predicted in the genomes of seven Cucurbitaceae crops, Arabidopsis thaliana, and rice using the Simple HMM Search plug-in in TBtools v2.300 software [31]. Candidate SRS genes were identified using the NCBI-Conserved Domain Database (CDD) (http: //www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi (accessed on 15 September 2024)).
2.2. Physicochemical Traits and Chromosomal Distribution of SRS in Cucurbitaceae
Physicochemical properties such as amino acid number, molecular weight, and isoelectric point of SRS family members of seven cucurbit crops were analysed using the ExPASy online software (https://web.expasy.org/protparam/ (accessed on 15 September 2024)). Chromosome physical mapping of SRS family members in seven Cucurbitaceae crops was performed using the mg2c online software (http://mg2c.iask.in/mg2c_v2.1/ (accessed on 15 September 2024)).
2.3. Analysis of Gene Structure, Conserved Motifs, and Evolution of the SRS Gene in Cucurbitaceae
The One Step Build an ML Tree plug-in and Simple MEME Wrapper v2.300 plug-in of Tbtools software were used for the SRS gene evolutionary analysis and motif analysis of Cucurbitaceae, respectively. The Gene Structure View v2.300 (Advanced) plug-in was used to merge and visualise the results. Phylogenetic analysis was performed using MEGA11, and the evolutionary tree was constructed using the neighbour-joining method [32]. The evolutionary tree was presented using the evolview online tool (https://evolgenius.info//evolview-v2/ (accessed on 15 September 2024)).
2.4. Analysis of Covariance of SRS Genes in Cucurbitaceae
Covariance analyses were performed for each of the seven Cucurbitaceae crops, Arabidopsis thaliana, and rice species using the One Step MCScanX plug-in v2.300 of Tbtools software. The results were combined for visualisation using the Multiple Synteny Plot plug-in v2.300.
2.5. Analysis of the Role of Cis-Elements in the Promoter of the SRS Genes in Cucurbitaceae
The GXF Sequences Extract plug-in v2.300 of Tbtools software was used to extract 2000 bp promoter sequences upstream of the transcription start site. These promoter sequences were submitted to the PlantCARE online analysis tool (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/ (accessed on 16 September 2024)) for cis-acting element prediction. Heat mapping was performed using the Heat Map plug-in v2.300.
2.6. RNA Extraction and qRT-PCR Analysis
In order to analyse the expression pattern of CmSRS genes in various organ tissues and the expression changes under drought and salt stress, RNA was extracted using the TIANGEN Plant Total RNA Extraction Kit (Beijing, China). cDNA was reverse-transcribed from quality-tested RNA using the TIANGEN FastKing One-Step De-genomic cDNA First Strand Synthesis Kit (Beijing, China). qRT-PCR was performed using the melon ACTIN gene (GenBank: AY859055.1) as an internal reference. The reactions were carried out using the SGExcel FastSYBR qPCR kit (Shanghai, China) on a 96-well reaction plate with cDNA as the template. The qRT-PCR reaction system consisted of 1 μL of 2× SGExcel FastSYBR Mixture, 1 μL of cDNA template, 0.4 μL each of forward and reverse primers, as well as ddH2O to a final volume of 20 μL. The PCR amplification program was as follows: 95 °C for 3 min, followed by 95 °C for 5 s and 60 °C for 20 s. The relative expression of the gene was calculated using the 2−△△Ct method. To ensure the accuracy and reliability of the experimental results, three biological replicates were included for each sample, and each PCR reaction was performed with three technical replicates. The primers are listed in Supplemental Table S1.
2.7. Transcriptome Data Analysis
Transcriptome sequencing data PRJEB15551 and PRJNA434538 from the NCBI database were used to analyse the expression of melon SRS genes under Fusarium spinosum and powdery mildew fungus infestation. Heat maps were drawn using TBtools.
2.8. Subcellular Localization
The fusion proteins were constructed by cloning the coding sequences of CmSRS1, CmSRS3, and CmSRS4, without the stop codon, into a Super1300 vector containing green fluorescent protein (GFP). Then, the fusion plasmid was transformed into Agrobacterium tumefaciens (GV3101). Fused CmSRS-GFP proteins were transiently expressed in 4-week-old Nicotiana benthamiana leaves. The GFP fluorescence signal was analysed by confocal microscopy. The primers are listed in Supplemental Table S2.
2.9. Interaction Network and Protein Structure Prediction of Melon SRS Proteins
Protein interaction network prediction was performed using the STRING database (STRING functional protein association networks (https://cn.string-db.org/cgi/input?sessionId=bXV51TlyIO7u&input_page_show_search=on (accessed on 17 September 2024)). The online website SOPMA (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma.html (accessed on 17 September 2024)) was used for protein secondary structure prediction. The prediction of the protein tertiary structure was completed using the SWISS-MODEL online website (http://swissmodel.expasy.org/ (accessed on 17 September 2024)).
3. Results
3.1. Identification and Physicochemical Properties of Seven SRS Genes in Cucurbitaceae
Using the HMM Search tool in Tbtools software, a total of 60 SRS family members were identified in the whole genomes of seven Cucurbitaceae crops in this study (Table 1), with the following distribution: 5 in melon (Cucumis melo), 8 in cucumber (Cucumis sativus), 8 in watermelon (Citrullus lanatus), 7 in bottle gourd (Lagenaria siceraria), 7 in wax gourd (Benincasa hispida), 13 in moschata pumpkin (Cucurbita moschata), and 12 in pumpkin (Cucurbita maxima) (Table 1). Predictive analyses of the physicochemical properties of these SRS family proteins revealed significant differences among the 60 Cucurbitaceae SRS proteins. Specifically, the lengths of the encoded amino acids ranged from 211 to 791 aa, the molecular weights ranged from 24,649.34~56,691.2 Da, the theoretical isoelectric points (pI) ranged from 5.98 to 9.19, and the instability coefficients ranged from 37.75 to 66.87. Among these, most of the SRS family members were unstable proteins (instability coefficients > 40), and the average hydrophilicity values ranged from −0.876 to −0.165, indicating that most of these proteins are hydrophobic (Table 1). In addition, the identified genes were named according to their positional order on the chromosome rather than following a uniform naming rule.

Table 1.
Physicochemical properties of SRS proteins in Cucurbitaceae.
To clarify the physical location of SRS genes in the genomes of these seven cucurbit crops, we mapped the chromosomal distribution of these genes (Figure S1). In melon, 5 SRS genes were distributed on chromosomes 1, 3, 4, 7, and 8; in cucumber, 8 SRS genes were distributed on chromosomes 1, 2, 3, 4, 5, 6, and 7; in watermelon, 8 SRS genes were distributed on chromosomes 1, 2, 3, 4, 5, 6, 7, 8, and 9; in bottle gourd, 7 SRS genes were distributed on chromosomes 1, 4, 6, 7, 8, and 10; in wax gourd, 7 SRS genes were distributed on chromosomes 1, 2, 3, 5, 7, 9, and 10; in moschata pumpkin, 13 SRS genes were distributed on chromosomes 1, 2, 3, 4, 5, 7, 11, 12, 18, 19, and 20; and in pumpkin, 12 SRS genes were distributed on chromosomes 1, 2, 3, 4, 5, 7, 11, 12, 18, 16, 19, and 20 (Figure S1). These results indicated that SRS genes were dispersed in the genomes of different Cucurbitaceae crops, and there were some differences in distribution among different species.
3.2. Analysis of Gene Structure and Conserved Structural Domains of SRS in Cucurbitaceae
In order to investigate the relationships among SRS family members of Cucurbitaceae crops in terms of structure, function, and evolution, a phylogenetic tree was constructed in this study, and the conserved motifs and gene structures of the family members were comprehensively analysed. Phylogenetic tree analysis revealed that the 60 Cucurbitaceae SRS family members could be classified into three major branches (Figure 1A), suggesting that they exhibit different evolutionary divergence paths. Further conserved motif analysis revealed that the motif numbers of cucurbit SRS family members ranged from 5 to 9 (Figure 1B). All cucurbit SRS proteins contained Motif1 and Motif2, with Motif1 containing the highly conserved RING structural domain characterized by the sequence CX2CX7CX4CX2CCX6C (“X” stands for any amino acid, and the number represents the number of amino acid residues), and Moitf2 containing the highly conserved IXGH structural domain (Figure 1B,D). Amino acid sequence alignment showed that all 60 Cucurbitaceae SRS family members contain the RING structural domain at the N-terminus and the IXGH structural domain at the C-terminus. Both structural domains are highly conserved during evolution (Figure 2). Gene structure analysis (Figure 1C) revealed that most of the cucurbit SRS family members lacked either the 5′UTR or 3′UTR region. All members, except for CmoSRS4, CmoSRS5, ClaSRS2, ClaSRS5, and ClaSRS8, contained 1 to 2 introns and 2 to 3 exons. Overall, most of the cucurbit SRS genes in the same branch were structurally similar, suggesting a high degree of conservation of gene structure and protein conserved motifs in the cucurbit SRS family during evolution. This result provides important clues for understanding the functional divergence and evolution of the SRS family in Cucurbitaceae crops.
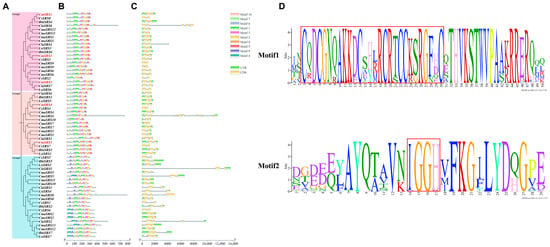
Figure 1.
Analysis of gene structure and protein conserved structural domains of SRS genes in seven Cucurbitaceae crops. (A) Phylogenetic tree of SRS genes in Cucurbitaceae. Light red, light brown, and light blue represent the three branches. Red font represents melon SRS genes. (B) Conserved motifs of cucurbit SRS family members. Different colours represent different motifs. (C) Cucurbitaceae SRS family member gene structures. Green and yellow colours represent UTR and CDS, respectively. (D) Cucurbitaceae SRS family members Motif1 and Motif2, with red boxes indicating RING and IXGH structural domains.

Figure 2.
Sequence comparison of SRS family proteins in melon. N- and C- denote the N- and C-termini, respectively, of the SRS amino acid sequences of seven Cucurbitaceae crops.
3.3. Evolutionary Analysis of SRS Family Genes in Cucurbitaceae
To further explore the kinship and evolutionary patterns among the SRS genes in Cucurbitaceae, a phylogenetic tree was constructed in this study using 11 SRS genes from Arabidopsis thaliana, 4 SRS genes from rice, 14 SRS genes from maize, and 60 SRS genes from seven Cucurbitaceae crops via the neighbour-joining method (Figure 3). Based on evolutionary relationships, we classified these genes into three subfamilies (Group 1~3). Among these subfamilies, the Group 3 contained the most members, with 8 SRS genes from eight dicotyledonous plants, which contained 5 CmSRS, 5 CsSRS, 5 ClaSRS, 7 CmaSRS, 7 CmoSRS, 3 LsiSRS, 4 BhiSRS, and 11 AtSRS. The Group 2 subfamily contained SRS genes from eight crops other than Arabidopsis thaliana and melon, with 3 CsSRS, 3 ClaSRS, 6 CmaSRS, 5 CmoSRS, 4 LsiSRS, 3 BhiSRS, 2 OsSRS, and 3 ZmSRS. The Group 1 subfamily represented only monocotyledonous SRS genes, with 11 ZmSRS and 2 OsSRS. This taxonomic pattern suggests that the SRS proteins in Group 1 and Group 3 may have undergone evolutionary divergence only after the completion of monocotyledonous and dicotyledonous plant differentiation, whereas the Group 2 subfamily may have formed at a much earlier evolutionary stage and display a much wider distribution of members.
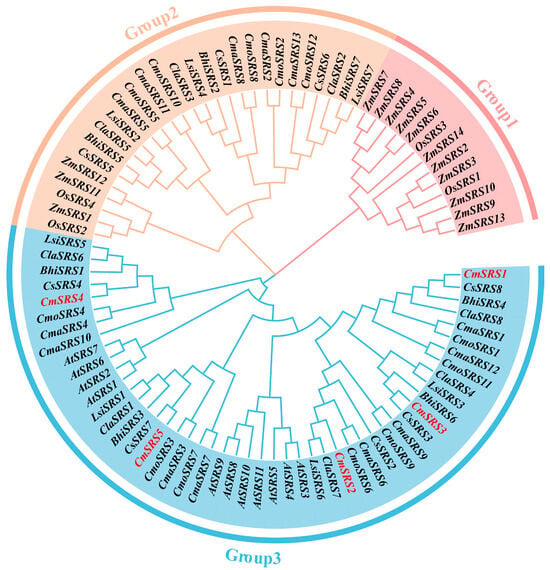
Figure 3.
Phylogenetic analysis of SRS genes in Arabidopsis thaliana, rice, maize, and seven Cucurbitaceae crops. Red font indicates melon SRS genes.
To better understand the amplification patterns of the SRS family during evolution, covariance analysis of melon and 6 other Cucurbitaceae crops (Figure 4A; Table S3) revealed that melon shares 16, 20, 14, 13, 14, and 14 homologous genes with pumpkin, moschata pumpkin, bottle gourd, wax gourd, cucumber, and watermelon, respectively. These cucurbit crops may have originated from common ancestral genes and retained similar gene structures and functions during evolution. Interestingly, there were covariations between CmSRS2, CmSRS3, CmSRS4, and CmSRS5 in melon and SRS genes of the other six Cucurbitaceae crops, with three pairs of covariations with cucumber, watermelon, wax gourd, and bottle gourd and four pairs with pumpkin. Arabidopsis thaliana and rice, as model species for dicotyledonous and monocotyledonous plants, respectively, represent two major taxa of the plant kingdom. Therefore, we performed a combined analysis of melon, Arabidopsis thaliana, and rice (Figure 4B). The results showed that there were 18 homologous genes between melon and Arabidopsis thaliana, whereas there was only 1 homologous gene with rice. This difference suggests that monocotyledons and dicotyledons may not share a large number of homologous genes prior to their divergence, which further emphasises the independent evolutionary pathways of the SRS family in different plant taxa. Gene duplication events are an important mechanism for gene family amplification, usually caused by genome-wide duplications or segmental duplications. Gene duplication significantly affects the diversity and conservation of gene functions. To explore gene duplication events in SRS family members in melon, we performed within-species covariance analysis in melon (Figure 4C). The results revealed four pairs of duplicated genes in melon: CmSRS2-CmSRS3, CmSRS5-CmSRS4, CmSRS5-CmSRS3, and CmSRS5-CmSRS2. CmSRS5 was involved in three segmental duplications, while CmSRS2 and CmSRS3 were each involved in two segmental duplications. These results suggest that segmental duplication events may be the main driver of SRS family amplification in melon. They also provide the basis for the generation and diversity of new gene functions.
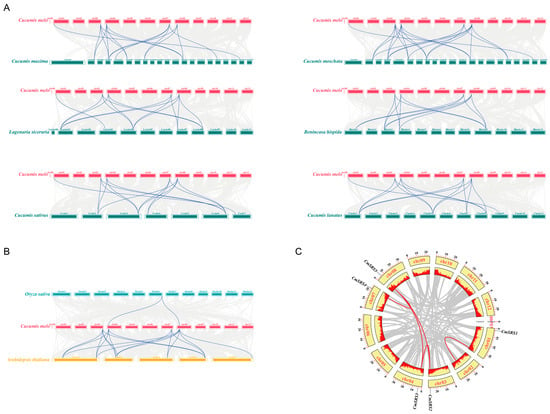
Figure 4.
Co-collinearity analysis of SRS genes: (A) co-collinearity analysis of SRS genes in melon and six other Cucurbitaceae crops; (B) co-linearity analysis of SRS genes in melon, Arabidopsis thaliana, and rice; (C) CmSRS gene duplication analysis. Red lines indicate gene duplication pairs.
3.4. Analysis of Cis-Acting Elements in the Promoter Sequence of Melon SRS Genes
Plants respond to biotic and abiotic stresses through complex regulatory mechanisms, many of which depend on cis-acting elements in gene promoter regions. To investigate the transcriptional regulation of the SRS genes and its potential functions, we predicted cis-acting elements in the promoter regions of 60 Cucurbitaceae SRS genes. We found core elements such as the CAAT-box and the TATA-box in all SRS genes, as well as 32 different cis-acting elements related to biotic and abiotic stress response, plant growth and development, and phytohormone response (Figure 5). Among them, 15 elements were related to growth and development, including circadian, O2-stie and GCN4_motif; 10 elements were related to biotic and abiotic stresses, including ARE, MBS, MYB, MYB, and MBS; 10 elements were related to drought response, including MYB, MBS, and MYB; 10 elements were related to plant growth and development; 10 elements were related to plant growth and development, including MYB, MYB, and MBS; 10 elements were related to plant growth and development, such as MYB; and 10 other elements and the action elements were related to phytohormone response, including ABRE (abscisic acid response element), TGA- element (growth hormone response element), CGTCA-motif, TGACG-motif (jasmonic acid response element), P-box (gibberellin response element), and TCA-element (salicylic acid response element).
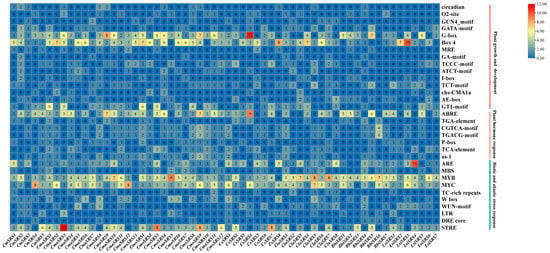
Figure 5.
Cis-acting elements in the promoters of SRS genes in seven Cucurbitaceae crops. Colours indicate the number of different cis-acting elements. Numbers indicate the statistical number of cis-acting elements.
Among the growth and development-related cis-acting elements, the light-responsive elements G-box and Box4 were widely distributed in most cucurbit SRS genes and were most abundant in CsSRS4 and LsiSRS3, respectively. Among the phytohormone-related acting elements, the ABRE was most abundantly distributed among the cucurbit SRS genes, especially in the melon (CmSRS) family, where all members contained this element. Among the biotic and abiotic stress response elements, the drought response element (MYC) appeared in 58 Cucurbitaceae crop SRS genes, whereas the defence and stress response element (MYB) was also widely distributed in this family of genes. Among the CmSRS genes of melon, all members except CmSRS3 contained MYC elements, and all members contained MYB elements, suggesting that the CmSRS genes may play an important role in drought and defence response in melon. In summary, the SRS genes of seven cucurbit crops exhibit important functions in physiological processes such as plant growth and development, hormone signal transduction, and stress response. The diversity of their cis-acting elements provides a basis for their regulation in a variety of biological processes.
3.5. Tissue-Specific Expression Analysis of SRS Genes in Melon
In order to investigate the expression patterns of melon SRS family members in different tissues, we analysed the tissues of roots, stems, leaves, stamens, and ovaries of melon by qRT-PCR. The results (Figure 6) showed that the expression levels of the five CmSRS family members in different tissues differed significantly. Among them, CmSRS1, CmSRS3 and CmSRS4 showed higher expression levels in the roots, whereas CmSRS2 and CmSRS5 showed higher expression in the leaves and ovaries, respectively. The expression of CmSRS1 was mainly observed in the roots, and expression was very low in the other tissues. CmSRS3 expression in the stems was significantly higher than that of the other four CmSRS members. These results suggest that melon SRS genes display tissue-specific expression patterns during their growth and development, which may play an important role in regulating the organ development and functional differentiation of melon.
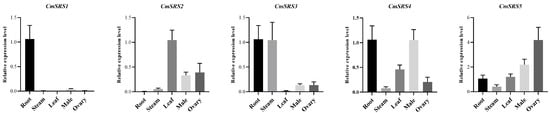
Figure 6.
Tissue-specific expression analysis of SRS genes in melon. Values are mean ± SD of three biological replicates.
3.6. Expression Analysis of SRS Family Genes in Melon Under Drought and Salt Stresses
Drought and salt stress are two of the most important environmental factors affecting plant growth and development, as they alter the crop’s access to nutrients and thus affect the final yield and quality [33,34,35]. In soybean, the GmSRS18 gene has been shown to be a negative regulator in response to drought stress [1]. To investigate whether melon SRS genes were induced by drought and salt stress, melon roots were treated with 20% PEG6000 and 100 mM NaCl (Figure 7A). The expression was analysed by qRT-PCR after 0, 12, 24, and 36 h treatments (Figure 7B). The results showed that the five CmSRS genes responded to both drought and salt stresses. Under drought stress, the expression of CmSRS1/2/3/4 showed an increasing trend at 12 h, declined at 24 h, and then increased again to the highest value at 36 h. In contrast, the expression pattern of CmSRS5 was the opposite, with the lowest expression at 36 h. Under salt stress, the expression levels of CmSRS1/2/3/4 all peaked at 12 h and showed a decreasing trend at 24 h. However, CmSRS2 and CmSRS4 showed an increasing trend at 36 h, whereas CmSRS1 and CmSRS3 continued to decrease. The expression pattern of CmSRS5 was in the opposite direction, with the lowest expression at 12 h. In addition, it was observed that melon already showed wilting after 12 h of treatment, suggesting that CmSRS genes may play an important regulatory role in melon response to drought and salt stress.
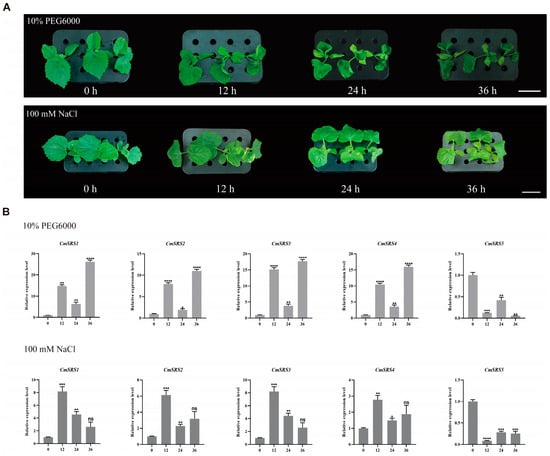
Figure 7.
Expression analysis of melon SRS genes under drought and salt stress. (A) Phenotypic observations of melon after 0, 12, 24, and 36 h of 20% PEG6000 treatment. (B) Expression analysis of 5 CmSRS genes under drought stress. Values are mean ± SD of three biological replicates. Asterisks indicate significant differences between 0 h and other time points (ns: no significance; * p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001).
3.7. Expression Analysis of SRS Family Genes in Melon Under Biotic Stresses
Powdery mildew and wilt are major fungal diseases in melon production, which seriously affect the growth, development, yield, and quality of melon. To investigate the response patterns of melon SRS genes under biotic stresses (wilt and powdery mildew), the expression responses of the disease-resistant melon variety NAD and the susceptible variety Charentais-T (CHT) to wilt and that of the variety Rochet to powdery mildew were analysed using the NCBI SRA public transcriptome database (Figure 8). After 24 h of infection with Fusarium spinosum, the expression of all CmSRS genes increased in the susceptible variety CHT, except for a slight decrease noted in the expression of CmSRS1, with a greater increase in CmSRS3. The expression levels of CmSRS2 and CmSRS5 increased, while the expression levels of CmSRS1, CmSRS3, and CmSRS4 declined in the disease-resistant variety NDA. After 48 h of infection, the expression levels of all CmSRS genes decreased in the susceptible variety CHT, except for CmSRS1, in which the expression level of CmSRS3 decreased to a greater extent. The expression levels of the five CmSRS genes increased in the disease-resistant variety NAD. The expression levels of all five CmSRS family genes increased 24 h after infestation with powdery mildew pathogens. At 48 h post infestation, the expression of CmSRS2 and CmSRS5 increased. Additionally, the expression of CmSRS3 and CmSRS4 decreased, while the expression of CmSRS1 remained unchanged. At 72 h post infestation, the expression levels of all five CmSRS genes decreased. These results indicated that melon SRS genes showed different expression patterns under wilt and powdery mildew stresses, and significant differences existed between resistant and susceptible varieties.
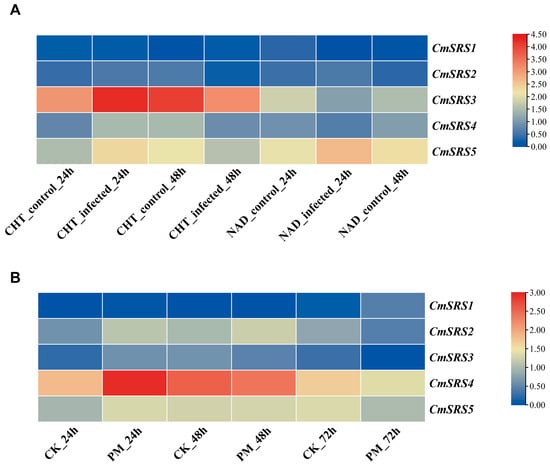
Figure 8.
Expression analysis of SRS genes in melon upon biotic stresses. (A) Expression analysis of CmSRS genes in CHT and NAD varieties infested with Fusarium spinosum for 24 h and 48 h. (B) Expression analysis of CmSRS genes infested with powdery mildew fungus for 24 h, 48 h and 72 h.
3.8. Analysis of Subcellular Localization of SRS Family Genes in Melon
Based on these results, we selected CmSRS1, CmSRS3, and CmSRS4, which are highly expressed in roots, and analysed their protein functions. To detect their subcellular localization, we constructed CmSRS proteins fused with green fluorescent protein (GFP) and transiently expressed them in Nicotiana benthamiana leaves (Figure 9). The results showed that GFP fluorescent signals were observed only in the nucleus for CmSRS3 and CmSRS4, which was caused by the nuclear input of transcription factors contributing to transcriptional activity. CmSRS1-GFP fluorescent signals were observed in both the nucleus and the cell membrane, with a predominant nuclear localisation, which is likely regulated post-translationally.
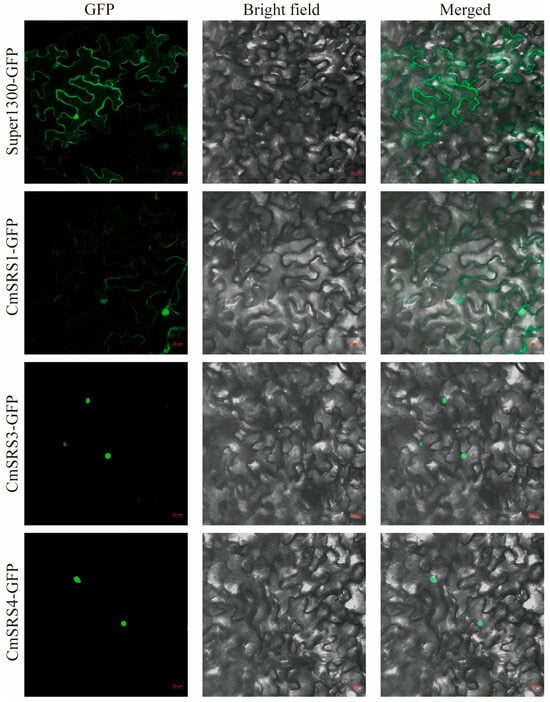
Figure 9.
Analysis of subcellular localization of CmSRS protein. Subcellular localization of CmSRS protein fused to GFP and transiently expressed in Nicotiana benthamiana leaf cells was observed by laser scanning confocal microscopy, using Super1300-GFP empty vector as a control. Bars = 20 μm.
3.9. Interaction Network and Structure Prediction of SRS Family Proteins in Melon
To further explore the biological functions of SRS proteins in melon, we searched and analysed their potential interacting proteins using the STRING database (Version 12.0). Based on the prediction, all five CmSRS proteins were found to interact with A0A5A7TZS9 (ethylene-inducible protein), A0A5A7VAA7 (calcium-binding EF-hand family protein), A0A5A7VME9 (RING-type E3 ubiquitinyltransferase), A0A5A7UT44 (F-box protein PP2-B10 analogue), and A0A5A7UXG1 (F-box protein PP2-B10 analogue isoform X1) (Figure 10A; Table S4). These proteins are involved in lutein biosynthesis (Figure 10B), implying that CmSRS proteins may have an important role in metabolic regulation in melon. Further protein secondary structure analysis showed (Figure 11A; Table S5) that the secondary structure of melon CmSRS family member proteins consisted of an α-helix, β-turn, an extended strand, and a random coil. The prediction results showed that a large number of irregular coils with a small number of α-helices and extended strands, along with a very small number of β-turns, were present in these five CmSRS proteins. In addition, tertiary structure analysis of melon SRS proteins showed (Figure 11B) that the three-dimensional structures of this family of proteins are highly similar, which may be related to their functional conservation.
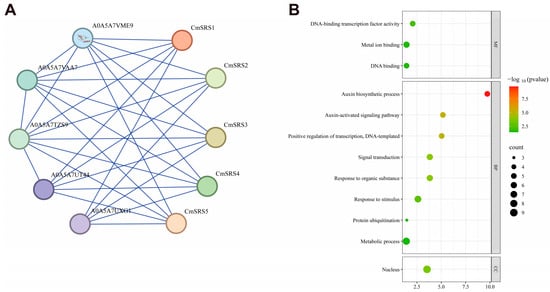
Figure 10.
Predicted interactions of melon SRS proteins: (A) predicted interactions of five CmSRS proteins; (B) GO enrichment analysis.
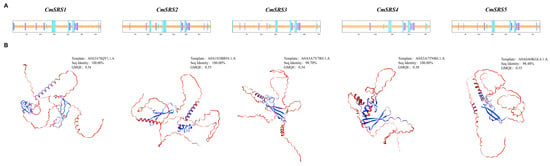
Figure 11.
Predicted structure of melon SRS proteins: (A) secondary structure of five CmSRS proteins; (B) tertiary structure of five CmSRS proteins.
4. Discussion
Thus far, the SRS genes have been identified in a variety of plants, including Arabidopsis thaliana [6], rice [2], cotton [19], maize [36], and cucumber [37], among others. Among them, the SRS genes in Arabidopsis thaliana are the most intensively and comprehensively studied, involving several biological processes such as root formation [12], leaf development [38], and floral organ development [39]. However, among cucurbit crops, the SRS genes have only been identified in cucumber. Therefore, this study systematically identified SRS genes in seven cucurbit crops via bioinformatics and focused on the expression pattern of SRS genes in melon to fill the gap in the study of SRS genes in Cucurbitaceae crops.
In this study, 5, 8, 8, 7, 7, 13, and 12 SRS genes were identified in seven Cucurbitaceae crops including melon, cucumber, watermelon, bottle gourd, wax gourd, moschata pumpkin, and pumpkin, respectively. These genes were randomly distributed on the chromosomes. All CmSRS family members contained the RING structural domains (CX2CX7CX4CX2C2X6C) and the IXGH structural domains, which is in agreement with the findings in Arabidopsis thaliana [6]. These results indicate the conservation of SRS proteins in these seven Cucurbitaceae crops. However, the parameters of the protein physicochemical properties such as amino acid number, isoelectric point, and instability coefficient varied among crops, which may be related to the structural diversity of SRS genes. Gene structure provides clues to the diversity of gene functions and plays a crucial role in the evolution of gene families [40]. It has been reported that members of the CqSRS family contain two to five exons, and genes within the same subfamily exhibit similar gene structures [41]. To explore the structural diversity of SRS genes in seven Cucurbitaceae crops, we analysed their gene structures and protein conserved motifs and found that 60 cucurbit SRS genes had two to three exons and one to two introns. In addition, all contain Motif1, with a RING structural domain, and Motif2, with an IXGH structural domain. We also found that genes in the same subfamily display similar protein motifs and gene structures, suggesting that they may have similar functions.
To better understand the evolutionary relationships of the SRS family, the phylogenies of seven Cucurbitaceae crops were compared with Arabidopsis thaliana, rice and maize. It was found that the genes of the SRS family in monocotyledonous and dicotyledonous plants belonged to different branches, which is similar to the findings in cucumber [37]. Gene duplication events play an important role in genome amplification and gene functional diversity [42]. In order to investigate the amplification pattern of SRS genes in seven Cucurbitaceae crops during the evolutionary process, by covariance analysis, we found that melon possesses 16, 20, 14, 13, 14, and 14 homologous genes with pumpkin, moschata pumpkin, bottle gourd, wax gourd, cucumber, and watermelon, respectively. Interestingly, all of the CmSRS2/3/4/5 in melon are covaried with SRS genes in six other Cucurbitaceae crops, such as cucumber, watermelon, wax gourd, and bottle gourd with three pairs of co-linear genes, and pumpkin with four pairs of co-linear genes. In addition, four pairs of duplicated genes were identified in CmSRS family members, of which CmSRS5 was involved in three segmental duplications, and CmSRS2 and CmSRS3 were each involved in two segmental duplications, suggesting that segmental duplication events may be the main drivers of gene amplification in the CmSRS family.
Gene promoter region action elements play an important role in plant defence against biotic and abiotic stresses [43]. A large number of elements involved in stress response and growth and development were found in the ZmSRS family genes of maize [36]. This suggests that SRS genes may play a role in pathways related to plant stress response mechanisms. The seven Cucurbitaceae SRS genes contain low temperature response elements (LTR), drought response elements (MBS and MYC), and defence and stress response elements (TC-rich repeats, MYB, and W-box), as well as a wide range of components related to SRS transcription factors. These factors are involved in the development of plant organs and tissues by regulating the synthesis and signal transduction of a variety of plant hormones. In Arabidopsis thaliana, AtLRP1 regulates root development [14], and AtSTY1 is involved in leaf and flower development [44,45,46]. In this study, the expression pattern of melon SRS family genes in different tissues was analysed by qRT-PCR. CmSRS1, CmSRS3, and CmSRS4 were highly expressed in the roots, whereas CmSRS2 and CmSRS5 were highly expressed in the leaves and ovaries, respectively. This suggests that the CmSRS genes are tissue-specific in the growth and development of different organs in melon. Plants are affected by biotic and abiotic factors during the growth cycle. In Arabidopsis thaliana, SRS5 gene expression can be significantly upregulated by pathogen induction, suggesting that AtSRS5 may be involved in regulating the immune response of plants [47]. In the present study, we found that the expression of five CmSRS genes was elevated after 48 h of infestation by NAD in wilt-resistant varieties and after 24 h of infestation by powdery mildew pathogens through the response of melon to wilt and powdery mildew diseases. In addition, abiotic stresses also induce the expression of SRS genes. For example, GmSRS18 in soybean is induced by drought, NaCl, and exogenous ABA. Its overexpression increases the sensitivity of transgenic Arabidopsis thaliana to drought and salt stresses [1]. We further investigated the expression pattern of CmSRS genes under drought stress. It is noteworthy that CmSRS1/2/3/4 showed an increasing trend at 12 h under drought stress, declined at 24 h, and increased again to its highest levels at 36 h, whereas under salt stress, CmSRS1/2/3/4 peaked at 12 h and declined at 24 h. These results indicate that the CmSRS genes are diverse in their response to drought and salt stress. Combined with the prediction of promoter cis-acting elements, we found that the all CmSRS genes contain drought-responsive elements (MYC), as well as stress-responsive elements (MYB). This suggests that melon SRS genes may play an important role in abiotic stress response.
5. Conclusions
In summary, in this study, a total of 60 SRS genes were identified from seven Cucurbitaceae crops. Their chromosomal distribution, protein physicochemical properties, gene structure and protein conserved motifs, conserved structural domains, evolutionary relationships, gene duplications, and cis-acting elements were systematically analysed. The results showed that the SRS genes of cucurbit crops were classified into three subfamilies. five SRS genes of melon and six other Cucurbitaceae crop SRS genes were duplicated, thus expanding the SRS gene family of Cucurbitaceae crops. In addition, CmSRS2/3/4/5 exhibited duplication events within melon species. Gene structure and protein conserved motif results indicated that the same subfamily displays similar conserved motifs and gene structures. Analysis of promoter cis-regulatory elements indicated that these SRS genes may be involved in hormone response, growth and development, and biotic and abiotic stress response in plants. In addition, most of the CmSRS genes are expressed in the roots, and a few are expressed in the leaves and ovaries. Drought and salt stresses often exacerbate diseases such as powdery mildew and wilt by indirectly weakening the physiological condition of melon plants and altering the rhizosphere environment. Transcriptome analysis of melon roots subjected to 20% PEG6000, 100 mM NaCl, wilt, and powdery mildew revealed that CmSRS gene expression was responsive to all four stress conditions. The subcellular localization and structure of CmSRS proteins were further investigated. This study provides insights into the functions of SRS genes in the growth and development of cucurbit crops and stress response, laying a solid foundation for subsequent functional validation and application studies.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biology14070891/s1. Supplemental Figure S1. Chromosomal distribution of SRS genes in seven cucurbit crops. Supplemental Table S1. RT-qPCR primers for CmSRS genes. Supplemental Table S2. Subcellular localization primers for CmSRS genes. Supplemental Table S3. Orthologous relationships of SRS genes in melon and six other cucurbit species. Supplemental Table S4. CmSRS protein interaction prediction information. Supplemental Table S5. Prediction of secondary structure of SRS gene family proteins in melon.
Author Contributions
Conceptualization, Z.X. and J.M.; methodology, H.M. and K.W.; software, H.M.; writing—original draft preparation, H.M. and K.W.; writing—review and editing, H.M., K.W. and Z.X.; visualization, H.M., K.W., Y.G., J.Y., X.W., M.H., T.L., J.M. and Z.X.; supervision, Z.X.; project administration, J.M.; funding acquisition, J.M. and Z.X. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported in part by the Corps Graduate Student Research and Innovation Program (BTYJXM-2024-K59), the Improvement of Local Varieties in Xinjiang and Selection and Breeding of Major New Hami Melon Varieties (2024A02007-1).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article and its additional files. Genomic sequences and gene annotation information of Cucumis melo, Cucumis sativus, Citrullus lanatus, Lagenaria siceraria, Benincasa hispida, Cucurbita moschata, and Cucurbita maxima were downloaded from http://www.cucurbitgenomics.org/. (accessed on 15 September 2024) Genomic sequences and gene annotation information of Arabidopsis thaliana, rice, and maize were downloaded from https://plants.ensembl.org/ (accessed on 15 September 2024).
Conflicts of Interest
Kexiang Wang was employed by the Hami Agricultural and Animal Husbandry Industry Development Investment Co. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
| aa | Amino Acid |
| ABA | Abscisic Acid |
| bp | Base Pair |
| BP | Biological Process |
| CC | Cellular Component |
| CDD | Conserved Domain Database |
| cDNA | Complementary DNA |
| CDS | Coding Sequence |
| CHT | Charentais-T |
| Da | Dalton |
| GA | Gibberellic Acid |
| HMM | Hidden Markov Model |
| IAA | Indole-3-Acetic Acid |
| LRP1 | Lateral Root Primordium1 |
| MeJA | Methyl Jasmonate |
| MEME | Motif-Based Sequence Analysis Tools |
| MF | Molecular Function |
| qRT-PCR | Quantitative Real-Time PCR |
| SA | Salicylic Acid |
| SHI | Short Internodes |
References
- Zhao, S.; Song, X.; Guo, L.; Zhang, X.; Zheng, W. Genome-wide analysis of the shi-related sequence family and functional identification of gmsrs18 involving in drought and salt stresses in soybean. Int. J. Mol. Sci. 2020, 21, 1810. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Xu, P.; Yu, D. Genome-wide identification and characterization of the shi-related sequence gene family in rice. Evol. Bioinform. 2020, 16, 1612687799. [Google Scholar] [CrossRef] [PubMed]
- Kaulen, H.; Pognonec, P.; Gregor, P.D.; Roeder, R.G. The xenopus b1 factor is closely related to the mammalian activator usf and is implicated in the developmental regulation of tfiiia gene expression. Mol. Cell. Biol. 1991, 11, 412–424. [Google Scholar] [PubMed]
- Elenbaas, B.; Dobbelstein, M.; Roth, J.; Shenk, T.; Levine, A.J. The mdm2 oncoprotein binds specifically to rna through its ring finger domain. Mol. Med. 1996, 2, 439–451. [Google Scholar] [CrossRef] [PubMed]
- Fridborg, I.; Kuusk, S.; Moritz, T.; Sundberg, E. The arabidopsis dwarf mutant shi exhibits reduced gibberellin responses conferred by overexpression of a new putative zinc finger protein. Plant Cell 1999, 11, 1019–1031. [Google Scholar] [CrossRef] [PubMed]
- Fridborg, I.; Kuusk, S.; Robertson, M.; Sundberg, E. The arabidopsis protein shi represses gibberellin responses in arabidopsis and barley. Plant Physiol. 2001, 127, 937–948. [Google Scholar] [CrossRef] [PubMed]
- Kuusk, S.; Sohlberg, J.J.; Magnus Eklund, D.; Sundberg, E. Functionally redundant shi family genes regulate arabidopsis gynoecium development in a dose-dependent manner. Plant J. 2006, 47, 99–111. [Google Scholar] [CrossRef] [PubMed]
- De Rybel, B.; Audenaert, D.; Xuan, W.; Overvoorde, P.; Strader, L.C.; Kepinski, S.; Hoye, R.; Brisbois, R.; Parizot, B.; Vanneste, S. A role for the root cap in root branching revealed by the non-auxin probe naxillin. Nat. Chem. Biol. 2012, 8, 798–805. [Google Scholar] [CrossRef] [PubMed]
- Eklund, D.M.; Cierlik, I.; Ståldal, V.; Claes, A.R.; Vestman, D.; Chandler, J.; Sundberg, E. Expression of arabidopsis short internodes/stylish family genes in auxin biosynthesis zones of aerial organs is dependent on a gcc box-like regulatory element. Plant Physiol. 2011, 157, 2069–2080. [Google Scholar] [CrossRef] [PubMed]
- Gomariz-Fernández, A.; Sánchez-Gerschon, V.; Fourquin, C.; Ferrándiz, C. The role of shi/sty/srs genes in organ growth and carpel development is conserved in the distant eudicot species Arabidopsis thaliana and Nicotiana benthamiana. Front. Plant Sci. 2017, 8, 814. [Google Scholar] [CrossRef] [PubMed]
- Eklund, D.M.; Ståldal, V.; Valsecchi, I.; Cierlik, I.; Eriksson, C.; Hiratsu, K.; Ohme-Takagi, M.; Sundström, J.F.; Thelander, M.; Ezcurra, I. The Arabidopsis thaliana stylish1 protein acts as a transcriptional activator regulating auxin biosynthesis. Plant Cell 2010, 22, 349–363. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.L.; Fedoroff, N.V. Lrp1, a gene expressed in lateral and adventitious root primordia of arabidopsis. Plant Cell 1995, 7, 735–745. [Google Scholar] [PubMed][Green Version]
- Kuusk, S.; Sohlberg, J.J.; Long, J.A.; Fridborg, I.; Sundberg, E. Sty1 and sty2 promote the formation of apical tissues during arabidopsis gynoecium development. Development 2002, 129, 4707–4717. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Yadav, S.; Singh, A.; Mahima, M.; Singh, A.; Gautam, V.; Sarkar, A.K. Auxin signaling modulates lateral root primordium 1 (lrp 1) expression during lateral root development in arabidopsis. Plant J. 2020, 101, 87–100. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.A.; Lütken, H.; Haugslien, S.; Blystad, D.; Torre, S.; Rolcik, J.; Rasmussen, S.K.; Olsen, J.E.; Clarke, J.L. Overexpression of the atshi gene in poinsettia, Euphorbia pulcherrima, results in compact plants. PLoS ONE 2013, 8, e53377. [Google Scholar] [CrossRef]
- Lütken, H.; Jensen, L.S.; Topp, S.H.; Mibus, H.; Müller, R.; Rasmussen, S.K. Production of compact plants by overexpression of atshi in the ornamental kalanchoë. Plant Biotechnol. J. 2010, 8, 211–222. [Google Scholar] [CrossRef] [PubMed]
- Duan, E.; Wang, Y.; Li, X.; Lin, Q.; Zhang, T.; Wang, Y.; Zhou, C.; Zhang, H.; Jiang, L.; Wang, J. Osshi1 regulates plant architecture through modulating the transcriptional activity of ipa1 in rice. Plant Cell 2019, 31, 1026–1042. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Nian, L.; Ain, N.U.; Liu, X.; Yang, Y.; Zhu, X.; Haider, F.U.; Lv, Y.; Bai, P.; Zhang, X. Genome-wide identification and expression profiling of the srs gene family in Melilotus albus reveals functions in various stress conditions. Plants 2022, 11, 3101. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Yu, L.; Zhang, S.; Gu, Q.; Wang, M. Genome-wide characterization of the short inter-nodes/stylish and shi-related sequence family in Gossypium hirsutum and functional identification of ghsrs21 under salt stress. Front. Plant Sci. 2023, 13, 1078083. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, H.; Renner, S.S. Phylogenetic relationships in the order cucurbitales and a new classification of the gourd family (cucurbitaceae). Taxon 2011, 60, 122–138. [Google Scholar] [CrossRef]
- Perkins-Veazie, P.; Collins, J.K.; Davis, A.R.; Roberts, W. Carotenoid content of 50 watermelon cultivars. J. Agric. Food Chem. 2006, 54, 2593–2597. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Liu, C.; Liu, Y.; Xu, Q.; Li, X.; Yang, S. New triterpenoids and other constituents from the fruits of Benincasa hispida (thunb.) Cogn. J. Agric. Food Chem. 2013, 61, 12692–12699. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T.; Juliet, P.A.; Matsui-Hirai, H.; Miyazaki, A.; Fukatsu, A.; Funami, J.; Iguchi, A.; Ignarro, L.J. L-citrulline and l-arginine supplementation retards the progression of high-cholesterol-diet-induced atherosclerosis in rabbits. Proc. Natl. Acad. Sci. USA 2005, 102, 13681–13686. [Google Scholar] [CrossRef] [PubMed]
- Collins, J.K.; Wu, G.; Perkins-Veazie, P.; Spears, K.; Claypool, P.L.; Baker, R.A.; Clevidence, B.A. Watermelon consumption increases plasma arginine concentrations in adults. Nutrition 2007, 23, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Grover, J.K.; Adiga, G.; Vats, V.; Rathi, S.S. Extracts of benincasa hispida prevent development of experimental ulcers. J. Ethnopharmacol. 2001, 78, 159–164. [Google Scholar] [CrossRef] [PubMed]
- Gu, M.; Fan, S.; Liu, G.; Guo, L.; Ding, X.; Lu, Y.; Zhang, Y.; Ji, G.; Huang, C. Extract of wax gourd peel prevents high-fat diet-induced hyperlipidemia in c57bl/6 mice via the inhibition of the pparγ pathway. Evid.-Based Complement. Altern. Med. 2013, 2013, 342561. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.T.; Li, H.Y.; Xu, J.C. Bioinformatics analysis of the heavy metal transporting atpase gene family in poplar genome. Plant Physiol. J 2014, 50, e900. [Google Scholar]
- FAO. Food and Agriculture Organization of the United Nations; FAO: Rome, Italy, 2018; p. 403. Available online: http://faostat.fao.org (accessed on 10 October 2024).
- Roy, S.J.; Tucker, E.J.; Tester, M. Genetic analysis of abiotic stress tolerance in crops. Curr. Opin. Plant Biol. 2011, 14, 232–239. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, M.; Wu, L. Breeding for salinity tolerance in plants. Crit. Rev. Plant Sci. 1994, 13, 17–42. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. Tbtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. Mega11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Bechtold, U.; Field, B. Molecular Mechanisms Controlling Plant Growth During Abiotic Stress; Oxford University Press: Oxford, UK, 2018; Volume 69, pp. 2753–2758. [Google Scholar]
- Ben Ammar, H.; Picchi, V.; Arena, D.; Treccarichi, S.; Bianchi, G.; Lo Scalzo, R.; Marghali, S.; Branca, F. Variation of bio-morphometric traits and antioxidant compounds of Brassica oleracea L. Accessions in relation to drought stress. Agronomy 2022, 12, 2016. [Google Scholar] [CrossRef]
- Raza, A. Eco-physiological and biochemical responses of rapeseed (Brassica napus L.) to abiotic stresses: Consequences and mitigation strategies. J. Plant Growth Regul. 2021, 40, 1368–1388. [Google Scholar] [CrossRef]
- He, B.; Shi, P.; Lv, Y.; Gao, Z.; Chen, G. Gene coexpression network analysis reveals the role of srs genes in senescence leaf of maize (Zea mays L.). J. Genet. 2020, 99, 3. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ahmad, B.; Zeng, S.; Lan, Y.; Hu, X.; Fu, L.; Hu, T.; Li, J.; Zhang, X.; Pan, Y. Genome-wide characterization of shi-related sequence gene family and its roles in response to Zn2+ stress in cucumber. Horticulturae 2024, 10, 1154. [Google Scholar] [CrossRef]
- Baylis, T.; Cierlik, I.; Sundberg, E.; Mattsson, J. Short internodes/stylish genes, regulators of auxin biosynthesis, are involved in leaf vein development in a Rabidopsis thaliana. New Phytol. 2013, 197, 737–750. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Lee, S.; Kim, Y.; Yun, D.; Woo, J.; Park, C. Activation tagging of an arabidopsis shi-related sequence gene produces abnormal anther dehiscence and floral development. Plant Mol. Biol. 2010, 74, 337–351. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Yang, S.; Shi, K.; Yang, L.; An, M.; Wang, F.; Qi, Y.; Feng, M.; Wang, M.; Geng, P. Genome-wide identification of the lrx gene family in cucurbitaceae and expression analysis under salt and drought stress in cucumber. Veg. Res. 2024, 4, e026. [Google Scholar] [CrossRef]
- Zhu, X.; Wang, B.; Wang, X.; Wei, X. Genome-wide identification, structural analysis and expression profiles of short internodes related sequence gene family in quinoa. Front. Genet. 2022, 13, 961925. [Google Scholar] [CrossRef] [PubMed]
- Maher, C.; Stein, L.; Ware, D. Evolution of arabidopsis microrna families through duplication events. Genome Res. 2006, 16, 510–519. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Liu, Y.; Zhou, J.; Zhao, S.; Zhang, X.; Min, D. Genome-wide analysis of the lectin receptor-like kinase family in foxtail millet (Setaria italica L.). Plant Cell Tissue Organ Cult. (PCTOC) 2016, 127, 335–346. [Google Scholar] [CrossRef]
- Sohlberg, J.J.; Myrenås, M.; Kuusk, S.; Lagercrantz, U.; Kowalczyk, M.; Sandberg, G.; Sundberg, E. Sty1 regulates auxin homeostasis and affects apical–basal patterning of the arabidopsis gynoecium. Plant J. 2006, 47, 112–123. [Google Scholar] [CrossRef] [PubMed]
- Ståldal, V.; Sohlberg, J.J.; Eklund, D.M.; Ljung, K.; Sundberg, E. Auxin can act independently of crc, lug, seu, spt and sty1 in style development but not apical-basal patterning of the arabidopsis gynoecium. New Phytol. 2008, 180, 798–808. [Google Scholar] [CrossRef] [PubMed]
- Ståldal, V.; Cierlik, I.; Chen, S.; Landberg, K.; Baylis, T.; Myrenås, M.; Sundström, J.F.; Eklund, D.M.; Ljung, K.; Sundberg, E. The Arabidopsis thaliana transcriptional activator stylish1 regulates genes affecting stamen development, cell expansion and timing of flowering. Plant Mol. Biol. 2012, 78, 545–559. [Google Scholar] [CrossRef] [PubMed]
- Barcala, M.; García, A.; Cabrera, J.; Casson, S.; Lindsey, K.; Favery, B.; García Casado, G.; Solano, R.; Fenoll, C.; Escobar, C. Early transcriptomic events in microdissected arabidopsis nematode-induced giant cells. Plant J. 2010, 61, 698–712. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).