Analysis of Composition, Structure, and Driving Factors of Root-Associated Endophytic Bacterial Communities of the Chinese Medicinal Herb Glycyrrhiza
Simple Summary
Abstract
1. Introduction
2. Results
2.1. Plant and Soil Properties
2.2. Diversity of Endophytic Bacterial Community
2.3. Differences in the Composition of Endophytic Bacterial Community
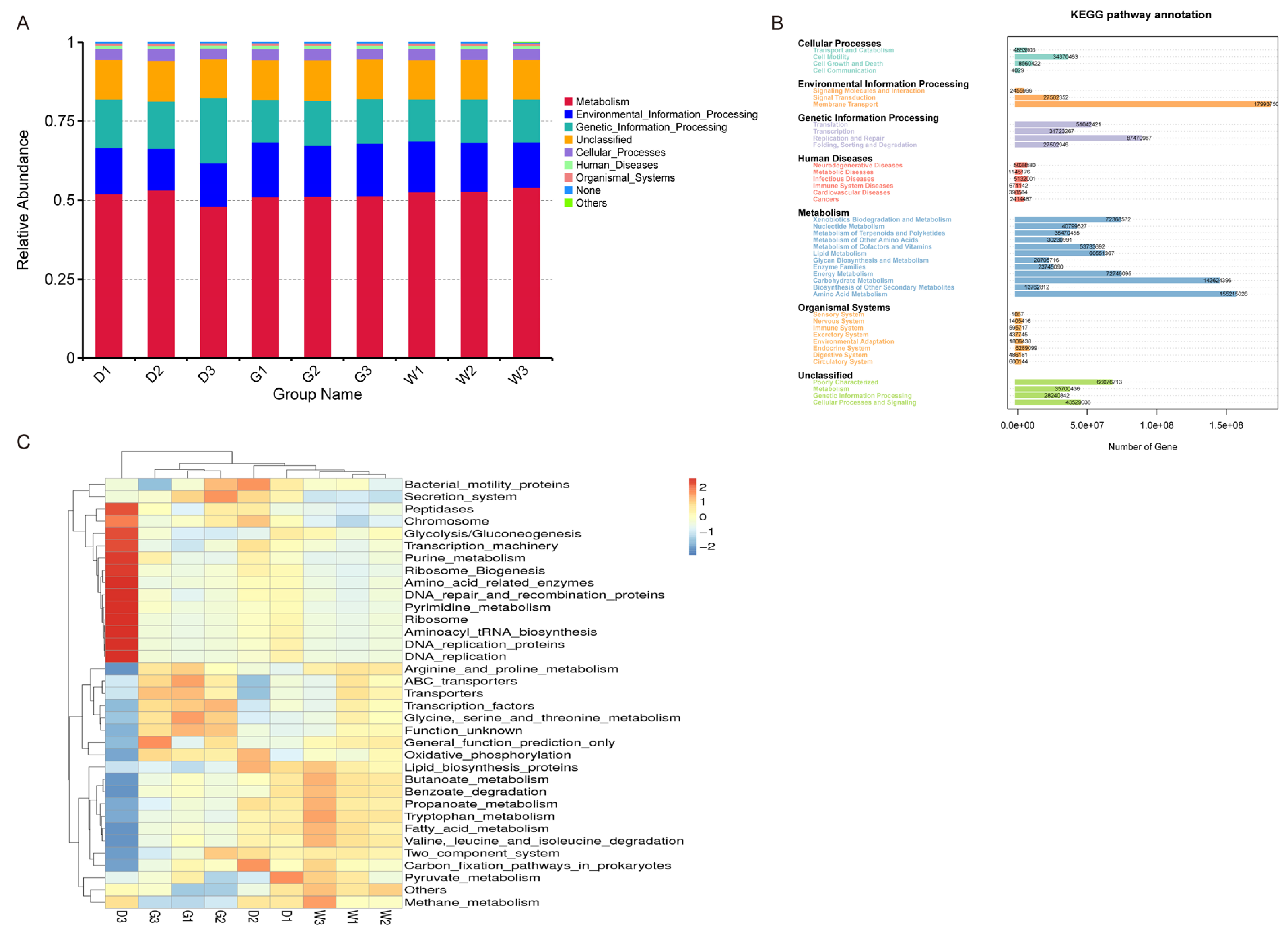
2.4. Prediction of Endophytic Bacterial Community Function
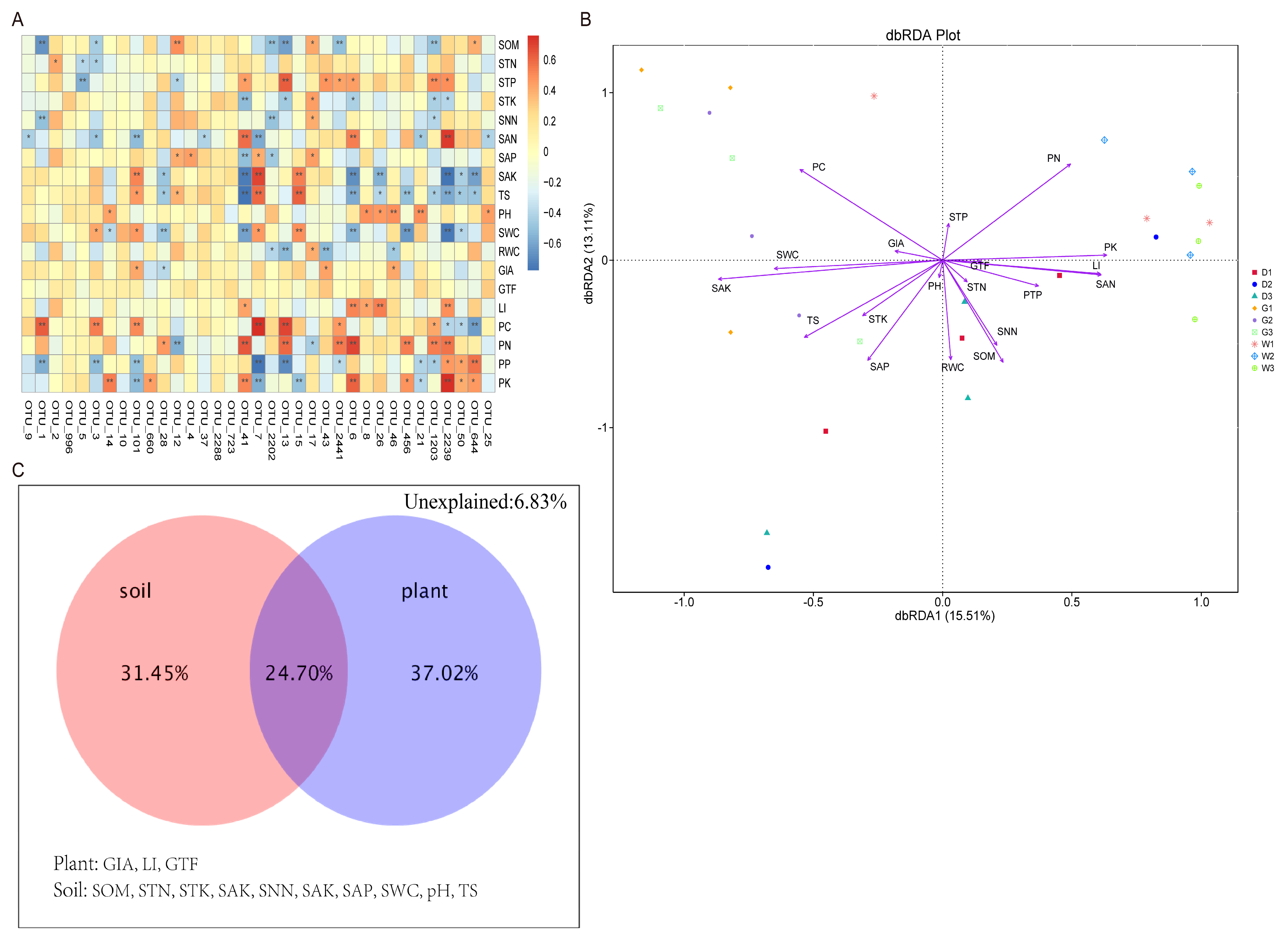
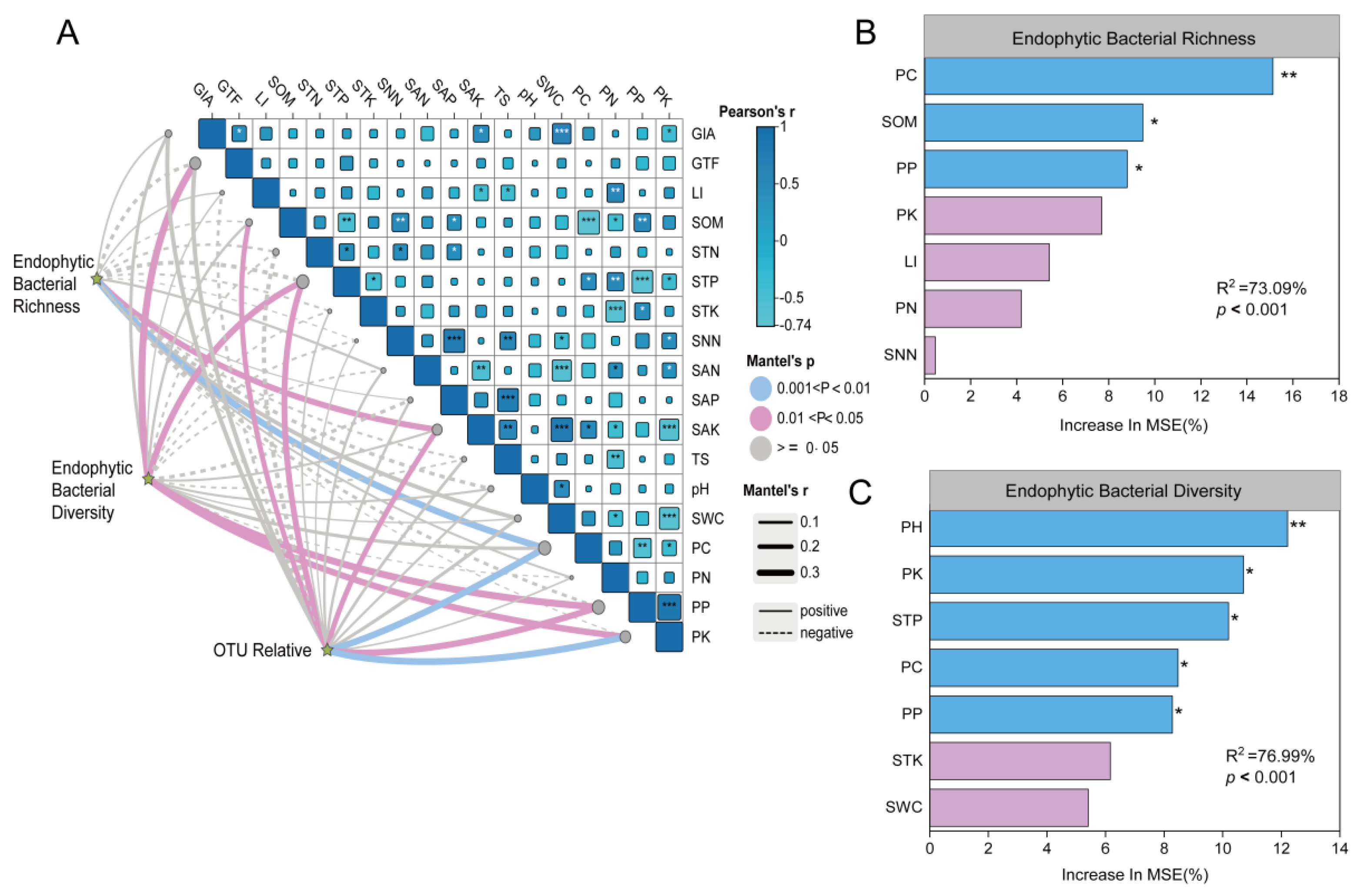
2.5. Relationship Between Plant and Soil Factors and Endophytic Bacterial Communities
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. Analysis of Soil Properties and Plant Nutrients
4.3. Determination of Secondary Metabolites in Roots
4.4. Microbial Community Analysis
4.5. Biological and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Egamberdieva, D.; Mamedov, N. Potential use of licorice in phytoremediation of salt affected soils. In Plants, Pollutants and Remediation; Springer: Dordrecht, The Netherlands, 2016; pp. 309–318. [Google Scholar]
- Yang, R.; Li, W.; Yuang, B.; Ma, M.Y.; Shan, Z.; Liu, C.; Liu, Y. Simultaneous determination of 18 α-glycyrrhizic acid and 18 β-glycyrrhizic acid in three licorice samples from different origin by HPLC. Chin. J. Pharm. Anal. 2016, 36, 1065–1071. [Google Scholar][Green Version]
- Rizzato, G.; Scalabrin, E.; Radaelli, M.; Capodaglio, G.; Piccolo, O. A new exploration of licorice metabolome. Food Chem. 2017, 221, 959–968. [Google Scholar] [CrossRef] [PubMed]
- Woo, C.K.; Jeong, E.B.; Eun, J.K.; Seok, H.C.; Tae, S.G. Antibacterial and antioxidative activities of licorice extracts fermented with Nuruk molds. Korean J. Food Preserv. 2018, 25, 830–836. [Google Scholar]
- Liu, Y.; Li, Y.; Luo, W.; Liu, S.; Chen, W.; Chen, C.; Jiao, S.; Wei, G. Soil potassium is correlated with root secondary metabolites and root-associated core bacteria in licorice of different ages. Plant Soil. 2020, 456, 61–79. [Google Scholar] [CrossRef]
- Hossein, H.; Marjan, N. Pharmacological Effects of Glycyrrhiza spp. Its Bioactive Constituents: Update Review. Phytother. Res. 2015, 29, 1868–1886. [Google Scholar]
- Wang, L.; Yang, R.; Yuan, B.; Liu, Y.; Liu, C. The antiviral and antimicrobial activities of licorice, a widely-used Chinese herb. Acta Pharm. Sin. B. 2015, 5, 310–315. [Google Scholar] [CrossRef]
- Hikaru, S.; Satoru, S.; Kiyoshi, O.; Masaharu, M.; Toshiyuki, O.; Hiroshi, S.; Toshiya, M. Triterpene Functional Genomics in Licorice for Identification of CYP72A154 Involved in the Biosynthesis of Glycyrrhizin. Plant Cell 2011, 23, 4112–4123. [Google Scholar]
- Vasisht, K.; Sharma, N.; Karan, M. Current Perspective in the International Trade of Medicinal Plants Material: An Update. Curr. Pharm. Des. 2016, 22, 4288–4336. [Google Scholar] [CrossRef]
- Miliute, I.; Buzaite, O.; Baniulis, D.; Stanys, V. Bacterial endophytes in agricultural crops and their role in stress tolerance: A review. Zemdirb. Agric. 2015, 102, 465–478. [Google Scholar] [CrossRef]
- Mushtaq, S.; Shafiq, M.; Tariq, M.R.; Sami, A.; Nawaz-ul-Rehman, M.S.; Bhatti, M.H.T.; Haider, M.S.; Sadiq, S. Interaction between bacterial endophytes and host plants. Front. Plant Sci. 2023, 13, 1092105. [Google Scholar]
- Papik, J.; Folkmanova, M.; Polivkova, M.; Suman, J.; Uhlik, O. The invisible life inside plants: Deciphering the riddles of endophytic bacterial diversity. Biotechnol. Adv. 2020, 44, 107614. [Google Scholar] [CrossRef] [PubMed]
- Cynthia, M.; Francisca, V.; Alex, E.V.; Aleydis, G.; Shrabana, S.; Cabeza, R.A.; Aparna, B. Plant-growth-promoting bacteria from rhizosphere of Chilean common bean ecotype (Phaseolus vulgaris L.) supporting seed germination and growth against salinity stress. Front. Plant Sci. 2022, 13, 1052263. [Google Scholar]
- Peeran, M.F. A Potential Source of Methyl-Eugenol from Secondary Metabolite of Rhizopus oryzae 6975. Int. J. Appl. Biol. Pharm. Technol. 2016, 7, 187–192. [Google Scholar]
- Naasko, K.I.; Naylor, D.; Graham, E.B.; Couvillion, S.P.; Danczak, R.; Tolic, N.; Nicora, C.; Fransen, S.; Tao, H.; Hofmockel, K.S.; et al. Influence of soil depth, irrigation, and plant genotype on the soil microbiome, metaphenome, and carbon chemistry. mBio 2023, 14, e0175823. [Google Scholar] [CrossRef]
- Vandenkoornhuyse, P.; Quaiser, A.; Duhamel, M.; Le, V.A.; Dufresne, A. The importance of the microbiome of the plant holobiont. New Phytol. 2015, 206, 1196–1206. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Rott, M.; Schlaeppi, K.; Ver Loren van Themaat, E.; Ahmadinejad, N.; Assenza, F.; Rauf, P.; Huettel, B.; Reinhardt, R.; Schmelzer, E.; et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 2012, 488, 91–95. [Google Scholar] [CrossRef]
- Jiao, S.; Zhang, Z.; Yang, F.; Lin, Y.; Chen, W.; Wei, G. Temporal dynamics of microbial communities in microcosms in response to pollutants. Mol. Ecol. 2017, 26, 923–936. [Google Scholar] [CrossRef]
- Jia, M.; Chen, L.; Xin, H.; Zheng, C.; Rahman, K.; Han, T.; Qin, L. A Friendly Relationship between Endophytic Fungi and Medicinal Plants: A Systematic Review. Front. Microbiol. 2016, 7, 906. [Google Scholar] [CrossRef]
- Martins, S.A.; Schurt, D.A.; Seabra, S.S.; Martins, S.J.; Ramalho, M.A.P.; Moreira, F.M.S.; Silva, J.C.P.; Silva, J.A.G.; Medeiros, F.H.V. Common bean (Phaseolus vulgaris L.) growth promotion and biocontrol by rhizobacteria under Rhizoctonia solani suppressive and conducive soils. Appl. Soil Ecol. 2018, 127, 129–135. [Google Scholar] [CrossRef]
- Carolina, C.; Marinella, D.L.; Vincenzo, L.; Ylenia, P.; Laura, P. Characterization of the endophytic bacterial community of Bituminaria bituminosa plant grown in vitro and its interaction with the plant extract. Front. Plant Sci. 2023, 13, 1076573. [Google Scholar]
- Tao, W.; Duan, J.; Zhao, R.; Li, X.; Yan, W.; Li, J.; Tang, Y. Comparison of three officinal Chinese pharmacopoeia species of Glycyrrhiza based on separation and quantification of triterpene saponins and chemometrics analysis. Food Chem. 2013, 141, 1681–1689. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Li, W.; Yuan, B.; Ren, G.; Wang, L.; Cheng, T.; Liu, Y. The genetic and chemical diversity in three original plants of licorice, Glycyrriza uralensis Fisch., Glycyrrhiza inflata Bat. and Glycyrrhiza glabra L. Pak. J. Pharm. Sci. 2018, 31, 525–535. [Google Scholar] [PubMed]
- Chen, C.; Zhong, C.; Gao, X.; Tan, C.; Bai, H.; Ning, K. Glycyrrhiza uralensis Fisch. Root-associated microbiota: The multifaceted hubs associated with environmental factors, growth status and accumulation of secondary metabolites. Environ. Microbiome 2022, 17, 23. [Google Scholar] [CrossRef]
- Hayashi, H. Molecular Biology of Secondary Metabolism: Case Study for Glycyrrhiza Plants. In Recent Advances in Plant Biotechnology; Springer: Berlin/Heidelberg, Germany, 2009; pp. 89–103. [Google Scholar]
- Han, Y.; Hou, Z.; Zhang, X.; He, Q.; Liang, Z. Multi-dimensional “projection”—The impact of abiotic stresses on the content of seven active compounds and expression of related genes in Glycyrrhiza uralensis Fisch. Environ. Exp. Bot. 2022, 197, 104846. [Google Scholar] [CrossRef]
- Golldack, D.; Li, C.; Mohan, H.; Probst, N. Tolerance to drought and salt stress in plants: Unraveling the signaling networks. Front. Plant 2014, 5, 151. [Google Scholar] [CrossRef]
- Ruiz-Lozano, J.M.; Aroca, R.; Zamarreño, Á.M.; Molina, S.; Andreo-Jiménez, B.; Porcel, R.; López-Ráez, J.A. Arbuscular mycorrhizal symbiosis induces strigolactone biosynthesis under drought and improves drought tolerance in lettuce and tomato. Plant Cell Environ. 2015, 39, 441–452. [Google Scholar] [CrossRef]
- Lou, J.; Ding, B.; Fang, M.; Xie, W.; Wang, X.; Wang, X.; Zhu, J. Glycyrrhizin Attenuates White Matter Injury by Inhibiting Neuroinflammation through the HMGB1/TLR4 Pathway. Mol. Neurobiol. 2024, 62, 1–18. [Google Scholar] [CrossRef]
- Li, L.; Zhang, Y.; Wang, Z.; Chen, X.; Fang, M. Glycyrrhizin attenuates renal inflammation in a mouse Con A-hepatitis model via the IL-25/M2 axis. Ren. Fail. 2024, 46, 2356023. [Google Scholar] [CrossRef]
- Nasrollahi, V.; Mirzaie-Asl, A.; Piri, K.; Nazeri, S.; Mehrabi, R. The effect of drought stress on the expression of key genes involved in the biosynthesis of triterpenoid saponins in liquorice (Glycyrrhiza glabra). Phytochemistry 2014, 103, 32–37. [Google Scholar] [CrossRef]
- Pant, B.D.; Pant, P.; Erban, A.; Huhman, D.; Kopka, J.; Scheible, W.R. Identification of primary and secondary metabolites with phosphorus status—Dependent abundance in Arabidopsis, and of the transcription factor PHR1 as a major regulator of metabolic changes during phosphorus limitation. Plant Cell Environ. 2015, 38, 172–187. [Google Scholar] [CrossRef]
- Li, Z.; Wen, W.; Qin, M.; He, Y.; Xu, D.; Li, L. Biosynthetic Mechanisms of Secondary Metabolites Promoted by the Interaction Between Endophytes and Plant Hosts. Front. Microbiol. 2022, 13, 928967. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, R.; Anand, G.; Gupta, P.; Mandal, S. Insight into the mechanisms of enhanced production of valuable terpenoids by arbuscular mycorrhiza. Phytochem. Rev. 2016, 16, 677–692. [Google Scholar] [CrossRef]
- Xie, W.; Hao, Z.; Yu, M.; Wu, Z.; Zhao, A.; Li, J.; Chen, B. Improved phosphorus nutrition by arbuscular mycorrhizal symbiosis as a key factor facilitating glycyrrhizin and liquiritin accumulation in Glycyrrhiza uralensis. Plant Soil 2018, 439, 243–257. [Google Scholar] [CrossRef]
- Franche, C.; Lindstrm, K.; Elmerich, C. Nitrogen-fixing bacteria associated with leguminous and non-leguminous plants. Plant Soil 2009, 321, 35–59. [Google Scholar] [CrossRef]
- Andrei, K.; Hélène, C.S.; Laurent, C.; Bernard, D.; Elodie, G. A Comprehensive Assessment of the Secretome Responsible for Host Adaptation of the Legume Root Pathogen Aphanomyces euteiches. J. Fungi 2022, 8, 88. [Google Scholar]
- Huang, W.; Long, C.; Lam, E. Roles of plant-associated microbiota in traditional herbal medicine. Trends Plant Sci. 2018, 23, 559–562. [Google Scholar] [CrossRef]
- Gao, Z.; Chen, J.; Liu, X.; Xue, C. The correlation between maize genetic polymorphisms and endophytic bacteria population in plant roots. Acta Ecol. Sin. 2017, 26, 1920–1923. [Google Scholar]
- Morella, N.M.; Weng, F.C.H.; Joubert, P.M.; Metcalf, C.J.E.; Lindow, S.; Koskella, B. Successive passaging of a plant-associated microbiome reveals robust habitat and host genotype-dependent selection. Proc. Natl. Acad. Sci. USA 2020, 117, 1148–1159. [Google Scholar] [CrossRef]
- Veach, A.M.; Morris, R.; Yip, D.Z.; Yang, Z.K.; Engle, N.L.; Cregger, M.A.; Schadt, C.W. Correction to: Rhizosphere microbiomes diverge among Populus trichocarpa plant-host genotypes and chemotypes, but it depends on soil origin. Microbiome 2021, 9, 21. [Google Scholar] [CrossRef]
- Saikkonen, K.; Wali, P.; Helander, M.; Faeth, S.H. Evolution of endophyte-plant symbioses. Trends Plant Sci. 2004, 9, 275–280. [Google Scholar] [CrossRef]
- Dong, L.; Cheng, R.; Xiao, L.; Wei, F.; Wei, G.; Xu, J.; Wang, Y.; Guo, Z.; Chen, Z.; Chen, S. Diversity and composition of bacterial endophytes among plant parts of Panax notoginseng. Chin. Med. 2018, 13, 41. [Google Scholar] [CrossRef] [PubMed]
- Webster, G.; Embley, T.M.; Prosser, J.I. Grassland Management Regimens Reduce Small-Scale Heterogeneity and Species Diversity of β-Proteobacterial Ammonia Oxidizer Populations. Appl. Environ. Microbiol. 2002, 68, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Chen, J.; Ren, S.; Shen, H.; Rong, B. Complete genome sequence of Mycobacterium Mya-zh01, an endophytic bacterium, promotes plant growth and seed germination isolated from flower stalk of Doritaenopsis. Arch. Microbiol. 2020, 202, 1965–1976. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Ren, G.; Zhou, N.; Qiao, Z.; Li, M.; Yin, Y.; Liu, C. A new simplified synthetic endophyte community regulates the synthesis of active ingredients in Glycyrrhiza uralensis Fisch. Ind. Crops Prod. 2025, 227, 120781. [Google Scholar] [CrossRef]
- Cheng, H.; Wu, B.; Wei, M.; Wang, S.; Rong, X.; Du, D.; Wang, C. Changes in community structure and metabolic function of soil bacteria depending on the type restoration processing in the degraded alpine grassland ecosystems in Northern Tibet. Sci. Total Environ. 2021, 755, 142619. [Google Scholar] [CrossRef]
- Paul, D.; Lade, H. Plant-growth-promoting rhizobacteria to improve crop growth in saline soils: A review. Agron. Sustain. Dev. 2014, 34, 737–752. [Google Scholar] [CrossRef]
- Diouf, D.; Samba-Mbaye, R.; Lesueur, D.; Ba, A.T.; Dreyfus, B.; Philippe, D.L.; Neyra, M. Genetic Diversity of Acacia seyal Del. Rhizobial Populations Indigenous to Senegalese Soils in Relation to Salinity and pH of the Sampling Sites. Microb. Ecol. 2007, 54, 553–566. [Google Scholar]
- Kristin, K.; Bernd, W.; Vera, K.; Franziska, W.; Heiko, N.; Ingo, S.; Rolf, D. Driving forces of soil bacterial community structure, diversity, and function in temperate grasslands and forests. Sci. Rep. 2016, 6, 33696. [Google Scholar]
- Guo, Y.; Chen, X.; Wu, Y.; Zhang, L.; Cheng, J.; Wei, G.; Lin, Y. Natural revegetation of a semiarid habitat alters taxonomic and functional diversity of soil microbial communities. Sci. Total Environ. 2018, 635, 598–606. [Google Scholar] [CrossRef]
- Li, J.; Yang, C.; Zhou, H.; Shao, X. Responses of plant diversity and soil microorganism diversity to water and nitrogen additions in the Qinghai-Tibetan Plateau. Glob. Ecol. Conserv. 2020, 22, e01003. [Google Scholar] [CrossRef]
- Wei, M.; Wang, S.; Xiao, H.; Wu, B.; Jiang, K.; Du, D.; Wang, C. Stand-alone or co-occurring invasive plant species do not modify the diversity of the soil N 2 -fixing bacterial community. Plant Ecol. Divers. 2020, 13, 277–287. [Google Scholar] [CrossRef]
- Zhou, Z.; Wang, C.; Luo, Y. Meta-analysis of the impacts of global change factors on soil microbial diversity and functionality. Nat. Commun. 2020, 11, 3072. [Google Scholar] [CrossRef]
- Wang, C.; Zhou, J.; Liu, J.; Du, D. Responses of soil N-fixing bacteria communities to invasive species over a gradient of simulated nitrogen deposition. Ecol. Eng. 2017, 98, 32–39. [Google Scholar] [CrossRef]
- Wang, C.; Wang, S.; Wu, B.; Wei, M.; Rong, X.; Li, Y.; Du, D. Ecological restoration treatments enhanced plant and soil microbial diversity in the degraded alpine steppe in Northern Tibet. Land Degrad. Dev. 2020, 32, 723–737. [Google Scholar] [CrossRef]
- Sun, Q.; Liu, W.; Gao, Y.; Li, J.; Yang, C. Spatiotemporal Variation and Climate Influence Factors of Vegetation Ecological Quality in the Sanjiangyuan National Park. Sustainability 2020, 12, 6634. [Google Scholar] [CrossRef]
- Dang, H.; Zhang, T.; Li, G.; Mu, Y.; Lv, X.; Wang, Z.; Zhuang, L. Root-associated endophytic bacterial community composition and structure of three medicinal licorices and their changes with the growing year. BMC Microbiol. 2020, 20, 1–18. [Google Scholar] [CrossRef]
- Bao, S. Soil Agro-Chemistrical Analysis; China Agriculture Press: Beijing, China, 2000; Volume 2030, pp. 30–107. [Google Scholar]
- Mago, T.; Salzberg, S.L. FLASH: Fast Length Adjustment of Short Reads to Improve Genome Assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Gregory, C.J.; Lauber, C.L.; Walters, W.A.; Donna, B.L.; Lozupone, C.A.; Turnbaugh, P.J.; Rob, K. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar]
- Bokulich, N.A.; Sathish, S.; Faith, J.J.; Dirk, G.; Gordon, J.I.; Rob, K.; Gregory, C.J. Quality-filtering vastly improves diversity estimates from illumina amplicon sequencing. Nat. Methods 2019, 10, 57–59. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Christopher, Q.; Rob, K. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Dirk, G.; Earl, A.M.; Mike, F.; Ward, D.V.; Georgia, G.; Birren, B.W. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011, 21, 494–504. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996. [Google Scholar] [CrossRef] [PubMed]
- Christian, Q.; Elmar, P.; Pelin, Y.; Jan, G.; Timmy, S.; Pablo, Y.; Oliver, G.F. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar]
- Edgar, R.C. Local homology recognition and distance measures in linear time using compressed amino acid alphabets. Nucleic Acids Res. 2004, 32, 380–385. [Google Scholar] [CrossRef]
- Li, B.; Zhang, X.; Guo, F.; Wu, W.; Zhang, T. Characterization of tetracycline resistant bacterial community in saline activated sludge using batch stress incubation with high-throughput sequencing analysis. Water Res. 2013, 47, 4207–4216. [Google Scholar] [CrossRef]
- Qin, J.; Li, Y.; Cai, Z.; Li, S.; Zhu, J.; Zhang, F.; Liang, S.; Zhang, W.; Guan, Y.; Shen, D.; et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012, 490, 55–60. [Google Scholar] [CrossRef]
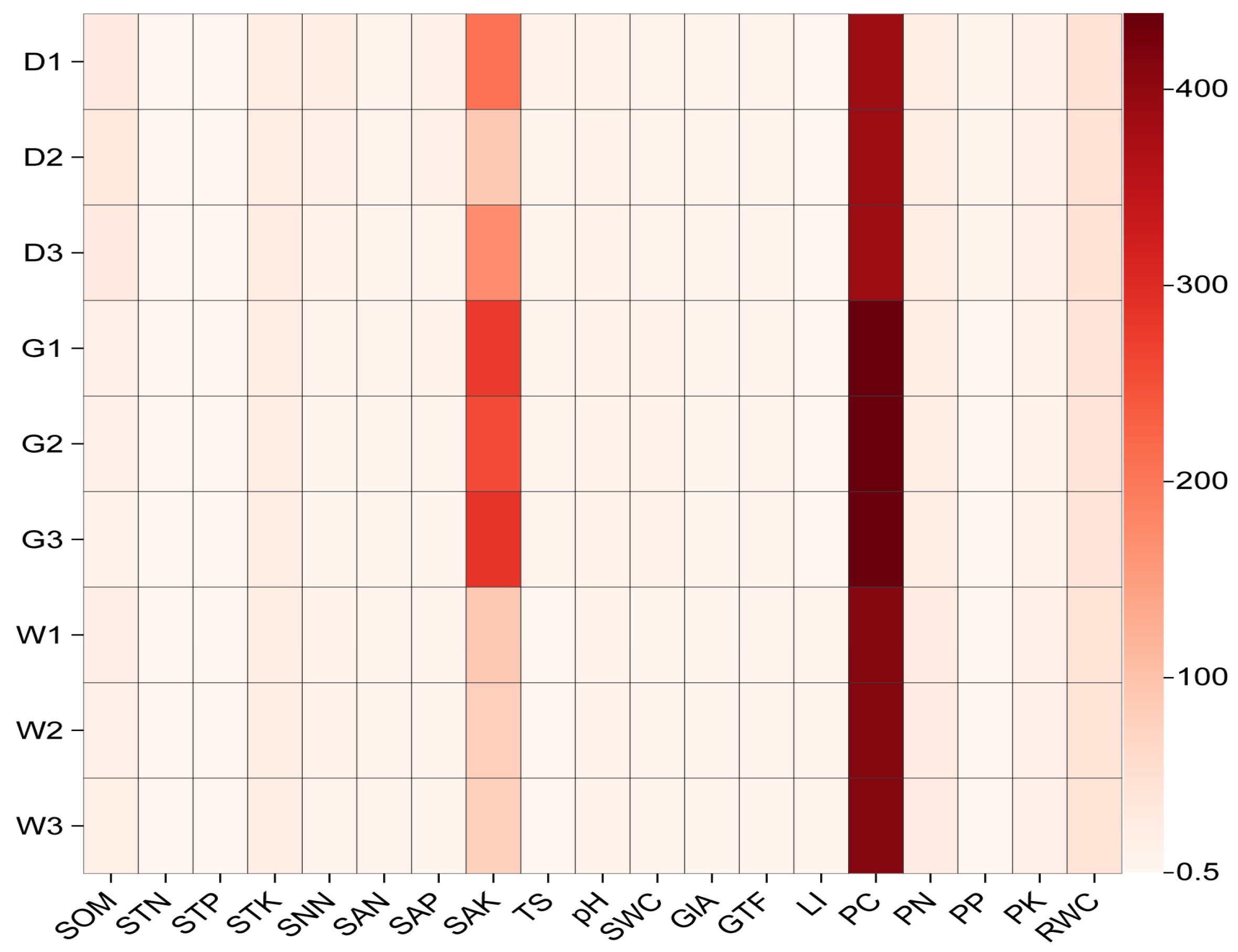
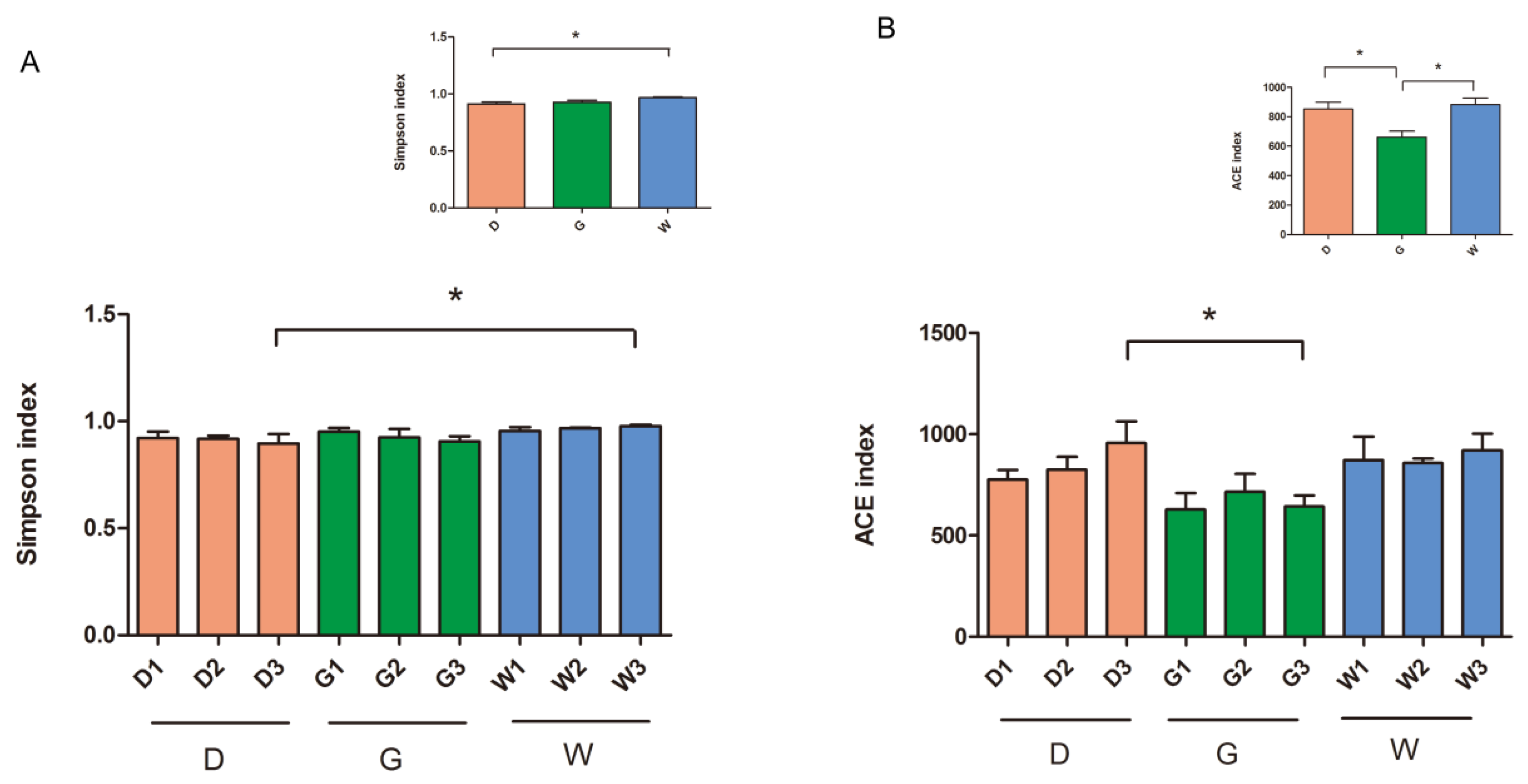

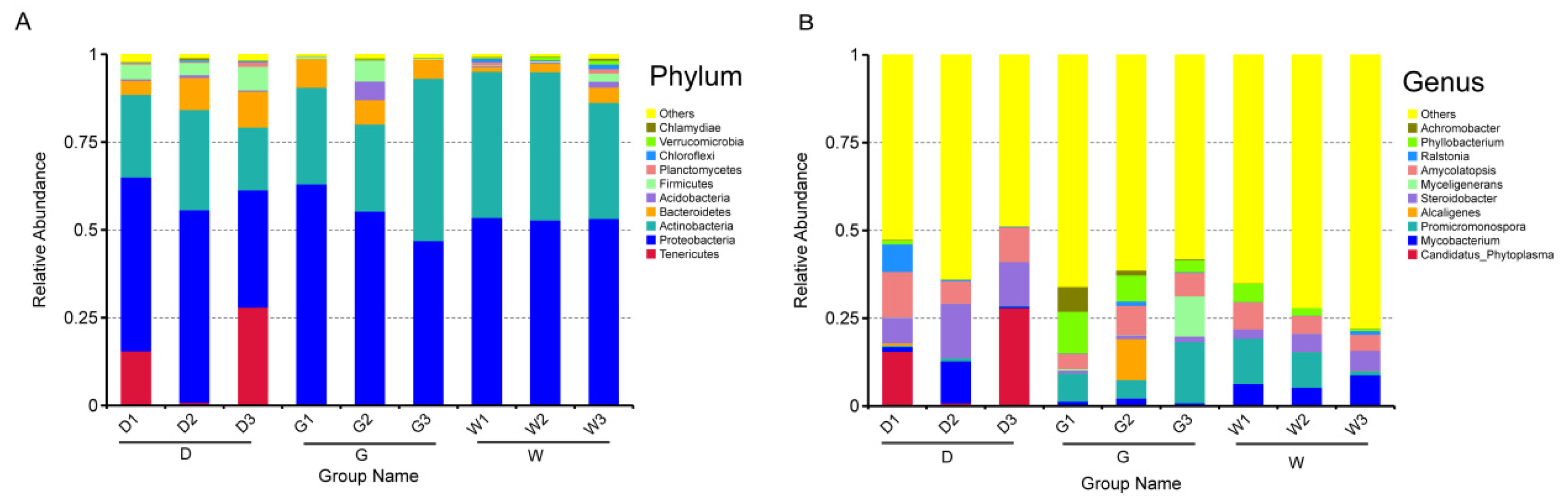
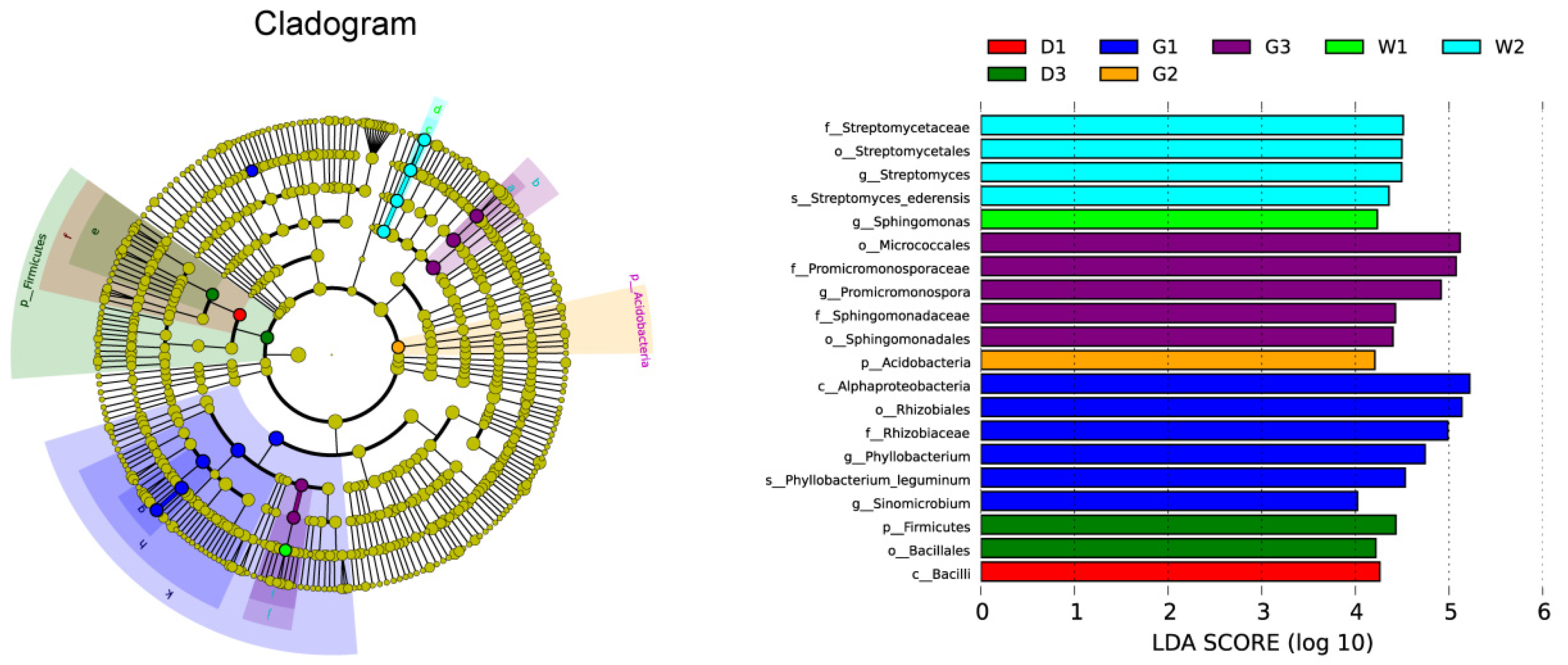
| Variables | D | G | W | |
|---|---|---|---|---|
| Plant Nutrients | PC (g/kg) | 390.640 ± 10.396 b | 441.021 ± 6.389 a | 412.617 ± 2.304 ab |
| PN (g/kg) | 17.959 ± 0.323 b | 21.021 ± 2.213 b | 24.886 ± 1.110 a | |
| PP (g/kg) | 1.460 ± 0.157 a | 1.223 ± 0.015 a | 1.274 ± 0.011 a | |
| PK (g/kg) | 12.257 ± 2.296 a | 9.021 ± 0.428 a | 12.330 ± 0.488 a | |
| Root Secondary Metabolite | GlA (%) | 2.031 ± 0.091 b | 2.751 ± 0.179 a | 2.073 ± 0.267 ab |
| GTF (%) | 4.522 ± 0.103 a | 4.582 ± 0.076 a | 4.597 ± 0.050 a | |
| LI (%) | 0.965 ± 0.110 b | 0.941 ± 0.133 b | 1.788 ± 0.319 a | |
| RWC (%) | 56.756 ± 1.762 a | 47.578 ± 1.165 b | 48.563 ± 1.080 b | |
| Soil Physicochemical Properties | SOM (g/kg) | 27.990 ± 1.981 a | 10.495 ± 0.817 b | 14.744 ± 1.533 b |
| STN (g/kg) | 0.762 ± 0.121 a | 0.693 ± 0.065 a | 0.832 ± 0.084 a | |
| STP (g/kg) | 0.537 ± 0.039 b | 0.666 ± 0.012 a | 0.712 ± 0.018 a | |
| STK (g/kg) | 21.864 ± 0.441 a | 20.771 ± 0.172 ab | 19.743 ± 0.278 b | |
| SNN (mg/kg) | 14.200 ± 2.443 a | 3.552 ± 0.616 b | 7.593 ± 2.387 ab | |
| SAN (mg/kg) | 4.869 ± 0.382 b | 3.326 ± 0.175 c | 6.021 ± 0.288 a | |
| SAP (mg/kg) | 9.678 ± 2.251 a | 5.291 ± 0.841 ab | 3.700 ± 0.963 b | |
| SAK (mg/kg) | 180.031 ± 22.146 b | 273.093 ± 19.963 a | 81.207 ± 9.539 c | |
| TS (g/kg) | 5.697 ± 1.716 a | 4.894 ± 0.694 ab | 1.033 ± 0.078 b | |
| PH | 8.450 ± 0.068 a | 8.831 ± 0.232 a | 8.534 ± 0.067 a | |
| SWC (%) | 4.923 ± 0.352 b | 7.983 ± 0.638 a | 3.583 ± 0.237 b |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; Ma, A.; Zhang, T.; Zhuang, L.; Dang, H. Analysis of Composition, Structure, and Driving Factors of Root-Associated Endophytic Bacterial Communities of the Chinese Medicinal Herb Glycyrrhiza. Biology 2025, 14, 856. https://doi.org/10.3390/biology14070856
Zhang Z, Ma A, Zhang T, Zhuang L, Dang H. Analysis of Composition, Structure, and Driving Factors of Root-Associated Endophytic Bacterial Communities of the Chinese Medicinal Herb Glycyrrhiza. Biology. 2025; 14(7):856. https://doi.org/10.3390/biology14070856
Chicago/Turabian StyleZhang, Zhilin, Aifang Ma, Tao Zhang, Li Zhuang, and Hanli Dang. 2025. "Analysis of Composition, Structure, and Driving Factors of Root-Associated Endophytic Bacterial Communities of the Chinese Medicinal Herb Glycyrrhiza" Biology 14, no. 7: 856. https://doi.org/10.3390/biology14070856
APA StyleZhang, Z., Ma, A., Zhang, T., Zhuang, L., & Dang, H. (2025). Analysis of Composition, Structure, and Driving Factors of Root-Associated Endophytic Bacterial Communities of the Chinese Medicinal Herb Glycyrrhiza. Biology, 14(7), 856. https://doi.org/10.3390/biology14070856






