Transcriptome and Cellular Evidence of Depot-Specific Function in Beef Cattle Intramuscular, Subcutaneous, and Visceral Adipose Tissues
Simple Summary
Abstract
1. Introduction
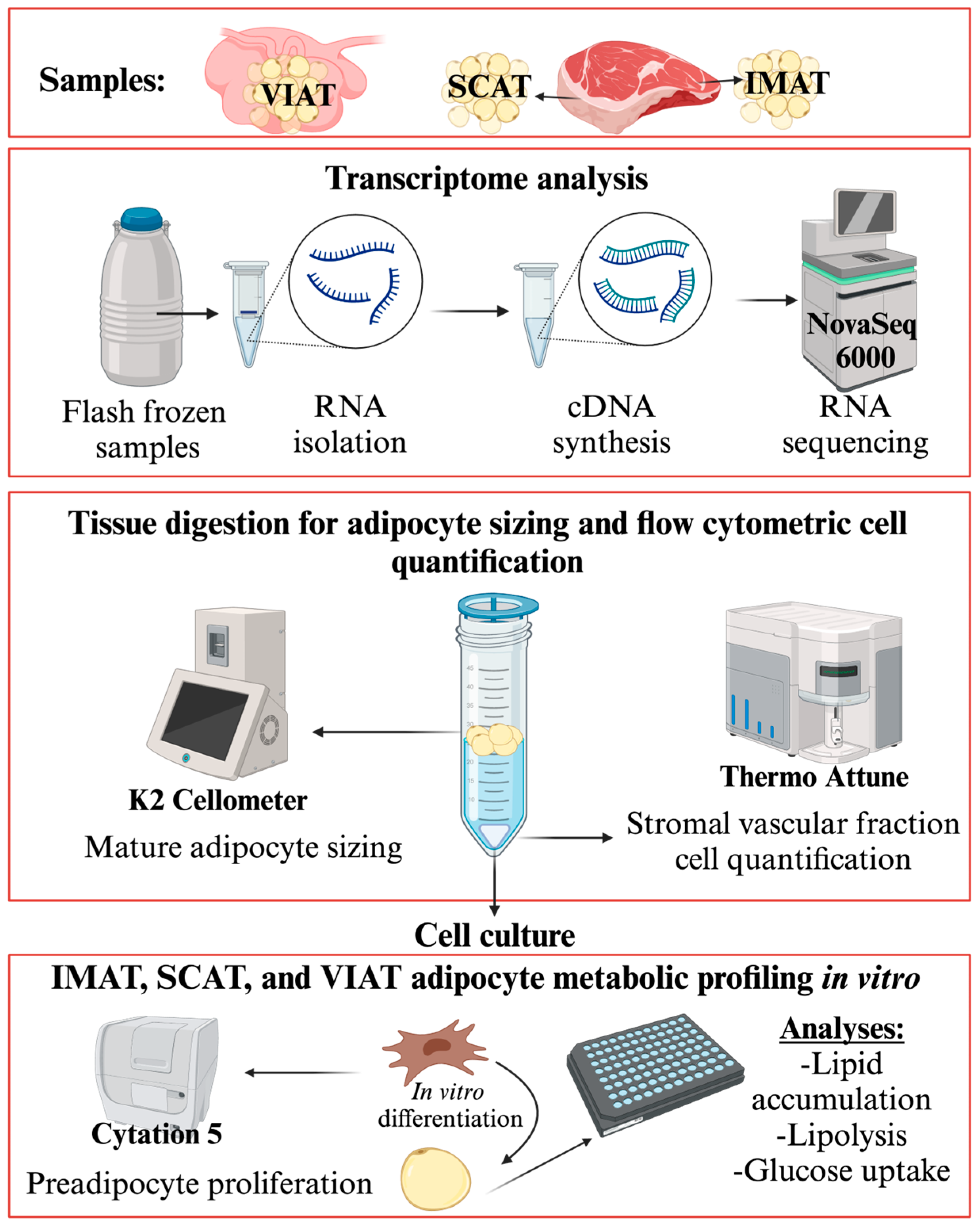
2. Materials and Methods
2.1. Adipose Tissue Collection and Study Design
2.2. Adipose Tissue Digestion
2.3. Adipocyte Culture
2.4. RNA Isolation
2.5. RNA Sequencing
2.6. Quantitative Real Time PCR
2.7. Adipogenic Function
2.7.1. Adipose Tissue Cellular Composition by Flow Cytometry
2.7.2. Preadipocyte Proliferation Assay
2.7.3. Mature Adipocyte Sizing
2.7.4. Adipocyte Lipid Accumulation
2.7.5. Adipocyte Insulin Response
2.8. Statistical Analyses
3. Results
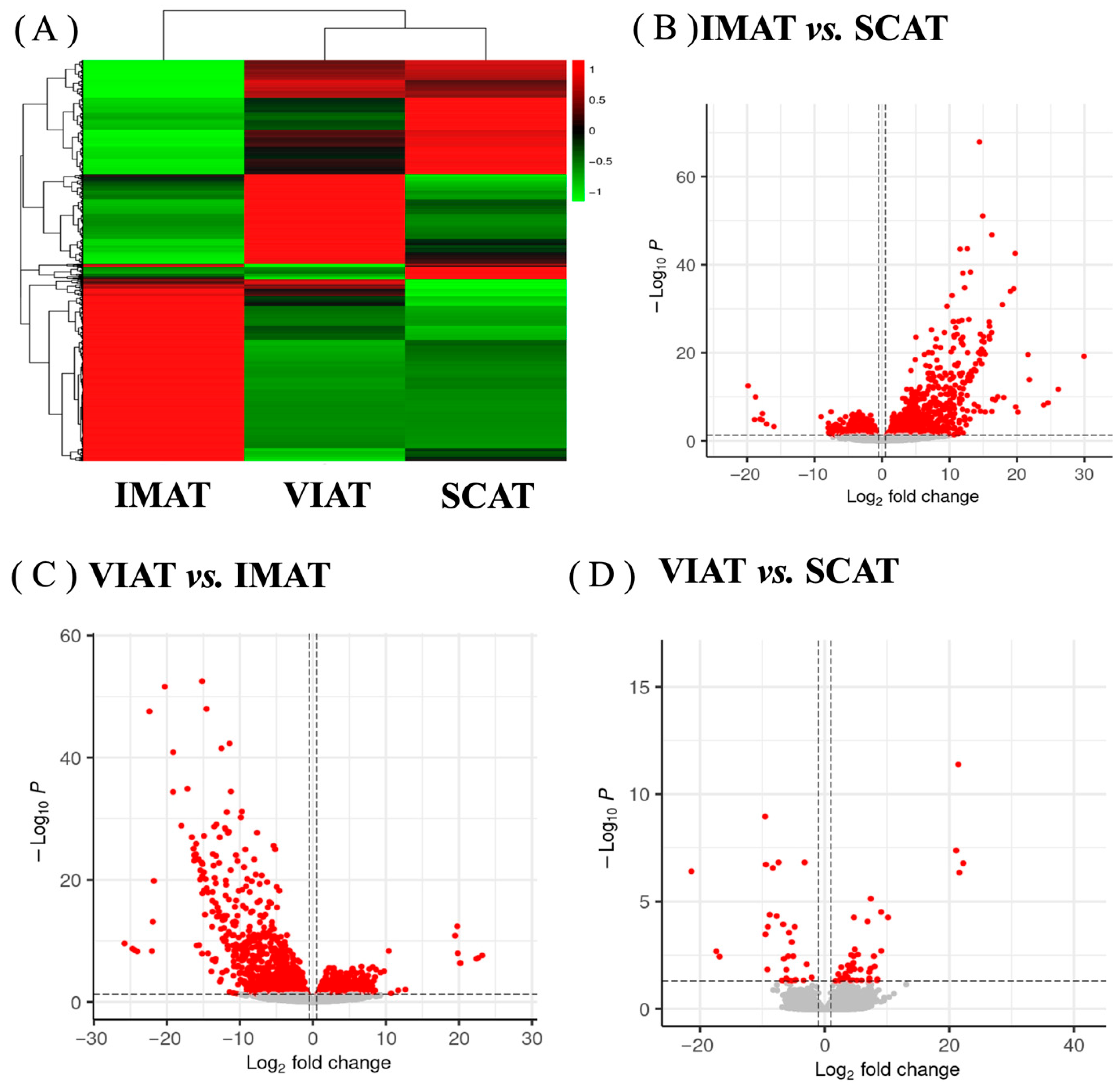
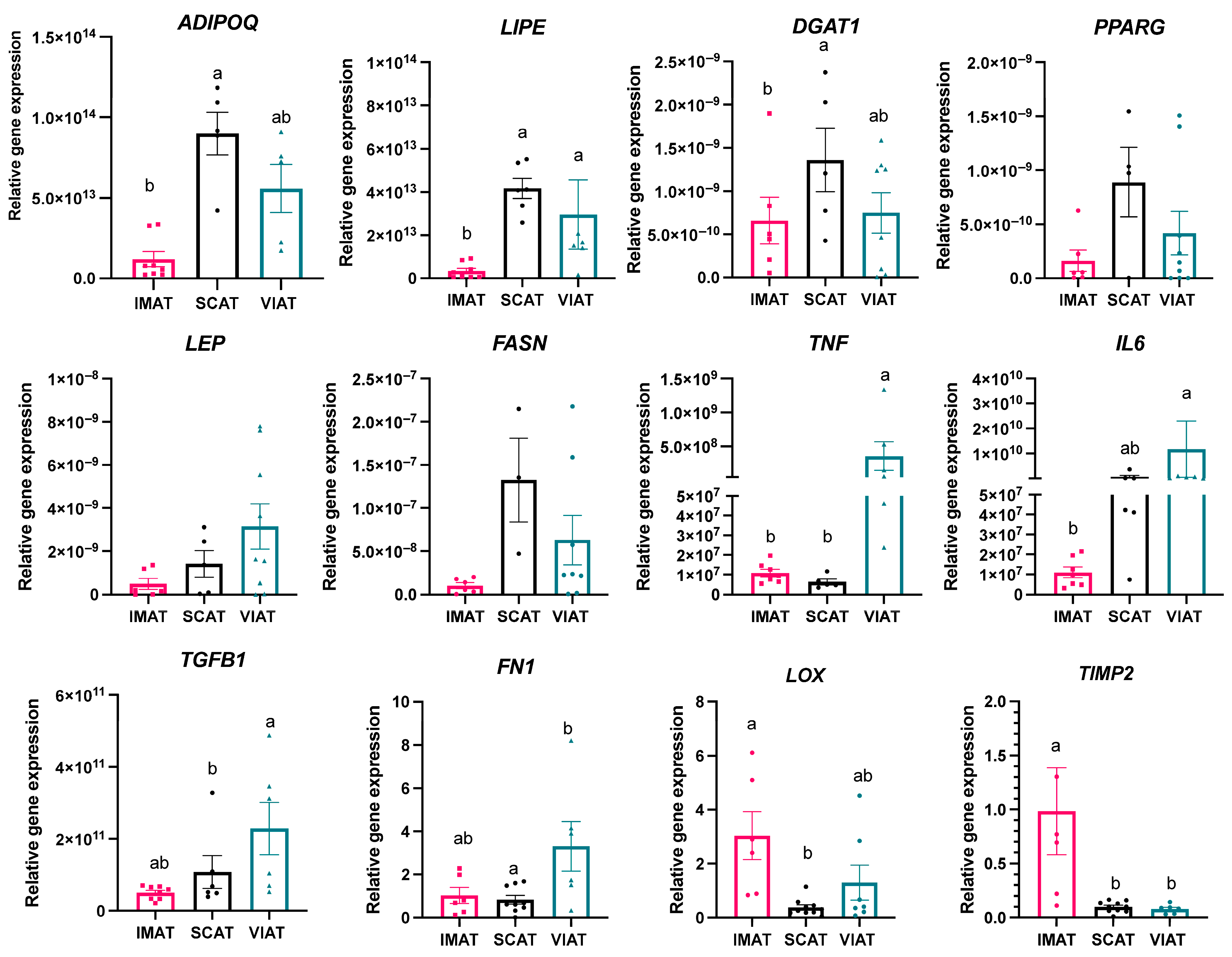
3.1. Transcriptome Analysis Reveals Differences in Key Genes Regulating Adipocyte Function, Extracellular Matrix Deposition, and Inflammation Among IMAT, SCAT, and VIAT
3.2. Enrichment Analysis Highlights Pathways of Lipid Metabolism and Immune Responses Differentiating IMAT, SCAT, and VIAT
3.3. Profiling of IMAT, SCAT, and VIAT Adipocytes Imply Depot-Specific Cell Function
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AT | Adipose Tissue |
| IMAT | Intramuscular adipose tissue; marbling |
| SCAT | Subcutaneous adipose tissue; backfat |
| VIAT | Visceral adipose tissue |
| FFA | Free Fatty Acids |
| LD | Longissimus Dorsi |
| KRB | Krebs Ringer Buffer |
| SVF | Stromal Vascular Fraction |
| PBS | Phosphate Buffer Solution |
| BSA | Bovine Serum Albumin |
| DMEM | Dulbecco’s Modified Eagle’s Medium |
| IBMX | Isobutyl-1-methylaxanthine |
| DEG | Differentially Expressed Genes |
| PCA | Principal Component Analysis |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| GO | Gene Ontology |
| PCR | Polymerase Chain Reaction |
| CT | Cycle Threshold |
| PFA | Paraformaldehyde |
| RFU | Relative fluorescent units |
| DAPI | diamidino-2-phenylindole |
References
- Nguyen, D.V.; Nguyen, O.C.; Malau-Aduli, A.E.O. Main regulatory factors of marbling level in beef cattle. Vet. Anim. Sci. 2021, 14, 100219. [Google Scholar] [CrossRef] [PubMed]
- Harris, C.L.; Wang, B.; Deavila, J.M.; Busboom, J.R.; Maquivar, M.; Parish, S.M.; McCann, B.; Nelson, M.L.; Du, M. Vitamin A administration at birth promotes calf growth and intramuscular fat development in Angus beef cattle. J. Anim. Sci. Biotechnol. 2018, 9, 55. [Google Scholar] [CrossRef]
- Hocquette, J.; Gondret, F.; Baéza, E.; Médale, F.; Jurie, C.; Pethick, D. Intramuscular fat content in meat-producing animals: Development, genetic and nutritional control, and identification of putative markers. Animal 2010, 4, 303–319. [Google Scholar] [CrossRef]
- USDA. United States Standards for Grades of Carcass Beef. 2018. Available online: https://www.ams.usda.gov/sites/default/files/media/CarcassBeefStandard.pdf (accessed on 6 July 2025).
- Wheeler, T.L.; Cundiff, L.V.; Koch, R.M. Effect of marbling degree on beef palatability in Bos taurus and Bos indicus cattle. J. Anim. Sci. 1994, 72, 3145–3151. [Google Scholar] [CrossRef]
- Cianzio, D.S.; Topel, D.G.; Whitehurst, G.B.; Beitz, D.C.; Self, H.L. Adipose Tissue Growth in Cattle Representing Two Frame Sizes: Distribution among Depots. J. Anim. Sci. 1982, 55, 305–312. [Google Scholar] [CrossRef]
- Irshad, A.; Kandeepan, G.; Kumar, S.; Ashish, K.; Vishnuraj, M.; Shukla, V. Factors influencing carcass composition of livestock: A review. J. Anim. Prod. Adv. 2012, 3, 177–186. [Google Scholar]
- Gotoh, T.; Albrecht, E.; Teuscher, F.; Kawabata, K.; Sakashita, K.; Iwamoto, H.; Wegner, J. Differences in muscle and fat accretion in Japanese Black and European cattle. Meat Sci. 2009, 82, 300–308. [Google Scholar] [CrossRef] [PubMed]
- USDA. USDA Beef Carcass Price Equivalent Index Value. USDA Market News, 9 June 2024. [Google Scholar]
- Gotoh, T.; Nishimura, T.; Kuchida, K.; Mannen, H. The Japanese Wagyu beef industry: Current situation and future prospects—A review. Asian-Australas. J. Anim. Sci. 2018, 31, 933. [Google Scholar] [CrossRef]
- Smith, S.B.; Gill, C.A.; Lunt, D.K.; Brooks, M.A. Regulation of Fat and Fatty Acid Composition in Beef Cattle. Asian-Australas. J. Anim. Sci. 2009, 22, 1225–1233. [Google Scholar] [CrossRef]
- Sheng, X.; Ni, H.; Liu, Y.; Li, J.; Zhang, L.; Guo, Y. RNA-seq analysis of bovine intramuscular, subcutaneous and perirenal adipose tissues. Mol. Biol. Rep. 2014, 41, 1631–1637. [Google Scholar] [CrossRef]
- Hudson, N.J.; Reverter, A.; Griffiths, W.J.; Yutuc, E.; Wang, Y.; Jeanes, A.; McWilliam, S.; Pethick, D.W.; Greenwood, P.L. Gene expression identifies metabolic and functional differences between intramuscular and subcutaneous adipocytes in cattle. BMC Genom. 2020, 21, 77. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Colón, G.C.; Grant, A.C.; Doumit, M.E.; Buskirk, D.D. Bovine intramuscular, subcutaneous, and perirenal stromal-vascular cells express similar glucocorticoid receptor isoforms, but exhibit different adipogenic capacity. J. Anim. Sci. 2009, 87, 1913–1920. [Google Scholar] [CrossRef]
- Chung, K.Y.; Kim, J.; Johnson, B.J. All-trans retinoic acid alters the expression of adipogenic genes during the differentiation of bovine intramuscular and subcutaneous adipocytes. J. Anim. Sci. Technol. 2021, 63, 1397. [Google Scholar] [CrossRef]
- May, S.; Savell, J.; Lunt, D.; Wilson, J.; Laurenz, J.; Smith, S. Evidence for preadipocyte proliferation during culture of subcutaneous and intramuscular adipose tissues from Angus and Wagyu crossbred steers. J. Anim. Sci. 1994, 72, 3110–3117. [Google Scholar] [CrossRef]
- Choi, S.H.; Silvey, D.T.; Johnson, B.J.; Doumit, M.E.; Chung, K.Y.; Sawyer, J.E.; Go, G.W.; Smith, S.B. Conjugated linoleic acid (t-10, c-12) reduces fatty acid synthesis de novo, but not expression of genes for lipid metabolism in bovine adipose tissue ex vivo. Lipids 2014, 49, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Gao, P.; Li, C.; Liu, Q.; Yao, Z.; Li, Y.; Zhang, X.; Sun, J.; Simintiras, C.; Welborn, M.; et al. A single-cell atlas of bovine skeletal muscle reveals mechanisms regulating intramuscular adipogenesis and fibrogenesis. J. Cachexia Sarcopenia Muscle 2023, 14, 2152–2167. [Google Scholar] [CrossRef] [PubMed]
- Ford, H.; Liu, Q.; Fu, X.; Strieder-Barboza, C. White Adipose Tissue Heterogeneity in the Single-Cell Era: From Mice and Humans to Cattle. Biology 2023, 12, 1289. [Google Scholar] [CrossRef]
- Diez, J.F.F.; Tegeler, A.P.; Flesher, C.G.; Michelotti, T.C.; Ford, H.; Hoque, M.N.; Bhattarai, B.; Benitez, O.J.; Christopher, G.F.; Strieder-Barboza, C. Extracellular matrix modulates depot-specific adipogenic capacity in adipose tissue of dairy cattle. J. Dairy Sci. 2024, 107, 9978–9996. [Google Scholar] [CrossRef]
- Sparks, B.B.; Ford, H.; Michelotti, T.C.; Strieder-Barboza, C. Adipose tissue oxylipin profile changes with subclinical ketosis and depot in postpartum dairy cows. J. Dairy Sci. 2025, 108, 781–791. [Google Scholar] [CrossRef]
- Hellemans, J.; Mortier, G.; De Paepe, A.; Speleman, F.; Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007, 8, R19. [Google Scholar] [CrossRef]
- Michelotti, T.C.; Kisby, B.R.; Flores, L.S.; Tegeler, A.P.; Fokar, M.; Crasto, C.; Menarim, B.C.; Loux, S.C.; Strieder-Barboza, C. Single-nuclei analysis reveals depot-specific transcriptional heterogeneity and depot-specific cell types in adipose tissue of dairy cows. Front. Cell Dev. Biol. 2022, 10, 1025240. [Google Scholar] [CrossRef] [PubMed]
- Vijay, J.; Gauthier, M.-F.; Biswell, R.L.; Louiselle, D.A.; Johnston, J.J.; Cheung, W.A.; Belden, B.; Pramatarova, A.; Biertho, L.; Gibson, M.J.N.M. Single-cell analysis of human adipose tissue identifies depot and disease specific cell types. Nat. Metab. 2020, 2, 97–109. [Google Scholar] [CrossRef]
- Ziemke, F.; Mantzoros, C.S. Adiponectin in insulin resistance: Lessons from translational research. Am. J. Clin. Nutr. 2010, 91, 258S–261S. [Google Scholar] [CrossRef] [PubMed]
- Dandekar, A.A.; Wallach, B.J.; Barthel, A.; Roth, R.A. Comparison of the signaling abilities of the cytoplasmic domains of the insulin receptor and the insulin receptor-related receptor in 3T3-L1 adipocytes. Endocrinology 1998, 139, 3578–3584. [Google Scholar] [CrossRef] [PubMed]
- Yoshino, J.; Patterson, B.W.; Klein, S. Adipose Tissue CTGF Expression is Associated with Adiposity and Insulin Resistance in Humans. Obesity 2019, 27, 957–962. [Google Scholar] [CrossRef] [PubMed]
- Pastel, E.; Price, E.; Sjöholm, K.; McCulloch, L.J.; Rittig, N.; Liversedge, N.; Knight, B.; Møller, N.; Svensson, P.-A.; Kos, K. Lysyl oxidase and adipose tissue dysfunction. Metabolism 2018, 78, 118–127. [Google Scholar] [CrossRef]
- Ou-yang, Y.; Dai, M.-m. Screening for genes, miRNAs and transcription factors of adipogenic differentiation and dedifferentiation of mesenchymal stem cells. J. Orthop. Surg. Res. 2023, 18, 46. [Google Scholar] [CrossRef]
- Huang, W.; Guo, Y.; Du, W.; Zhang, X.; Li, A.; Miao, X. Global transcriptome analysis identifies differentially expressed genes related to lipid metabolism in Wagyu and Holstein cattle. Sci. Rep. 2017, 7, 5278. [Google Scholar] [CrossRef]
- Chakrabarty, K.; Romans, J.R. Lipogenesis in the adipose cells of the bovine (Bos taurus) as related to their intramuscular fat content. Comp. Biochem. Physiol. 1972, 41, 603–615. [Google Scholar] [CrossRef]
- Wang, S.; Liu, J.; Zhao, W.; Wang, G.; Gao, S. Selection of candidate genes for differences in fat metabolism between cattle subcutaneous and perirenal adipose tissue based on RNA-seq. Anim. Biotechnol. 2023, 34, 633–644. [Google Scholar] [CrossRef]
- Chriett, S.; Lindqvist, A.; Shcherbina, L.; Edlund, A.; Abels, M.; Asplund, O.; López, J.M.; Ottosson-Laakso, E.; Hatem, G.; Prasad, R. SCRT1 is a novel beta cell transcription factor with insulin regulatory properties. Mol. Cell. Endocrinol. 2021, 521, 111107. [Google Scholar] [CrossRef] [PubMed]
- Duan, X.; Norris, D.M.; Humphrey, S.J.; Yang, P.; Cooke, K.C.; Bultitude, W.P.; Parker, B.L.; Conway, O.J.; Burchfield, J.G.; Krycer, J.R.; et al. Trafficking regulator of GLUT4-1 (TRARG1) is a GSK3 substrate. Biochem. J. 2022, 479, 1237–1256. [Google Scholar] [CrossRef] [PubMed]
- Kersten, S. Mechanisms of nutritional and hormonal regulation of lipogenesis. EMBO Rep. 2001, 2, 282–286. [Google Scholar] [CrossRef]
- Tokach, R.J.; Chung, K.Y.; Johnson, B.J. Factors Affecting Intramuscular Adipose Tissue Development in Beef Cattle. 2010. Available online: https://cabcattle.com/wp-content/uploads/Texas-Tech-white-paper.pdf (accessed on 6 July 2025).
- McGrattan, P.D.; Wylie, A.R.G.; Nelson, J. Tissue-specific differences in insulin binding affinity and insulin receptor concentrations in skeletal muscles, adipose tissue depots and liver of cattle and sheep. Anim. Sci. 2000, 71, 501–508. [Google Scholar] [CrossRef]
- Kenéz, Á.; Bäßler, S.C.; Jorge-Smeding, E.; Huber, K. Ceramide metabolism associated with chronic dietary nutrient surplus and diminished insulin sensitivity in the liver, muscle, and adipose tissue of cattle. Front. Physiol. 2022, 13, 958837. [Google Scholar] [CrossRef]
- Purslow, P.P. New developments on the role of intramuscular connective tissue in meat toughness. Annu. Rev. Food Sci. Technol. 2014, 5, 133–153. [Google Scholar] [CrossRef]
- Cui, H.-x.; Luo, N.; Guo, L.-p.; Liu, L.; Xing, S.-y.; Zhao, G.-p.; Wen, J. TIMP2 promotes intramuscular fat deposition by regulating the extracellular matrix in chicken. J. Integr. Agric. 2023, 22, 853–863. [Google Scholar] [CrossRef]
- Unamuno, X.; Gómez-Ambrosi, J.; Becerril, S.; Álvarez-Cienfuegos, F.J.; Ramírez, B.; Rodríguez, A.; Ezquerro, S.; Valentí, V.; Moncada, R.; Mentxaka, A.; et al. Changes in mechanical properties of adipose tissue after bariatric surgery driven by extracellular matrix remodelling and neovascularization are associated with metabolic improvements. Acta Biomater. 2022, 141, 264–279. [Google Scholar] [CrossRef] [PubMed]
- Iwayama, T.; Steele, C.; Yao, L.; Dozmorov, M.G.; Karamichos, D.; Wren, J.D.; Olson, L.E. PDGFRα signaling drives adipose tissue fibrosis by targeting progenitor cell plasticity. Genes Dev. 2015, 29, 1106–1119. [Google Scholar] [CrossRef]
- Guvendiren, M.; Burdick, J.A. Stiffening hydrogels to probe short-and long-term cellular responses to dynamic mechanics. Nat. Commun. 2012, 3, 792. [Google Scholar] [CrossRef]
- Young, D.A.; Choi, Y.S.; Engler, A.J.; Christman, K.L. Stimulation of adipogenesis of adult adipose-derived stem cells using substrates that mimic the stiffness of adipose tissue. Biomaterials 2013, 34, 8581–8588. [Google Scholar] [CrossRef] [PubMed]
- Strieder-Barboza, C.; Baker, N.A.; Flesher, C.G.; Karmakar, M.; Patel, A.; Lumeng, C.N.; O’Rourke, R.W. Depot-specific adipocyte-extracellular matrix metabolic crosstalk in murine obesity. Adipocyte 2020, 9, 189–196. [Google Scholar] [CrossRef]
- Strieder-Barboza, C.; Baker, N.A.; Flesher, C.G.; Karmakar, M.; Neeley, C.K.; Polsinelli, D.; Dimick, J.B.; Finks, J.F.; Ghaferi, A.A.; Varban, O.A.; et al. Advanced glycation end-products regulate extracellular matrix-adipocyte metabolic crosstalk in diabetes. Sci. Rep. 2019, 9, 19748. [Google Scholar] [CrossRef]
- Schrauwen, P.; Hesselink, M. UCP2 and UCP3 in muscle controlling body metabolism. J. Exp. Biol. 2002, 205, 2275–2285. [Google Scholar] [CrossRef] [PubMed]
- Roberts, S.; Lancaster, P.; DeSilva, U.; Horn, G.; Krehbiel, C. Coordinated gene expression between skeletal muscle and intramuscular adipose tissue in growing beef cattle. J. Anim. Sci. 2015, 93, 4302–4311. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Liu, T.; Peng, P.; Fu, Y.; Shi, S.; Liang, S.; Chen, X.; Wang, K.; Zhou, R. Integrated Transcriptomic Analysis of Liver and Muscle Tissues Reveals Candidate Genes and Pathways Regulating Intramuscular Fat Deposition in Beef Cattle. Animals 2025, 15, 1306. [Google Scholar] [CrossRef] [PubMed]
- Moisá, S.J.; Ji, P.; Drackley, J.K.; Rodriguez-Zas, S.L.; Loor, J.J. Transcriptional changes in mesenteric and subcutaneous adipose tissue from Holstein cows in response to plane of dietary energy. J. Anim. Sci. Biotechnol. 2017, 8, 85. [Google Scholar] [CrossRef]
- Contreras, G.A.; Kabara, E.; Brester, J.; Neuder, L.; Kiupel, M. Macrophage infiltration in the omental and subcutaneous adipose tissues of dairy cows with displaced abomasum. J. Dairy Sci. 2015, 98, 6176–6187. [Google Scholar] [CrossRef]
- Mathis, D. Immunological goings-on in visceral adipose tissue. Cell Metab. 2013, 17, 851–859. [Google Scholar] [CrossRef]
- Deiuliis, J.A.; Oghumu, S.; Duggineni, D.; Zhong, J.; Rutsky, J.; Banerjee, A.; Needleman, B.; Mikami, D.; Narula, V.; Hazey, J. CXCR3 modulates obesity-induced visceral adipose inflammation and systemic insulin resistance. Obesity 2014, 22, 1264–1274. [Google Scholar] [CrossRef]
- Wang, Y.H.; Bower, N.; Reverter, A.; Tan, S.; De Jager, N.; Wang, R.; McWilliam, S.; Cafe, L.; Greenwood, P.; Lehnert, S. Gene expression patterns during intramuscular fat development in cattle. J. Anim. Sci. 2009, 87, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Yu, S.; Guo, J.; Wang, J.; Mei, C.; Abbas Raza, S.H.; Cheng, G.; Zan, L. Comprehensive Analysis of Transcriptome and Metabolome Reveals Regulatory Mechanism of Intramuscular Fat Content in Beef Cattle. J. Agric. Food Chem. 2024, 72, 2911–2924. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Zhu, Z.; Sun, H.-Z.; Zhao, K.; Dugan, M.E.R.; Bruce, H.; Fitzsimmons, C.; Li, C.; Guan, L.L. Breed dependent regulatory mechanisms of beneficial and non-beneficial fatty acid profiles in subcutaneous adipose tissue in cattle with divergent feed efficiency. Sci. Rep. 2022, 12, 4612. [Google Scholar] [CrossRef] [PubMed]
- Schoettl, T.; Fischer, I.P.; Ussar, S. Heterogeneity of adipose tissue in development and metabolic function. J. Exp. Biol. 2018, 221, jeb162958. [Google Scholar] [CrossRef]
- Goodpaster, B.H.; He, J.; Watkins, S.; Kelley, D.E. Skeletal muscle lipid content and insulin resistance: Evidence for a paradox in endurance-trained athletes. J. Clin. Endocrinol. Metab. 2000, 85, 2444–2450. [Google Scholar] [CrossRef]
- Addison, O.; Marcus, R.L.; Lastayo, P.C.; Ryan, A.S. Intermuscular fat: A review of the consequences and causes. Int. J. Endocrinol. 2014, 2014, 309570. [Google Scholar] [CrossRef]


| Gene Symbols | TaqMan Gene Expression Assay | NCBI RefSeq | Amplicon Length (Base Pairs) | Exon Boundary ** |
|---|---|---|---|---|
| ADIPOQ | Bt0329341_s1 | NM_174742.2 | 121 | 3 |
| LIPE | Bt0325697_m1 | NM_001080220.1 | 62 | 8–9 |
| DGAT1 | Bt03251718_g1 | NM_174693.2 | 63 | 16–17 |
| PPARG | Bt03217547_m1 | NM_181024.2 | 85 | 6–7 |
| LEP | Bt03211909_m1 | NM_173928.2 | 88 | 2–3 |
| FASN | Bt03210481_m1 | NM_001012669.1 | 57 | 37–38 |
| IL6 | Bt03211905_m1 | NM_173923.2 | 115 | 4–5 |
| TNF | Bt03259156_m1 | NM_173966.3 | 69 | 3–4 |
| TGFB1 | Bt04259484_m1 | NM_001166068.1 | 60 | 1–2 |
| FN1 | Bt00415008_m1 | NM_001163778.1 | 56 | 43–44 |
| LOX | Bt03259128_m1 | NM_173932.4 | 72 | 2–3 |
| TIMP2 | Bt03231007_m1 | NM_174472.4 | 88 | 4–5 |
| EIF3K * | Bt03226565_m1 | NM_001034489.2 | 57 | 1–2 |
| RPS9 * | Bt03272016_m1 | NM_001101152.2 | 62 | 3–4 |
| Upregulated Genes IMAT vs. SCAT | |||
|---|---|---|---|
| Gene Symbol | Gene Description | Log2Fold Change | p-Adjusted |
| TNNC2 | Troponin C2 | 14.42 | 1.35 × 10−68 |
| CKM | Creatine Kinase, M-Type | 14.91 | 8.44 × 10−52 |
| ACTA1 | Actin alpha 1 | 16.26 | 1.60 × 10−47 |
| ATP2A1 | ATPase sarcoplasmic endoplasmic reticulum Ca2+ transporting 1 | 12.65 | 2.43 × 10−44 |
| ACTN3 | Actinin alpha 3 | 11.58 | 2.90 × 10−44 |
| MYLPF | Myosin Light Chain | 19.75 | 2.79 × 10−43 |
| TTN | Titin | 13.09 | 4.60 × 10−39 |
| PYGM | Glycogen Phosphorylase | 11.99 | 8.06 × 10−39 |
| MB | Myoglobin | 12.25 | 1.80 × 10−35 |
| MYL1 | Myosin Light Chain 1 | 19.51 | 2.75 × 10−35 |
| Downregulated IMAT vs. SCAT | |||
| Gene Symbol | Gene Description | Log2Fold Change | p-Adjusted |
| SCRT1 | Scratch Family Transcriptional Repressor 1 | −19.84 | 3.18 × 10−13 |
| KRT38 | Keratin 38 | −18.76 | 1.03 × 10−10 |
| MLC1 | Modulator Of VRAC Current 1 | −7.56 | 2.49 × 10−7 |
| ADTRP | Androgen-Dependent TFPI Regulating Protein | −3.39 | 2.83 × 10−7 |
| LASP1 | LIM And SH3 Protein 1 | −1.71 | 1.41 × 10−6 |
| LOC100849237 | SNU13 Homolog, Small Nuclear Ribonucleoprotein pseudogene | −4.34 | 7.84 × 10−7 |
| SLC25A35 | Solute Carrier Family 25 Member 35 | −3.72 | 9.63 × 10−7 |
| BAIAP2L1 | BAR/IMD Domain Containing Adaptor Protein 2 Like 1 | −3.24 | 1.03 × 10−6 |
| XPNPEP2 | X-Prolyl Aminopeptidase 2 | −4.43 | 1.34 × 10−6 |
| SYN2 | Synapsin II | −2.92 | 1.36 × 10−6 |
| Upregulated VIAT vs. IMAT | |||
|---|---|---|---|
| Gene Symbol | Gene Description | Log2Fold Change | p-Adjusted |
| SCRT1 | Scratch Family Transcriptional Repressor 1 | 21.13 | 1.65 × 10−14 |
| KRT38 | Keratin 38 | 19.481 | 1.34 × 10−11 |
| LOC101906766 | Ig lambda chain V-II region BUR-like | 24.79 | 1.54 × 10−9 |
| NKX3−2 | NK3 Homeobox 2 | 10.38 | 3.10 × 10−9 |
| CRYBG1 | Crystallin Beta-Gamma Domain Containing 1 | 2.19 | 2.66 × 10−6 |
| PRKAR2B | Protein Kinase CAMP-Dependent Type II Regulatory Subunit Beta | 2.38 | 2.79 × 10−6 |
| ABO | Alpha 1-3-N-Acetylgalactosaminyltransferase And Alpha 1-3-Galactosyltransferase | 6.64 | 2.90 × 10−6 |
| SYN2 | Synapsin II | 2.86 | 3.27 × 10−6 |
| TDRKH | Tudor And KH Domain Containing | 5.42 | 3.29 × 10−6 |
| LOC615733 | cDNA ORF clone | 5.54 | 5.78 × 10−6 |
| Downregulated VIAT vs. IMAT | |||
| Gene Symbol | Gene Description | Log2Fold Change | p-Adjusted |
| CKM | Creatine Kinase, M-Type | −15.19 | 1.58 × 10−56 |
| TNNC2 | Troponin C2 | −20.29 | 2.31 × 10−53 |
| MB | Myoglobin | −14.59 | 9.79 × 10−52 |
| ACTA1 | Actin alpha 1 | −22.39 | 2.04 × 10−50 |
| ACTN3 | Actinin alpha 3 | −11.41 | 1.03 × 10−44 |
| PYGM | Glycogen Phosphorylase, muscle-associated | −12.52 | 3.24 × 10−44 |
| MYLPF | Myosin Light Chain, Phosphorylatable | −19.14 | 1.21 × 10−43 |
| ATP2A1 | ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 | −11.22 | 1.00 × 10−36 |
| TTN | Titin | −11.79 | 2.38 × 10−33 |
| CASQ1 | Calsequestrin 2 | −9.73 | 4.62 × 10−33 |
| Upregulated Genes VIAT vs. SCAT | |||
|---|---|---|---|
| Gene Symbol | Gene Description | Log2Fold Change | p-Adjusted |
| CDH16 | Cadherin 16 | 20.12 | 3.71 × 10−10 |
| UCP3 | Uncoupling Protein 3 | 20.01 | 2.21 × 10−7 |
| FRAS1 | Fraser Extracellular Matrix Complex Subunit 1 | 21.02 | 1.11 × 10−6 |
| DSG2 | Desmoglein 2 | 20.48 | 2.86 × 10−6 |
| DSP | Desmoplakin | 7.39 | 2.86 × 10−6 |
| LOC505033 | Cystatin 9 Like | 10.16 | 3.88 × 10−5 |
| SLIT2 | Slit Guidance Ligand 2 | 4.68 | 3.88 × 10−5 |
| MYBPC1 | Myosin Binding Protein C1 | 17.25 | 3.98 × 10−5 |
| MYH8 | Myosin Heavy Chain 8 | 20.02 | 5.63 × 10−5 |
| NKX3-2 | NK3 Homeobox 2 | 6.87 | 5.63 × 10−5 |
| Downregulated VIAT vs. SCAT | |||
| Gene Symbol | Gene Description | Log2Fold Change | p-Adjusted |
| HOXC10 | Homeobox C10 | −9.54 | 1.06 × 10−9 |
| EN1 | Engrailed Homeobox 1 | −7.39 | 4.68 × 10−8 |
| ZIC1 | Zic Family Member 1 | −9.43 | 2.05 × 10−7 |
| PAX3 | Paired Box 3 | −21.44 | 2.21 × 10−7 |
| HOXD8 | Homeobox D8 | −3.21 | 2.21 × 10−7 |
| INSC | INSC Spindle Orientation Adaptor | −8.31 | 7.84 × 10−7 |
| ZIC4 | Zic Family Member 4 | −8.81 | 3.05 × 10−5 |
| ANGPTL5 | Angiopoietin Like 5 | −9.49 | 1.99 × 10−4 |
| HOXC9 | Homeobox C9 | −4.8 | 7.60 × 10−5 |
| MAB21L1 | Mab-21 Like 1 | −5.73 | 3.53 × 10−4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tegeler, A.P.; Ford, H.R.; Fiallo-Diez, J.F.; Michelotti, T.C.; Johnson, B.J.; Benitez, O.J.; Woerner, D.R.; Strieder-Barboza, C. Transcriptome and Cellular Evidence of Depot-Specific Function in Beef Cattle Intramuscular, Subcutaneous, and Visceral Adipose Tissues. Biology 2025, 14, 848. https://doi.org/10.3390/biology14070848
Tegeler AP, Ford HR, Fiallo-Diez JF, Michelotti TC, Johnson BJ, Benitez OJ, Woerner DR, Strieder-Barboza C. Transcriptome and Cellular Evidence of Depot-Specific Function in Beef Cattle Intramuscular, Subcutaneous, and Visceral Adipose Tissues. Biology. 2025; 14(7):848. https://doi.org/10.3390/biology14070848
Chicago/Turabian StyleTegeler, Alexandra P., Hunter R. Ford, Jean Franco Fiallo-Diez, Tainara C. Michelotti, Bradley J. Johnson, Oscar J. Benitez, Dale R. Woerner, and Clarissa Strieder-Barboza. 2025. "Transcriptome and Cellular Evidence of Depot-Specific Function in Beef Cattle Intramuscular, Subcutaneous, and Visceral Adipose Tissues" Biology 14, no. 7: 848. https://doi.org/10.3390/biology14070848
APA StyleTegeler, A. P., Ford, H. R., Fiallo-Diez, J. F., Michelotti, T. C., Johnson, B. J., Benitez, O. J., Woerner, D. R., & Strieder-Barboza, C. (2025). Transcriptome and Cellular Evidence of Depot-Specific Function in Beef Cattle Intramuscular, Subcutaneous, and Visceral Adipose Tissues. Biology, 14(7), 848. https://doi.org/10.3390/biology14070848






