Comparative Analysis of the Mitochondrial Genomes of Five Species of Anabropsis (Orthoptera: Anostostomatidae) and the Phylogenetic Implications of Anostostomatidae
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Specimen Extraction and Sequencing
2.2. Mitochondrial Genome Sequence Assembly and Analysis
2.3. Construction of Phylogenetic Trees
3. Results and Discussion
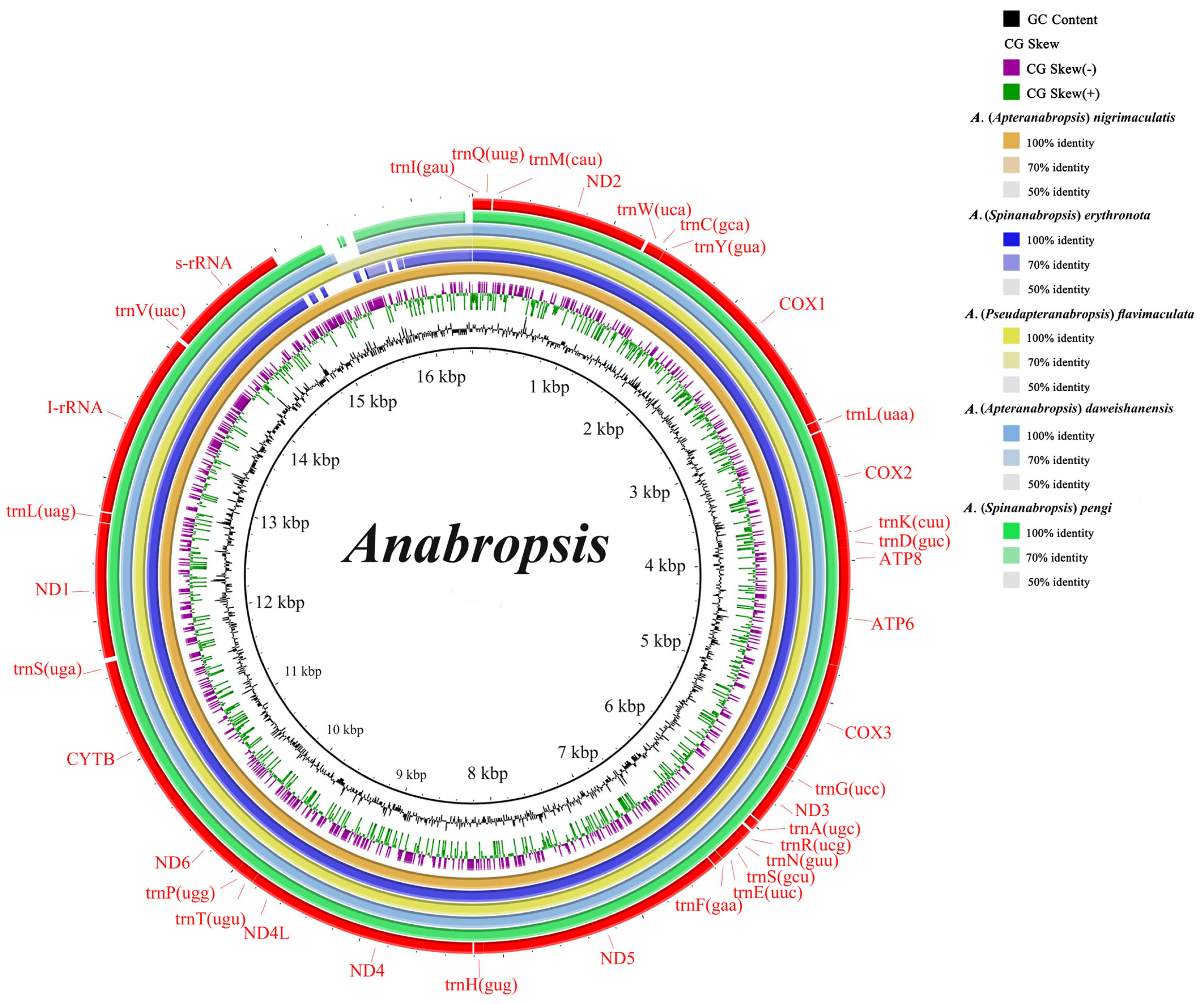
3.1. Mitochondrial Genome
3.2. Mitochondrial Gene Interval and Overlapping Regions
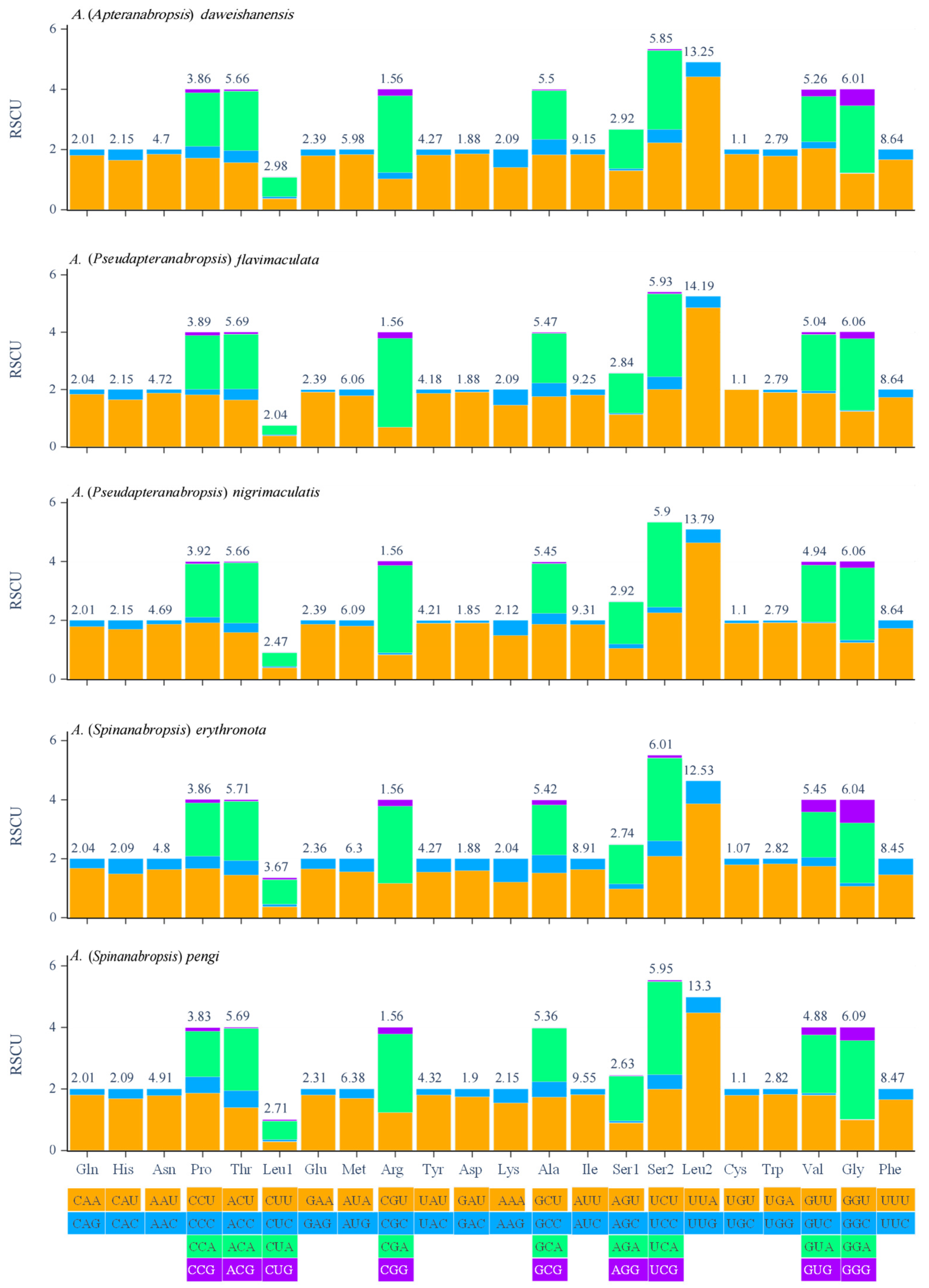
3.3. Protein Coding Genes and Codon Usages
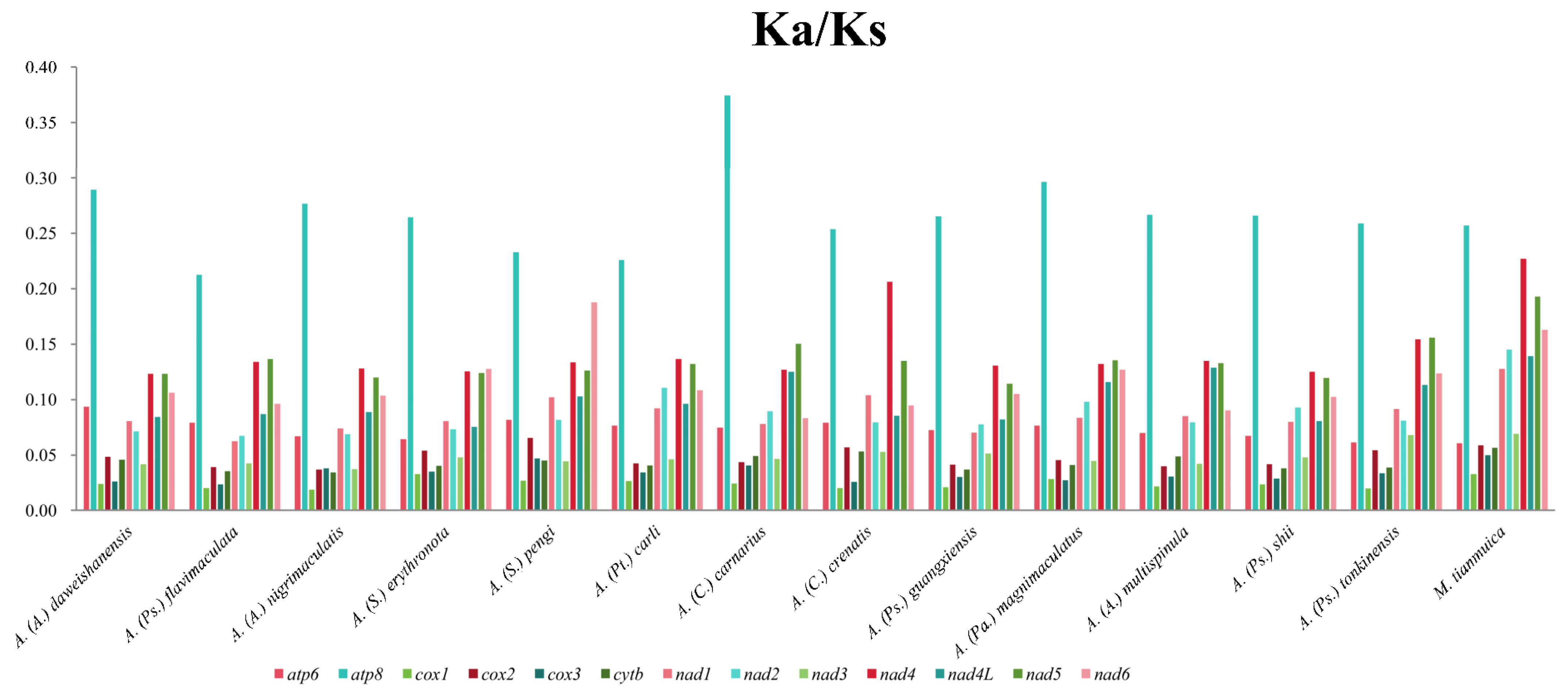
3.4. The Rate of Evolution of 13 PCGs
3.5. Substitution Saturation Tests and Nucleotide Heterogeneity
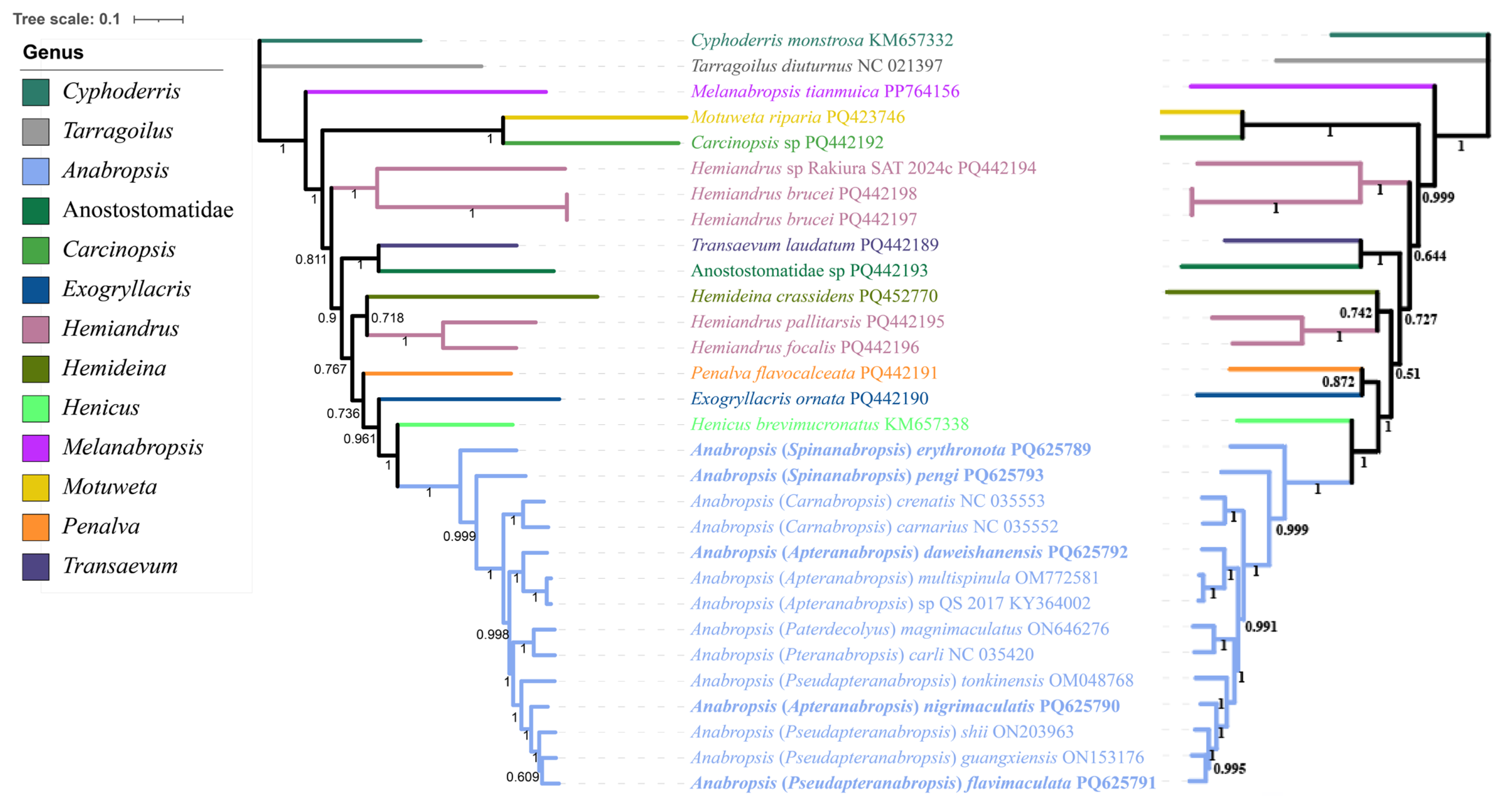
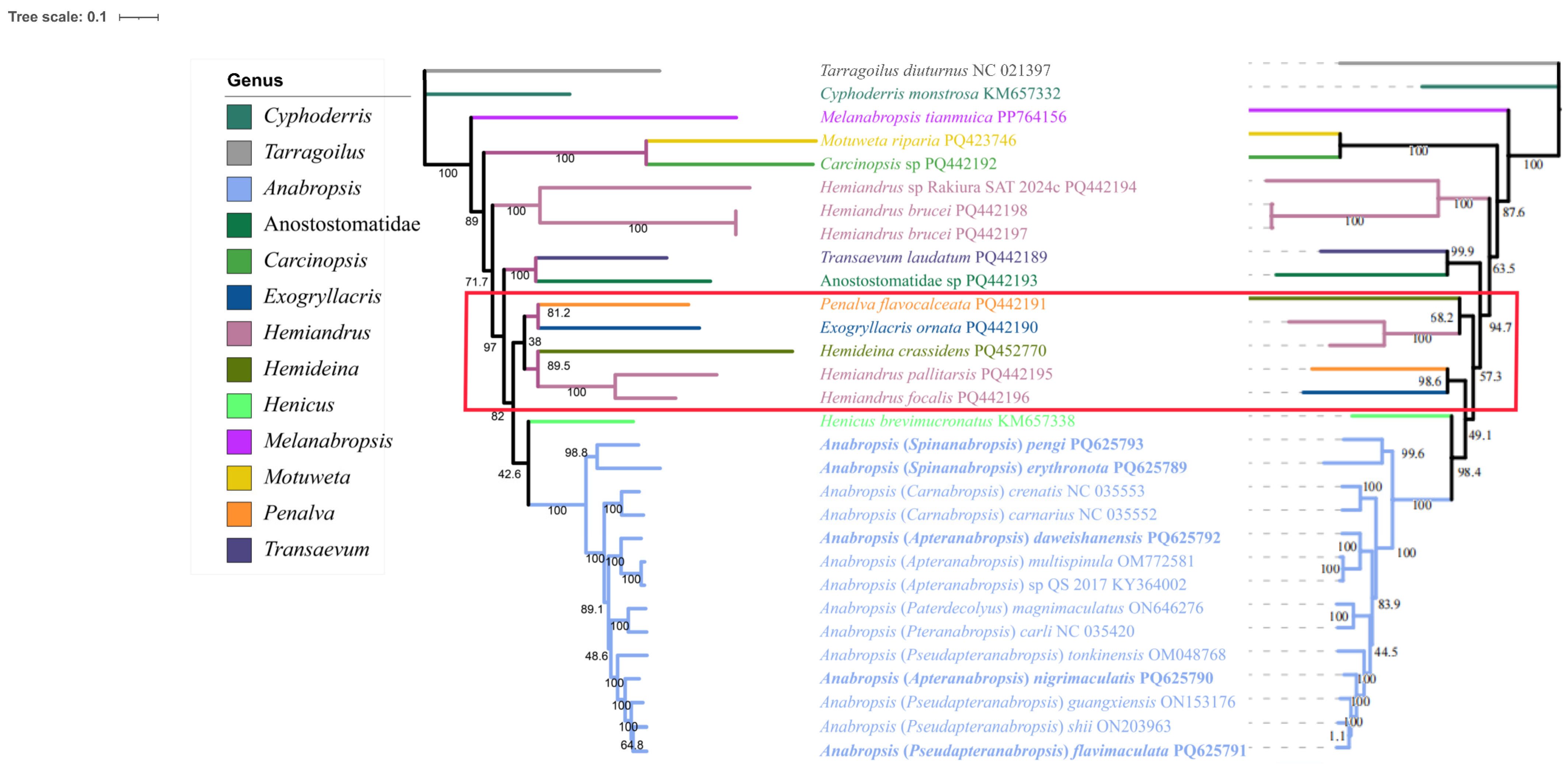
3.6. Phylogenetic Analysis of Anostostomatidae
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cigliano, M.M.; Braun, H.; Eades, D.C.; Otte, D. Orthoptera Species File. Version 5.0/5.0. [12–10]. 2024. Available online: http://Orthoptera.SpeciesFile.org (accessed on 13 May 2025).
- Lu, X.; Liu, J.; Bian, X. Study on the Chinese Subfamily Anostostomatinae (Orthoptera: Anostostomatidae) IV: One new recorded species Anabropsis (Apteranabropsis) tonkinensis Rehn, 1906. Zootaxa 2022, 5100, 349–360. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Zhang, Q.; Bian, X. Study on the Chinese Subfamily Anabropsinae (Orthoptera: Anostostomatidae) VI: One new species of Anabropsis (Pteranabropsis) from Yunnan Province. Zootaxa 2022, 5178, 178–192. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Lin, L.I.; Liu, J.; Liang, L.L.; Bian, X. Study of the Subfamily Anabropsinae (Orthoptera: Anostostomatidae) in China V: Two new species of Anabropsis (Apteranabropsis) from Guangxi and phylogenetic analysis of the genus Anabropsis. Zootaxa 2022, 5141, 227–248. [Google Scholar] [CrossRef] [PubMed]
- Bian, X. Study of the Chinese Anostostomatidae (Orthoptera: Ensifera) VIII: One new species Anabropsis (Apteranabropsis) from Chongqing. Zootaxa 2024, 5443, 446–450. [Google Scholar] [CrossRef]
- Pang, S.Y.; Lu, X.Y.; Bian, X. Study of the Chinese Members of the Subfamily Anabropsinae (Orthoptera: Anostostomatidae) VII: Description of two new subgenera and five new species of Anabropsis from China. Zootaxa 2023, 5318, 253–267. [Google Scholar] [CrossRef]
- Pang, S.; Yang, R.; Bian, X. Study on the Chinese Anostostomatidae (Orthoptera: Ensifera) X: New descriptions of Anabropsis (Pseudapteranabropsis) Pang, Lu & Bian, 2023 from Guangxi. Zootaxa 2024, 5506, 272–280. [Google Scholar] [CrossRef]
- Xu, H.; Shi, F. Two new species of the genus Anabropsis Rehn, 1901 (Orthoptera: Anostostomatidae) from Yunnan, China. Zootaxa 2024, 5523, 387–395. [Google Scholar] [CrossRef]
- Bian, X. Study of the Chinese Anostostomatidae (Orthoptera: Ensifera) IX: New additions of Anabropsis (Carnabropsis) from Yunnan Province. Zootaxa 2024, 5463, 291–297. [Google Scholar] [CrossRef]
- Rentz, D.C.F.; Weissman, D.B. The origins and affinities of the Orthoptera of the Channel Islands and adjacent mainland California. Part I. The genus Cnemotettix. Proc. Acad. Nat. Sci. Phila. 1973, 125, 89–120. [Google Scholar]
- Gorochov, A.V. System and phylogeny of the recent Orthoptera of the superfamilies Hagloidea and Stenopelmatoidea with a description of new taxa. Communication 1. Zool. Zhurnal 1988, 67, 353–366. [Google Scholar]
- Gorochov, A.V. System and phylogeny of the recent Orthoptera of the superfamilies Hagloidea and Stenopelmatoidea with a description of new taxa. Communication 2. Zool. Zhurnal 1988, 67, 518–529. [Google Scholar]
- Griffini, A. Sopra alcuni Grillacridi e Stenopelmatidi della collezione Pantel. Atti Della Soc. Ital. Sci. Nat. Mus. Civ. Stor. Nat. Milano 1913, 52, 61–104. [Google Scholar]
- Johns, P.M. The Gondwanaland Weta: Family Anostostomatidae (formerly in Stenopelmatidae, Henicidae or Mimnermidae): Nomenclatural problems, world checklist, new genera and species. J. Orthoptera Res. 1997, 6, 125–138. [Google Scholar] [CrossRef]
- Gorochov, A.V. New taxa of Anostostomatidae and Prophalangpsidae (Orthoptera). Zoosyst. Ross. 2001, 9, 299–315. [Google Scholar]
- Shi, F.M.; Bian, X. A new addition to the subfamily Anabropsinae (Orthoptera: Anostostomatidae) from China. Zootaxa 2016, 4079, 597–600. [Google Scholar] [CrossRef]
- Ingrisch, S. Review of the genus Pteranabropsis (Anostostomatidae: Anabropsinae) with description of six new species. J. Orthoptera Res. 2019, 28, 107–124. [Google Scholar] [CrossRef]
- Gorochov, A.V. The families Stenopelmatidae and Anostostomatidae (Orthoptera). 1. Higher classification, new and little known taxa. Entomol. Rev. 2020, 100, 1106–1151. [Google Scholar] [CrossRef]
- Gorochov, A.V.; Cadena-Castañeda, O.J. New and little known Stenopelmatoidea (Orthoptera: Ensifera) from America. Zoosystematica Ross. 2016, 25, 98–143. [Google Scholar] [CrossRef]
- Zhou, Z.; Zhao, L.; Liu, N.; Guo, H.; Guan, B.; Di, J.; Shi, F. Towards a higher-level Ensifera phylogeny inferred from mitogenome sequences. Mol. Phylogenet. Evol. 2017, 108, 22–33. [Google Scholar] [CrossRef]
- López-López, A.; Vogler, A.P. The mitogenome phylogeny of Adephaga (Coleoptera). Mol. Phylogenet. Evol. 2017, 114, 166–174. [Google Scholar] [CrossRef]
- Wolstenholme, D.R.; Koike, K.; Renger, H.C. Oncology 1970. A. Cellular and Molecular Mechanisms of Carcinogenesis; B. Regulations of Gene Expression (Proc. Xth International Cancer, Congress); Clark, D.R.L., Cumley, R.W., McCoy, J.E., Copeland, M.M., Eds.; Yew Book Medical Publishers: Chicago, IL, USA, 1971; pp. 627–648. [Google Scholar]
- Wolstenholme, D.R. Animal mitochondrial DNA: Structure and evolution. Int. Rev. Cytol. 1992, 141, 173–216. [Google Scholar] [CrossRef] [PubMed]
- Taanman, J.W. The mitochondrial genome: Structure, transcription, translation and replication. Biochim. Biophys. Acta 1999, 1410, 103–123. [Google Scholar] [CrossRef] [PubMed]
- Avise, J.C. Phylogeography: The History and Formation of Species; Harvard University Press: Cambridge, MA, USA, 2000. [Google Scholar]
- Zardoya, R.; Meyer, A. Phylogenetic performance of mitochondrial proteincoding genes in resolving relationships among vertebrates. Mol. Phylogenet. Evol. 1996, 13, 933–942. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Amédégnato, C.; Cigliano, M.M.; Desutter-Grandcolas, L.; Heads, S.W.; Huang, Y.; Otte, D.; Whiting, M.F. 300 million years of diversification: Elucidating the patterns of orthopteran evolution based on comprehensive taxon and gene sampling. Cladistics 2015, 31, 621–651. [Google Scholar] [CrossRef]
- Trewick, S.A.; Taylor-Smith, B.L.; Morgan-Richards, M. Wētā Aotearoa-Polyphyly of the New Zealand Anostostomatidae (Insecta: Orthoptera). Insects 2024, 15, 787. [Google Scholar] [CrossRef]
- Matvienko, M. Genomics Workbench and other products. Qiagen Bioinformatics Workshop at PAG 2015. Plant Anim. Genome 2015, 1, 1–42. [Google Scholar]
- Blankenberg, D.; Coraor, N.; Von Kuster, G.; Taylor, J.; Nekrutenko, A. Integrating diverse databases into an unified analysis framework: A Galaxy approach. Database 2011, 2011, bar011. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. 2021. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Alikhan, N.F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Kück, P.; Meid, S.A.; Groß, C.; Wägele, J.W.; Misof, B. AliGROOVE–visualization of heterogeneous sequence divergence within multiple sequence alignments and detection of inflated branch support. BMC Bioinform. 2014, 15, 294. [Google Scholar] [CrossRef]
- Xia, X.; Xie, Z. DAMBE: Software package for data analysis in molecular biology and evolution. J. Hered. 2001, 92, 371–373. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Ojala, D.; Merkel, C.; Gelfand, R.; Attardi, G. The tRNA genes punctuate the reading of genetic information in human mitochondrial DNA. Cell 1980, 22, 393–403. [Google Scholar] [CrossRef]
- Ojala, D.; Montoya, J.; Attardi, G. tRNA punctuation model of RNA processing in human mitochondria. Nature 1981, 290, 470–474. [Google Scholar] [CrossRef]
- Bibb, M.J.; Van Etten, R.A.; Wright, C.T.; Walberg, M.W.; Clayton, D.A. Sequence and gene organization of mouse mitochondrial DNA. Cell 1981, 26, 167–180. [Google Scholar] [CrossRef]
- Clary, D.O.; Wolstenholme, D.R. The mitochondrial DNA molecule of Drosophila yakuba: Nucleotide sequence, gene organization, and genetic code. J. Mol. Evol. 1985, 22, 252–271. [Google Scholar] [CrossRef]
- Bae, J.S.; Kim, I.; Sohn, H.D.; Jin, B.R. The mitochondrial genome of the firefly, Pyrocoelia rufa: Complete DNA sequence, genome organization, and phylogenetic analysis with other insects. Mol. Phylogenet. Evol. 2004, 32, 978–985. [Google Scholar] [CrossRef]
- Jiao, H.; Ding, M.; Zhao, H. Sequence and organization of complete mitochondrial genome of the firefly, Aquatica leii (Coleoptera: Lampyridae). Mitochondrial DNA 2015, 26, 775–776. [Google Scholar] [CrossRef]
- Nikolaou, C.; Almirantis, Y. A study on the correlation of nucleotide skews and the positioning of the origin of replication: Different modes of replication in bacterial species. Nucleic Acids Res. 2005, 33, 6816–6822. [Google Scholar] [CrossRef]
- Lin, L.L.; Li, X.J.; Zhang, H.L.; Zheng, Z.M. Mitochondrial genomes of three Tetrigoidea species and phylogeny of Tetrigoidea. PeerJ 2017, 5, e4002. [Google Scholar] [CrossRef]
- Cameron, S.L.; Lo, N.; Bourguignon, T.; Svenson, G.J.; Evans, T.A. A mitochondrial genome phylogeny of termites (Blattodea: Termitoidae): Robust support for interfamilial relationships and molecular synapomorphies define major clades. Mal. Pbylogenet. Evol. 2012, 65, 162–173. [Google Scholar] [CrossRef] [PubMed]
- Nelson, L.A.; Lambkin, C.L.; Batterham, P.; Wallman, J.F.; Dowton, M.; Whiting, M.F.; Yeates, D.K.; Cameron, S.L. Beyond barcoding: A mitochondrial genomics approach to molecular phylogenetics and diagnostics of blowflies (Diptera: Calliphoridae). Gene 2012, 511, 131–142. [Google Scholar] [CrossRef] [PubMed]
| Specimens | Date of Collection | Collection Site | Longitude (E) | Latitude (N) | GB Numbers |
|---|---|---|---|---|---|
| A. (Pseudapteranabropsis) nigrimaculatis | 2 August 2022 | Maguan, Yunnan | 104.0004 | 22.5115 | PQ625790 |
| A. (Spinanabropsis) erythronota | 2 August 2022 | Maguan, Yunnan | 104.0004 | 22.5115 | PQ625789 |
| A. (Pseudapteranabropsis) flavimaculata | 20 August 2022 | Pinglong Mountain, Guangxi | 109.8680 | 22.8403 | PQ625791 |
| A. (Apteranabropsis) daweishanensis | 23 May 2021 | Dawei Montain, Yunnan | 101.5128 | 23.1467 | PQ625792 |
| A. (Spinanabropsis) pengi | 13 August 2021 | Yakou, Yunnan | 99.1114 | 23.1722 | PQ625793 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, T.; Pang, S.; Wang, W.; Luo, T.; Qin, Y.; Bian, X.; Zhang, B. Comparative Analysis of the Mitochondrial Genomes of Five Species of Anabropsis (Orthoptera: Anostostomatidae) and the Phylogenetic Implications of Anostostomatidae. Biology 2025, 14, 772. https://doi.org/10.3390/biology14070772
Yu T, Pang S, Wang W, Luo T, Qin Y, Bian X, Zhang B. Comparative Analysis of the Mitochondrial Genomes of Five Species of Anabropsis (Orthoptera: Anostostomatidae) and the Phylogenetic Implications of Anostostomatidae. Biology. 2025; 14(7):772. https://doi.org/10.3390/biology14070772
Chicago/Turabian StyleYu, Tingting, Siyu Pang, Wenjing Wang, Ting Luo, Yanting Qin, Xun Bian, and Bin Zhang. 2025. "Comparative Analysis of the Mitochondrial Genomes of Five Species of Anabropsis (Orthoptera: Anostostomatidae) and the Phylogenetic Implications of Anostostomatidae" Biology 14, no. 7: 772. https://doi.org/10.3390/biology14070772
APA StyleYu, T., Pang, S., Wang, W., Luo, T., Qin, Y., Bian, X., & Zhang, B. (2025). Comparative Analysis of the Mitochondrial Genomes of Five Species of Anabropsis (Orthoptera: Anostostomatidae) and the Phylogenetic Implications of Anostostomatidae. Biology, 14(7), 772. https://doi.org/10.3390/biology14070772






