Transcriptomic Analysis Reveals the Role of Long Non-Coding RNAs in Response to Drought Stress in Tibetan Hulless Barley
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. The Plant Materials and Growth Conditions
2.2. Comprehensive Transcriptome Alignment and Assembly
2.3. Workflow for Systematic LncRNA Annotation
2.4. Prediction of Trans-/Cis-Regulatory Target Genes of LncRNAs in Hulless Barley
2.5. Differential Expression Analysis
2.6. Drought Stress Treatments
2.7. Total RNA Extraction, cDNA Construction, and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) Validation
3. Result
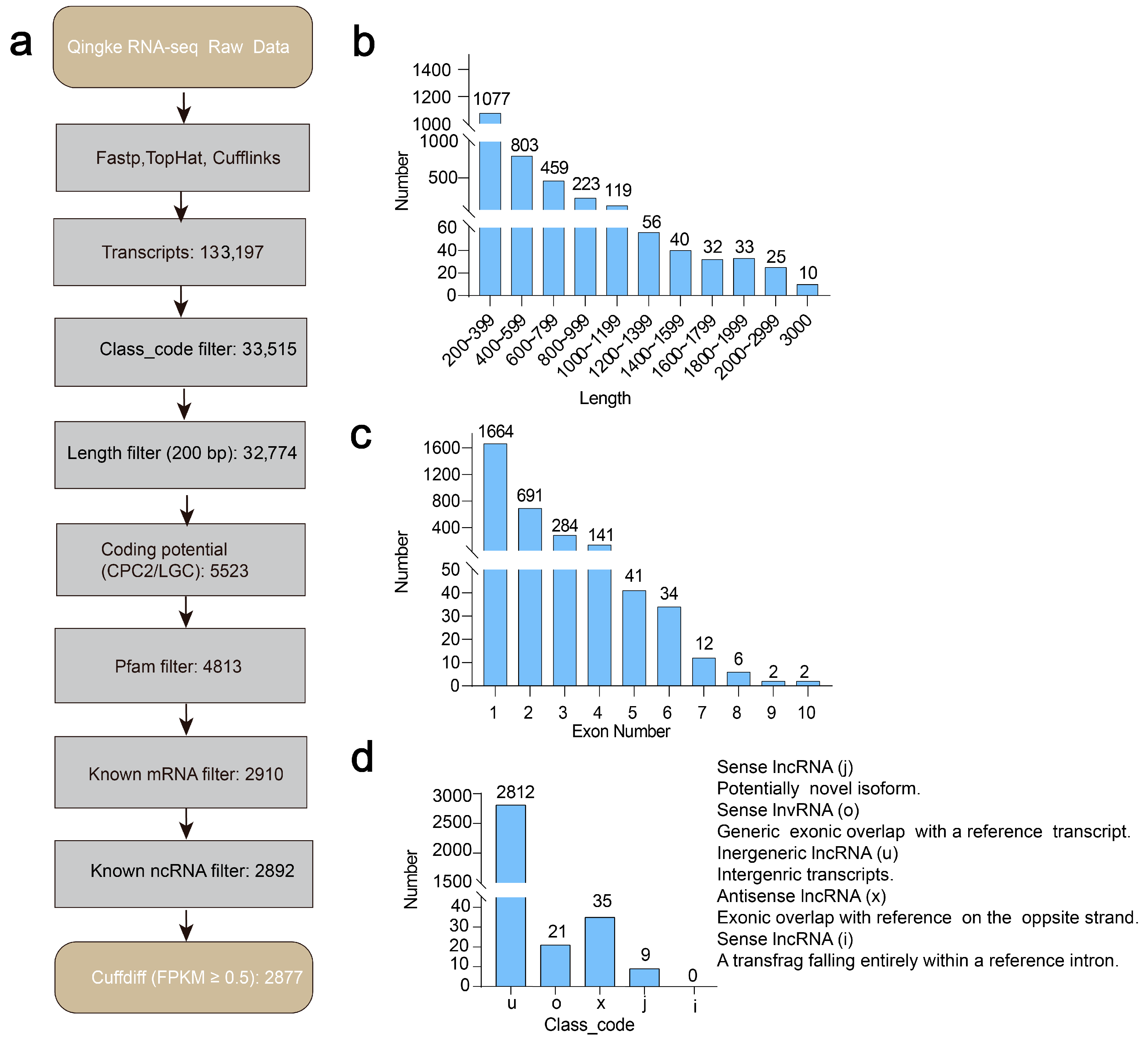
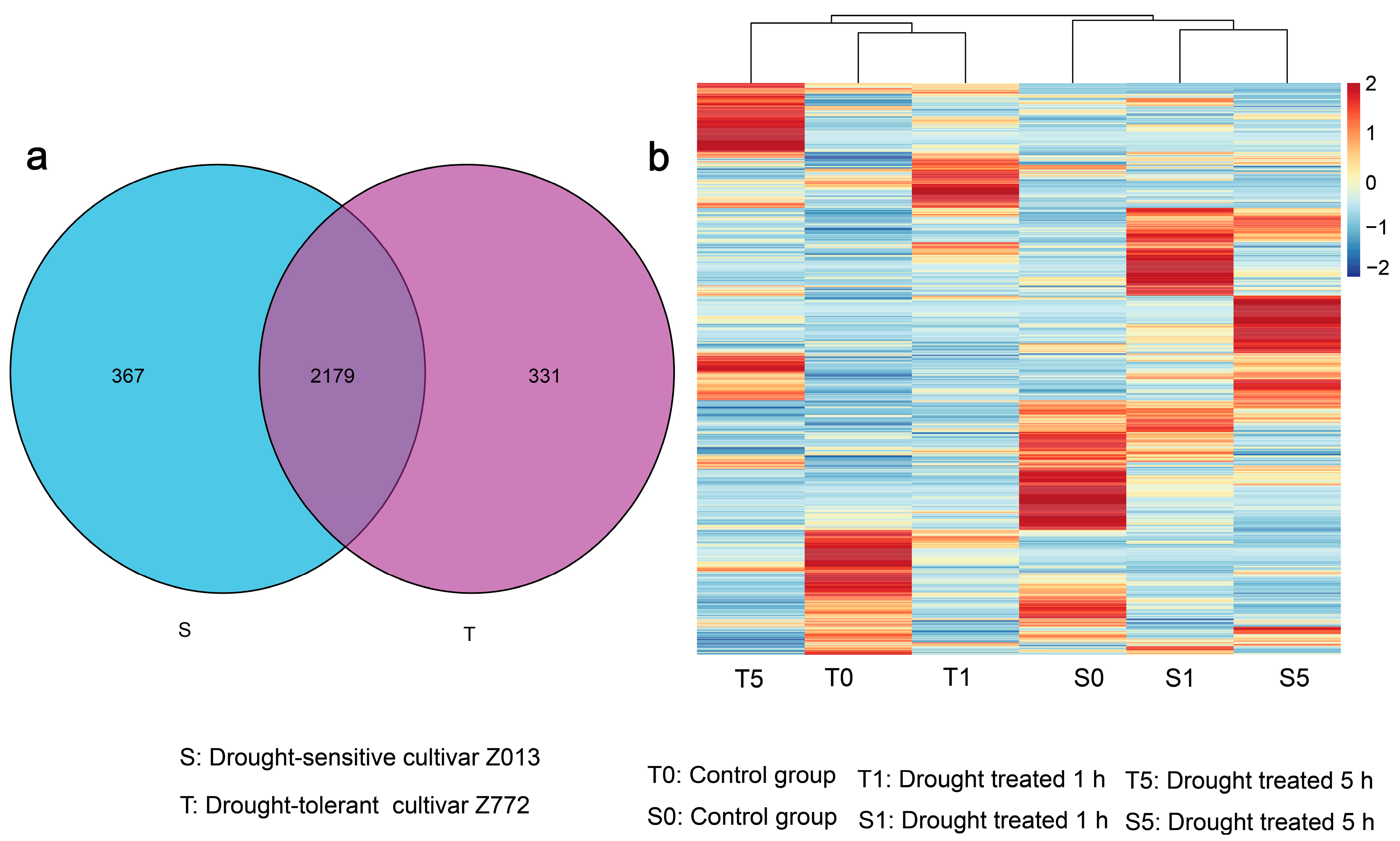
3.1. Genome-Wide Scale Transcriptional Signatures of LncRNAs in Two Tibetan Hulless Barley Cultivars Under Drought Conditions
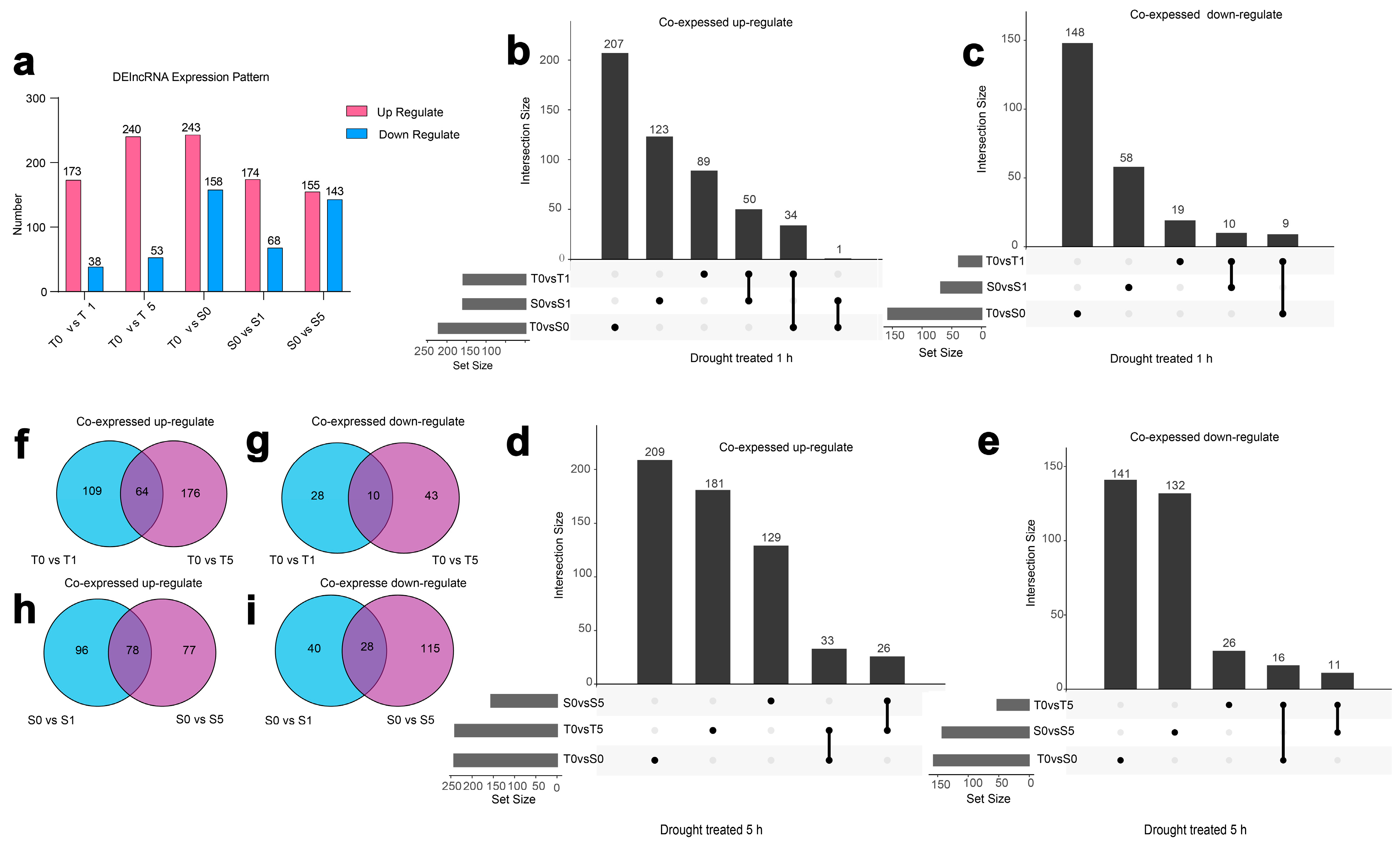
3.2. Characterization of DElncRNAs in Two Hulless Barley Cultivars Under Different Drought Treatment Times
3.3. Exploring the Regulatory Mechanisms (Trans- and Cis-) of LncRNAs Based on the Distances Between PCGs and LncRNAs
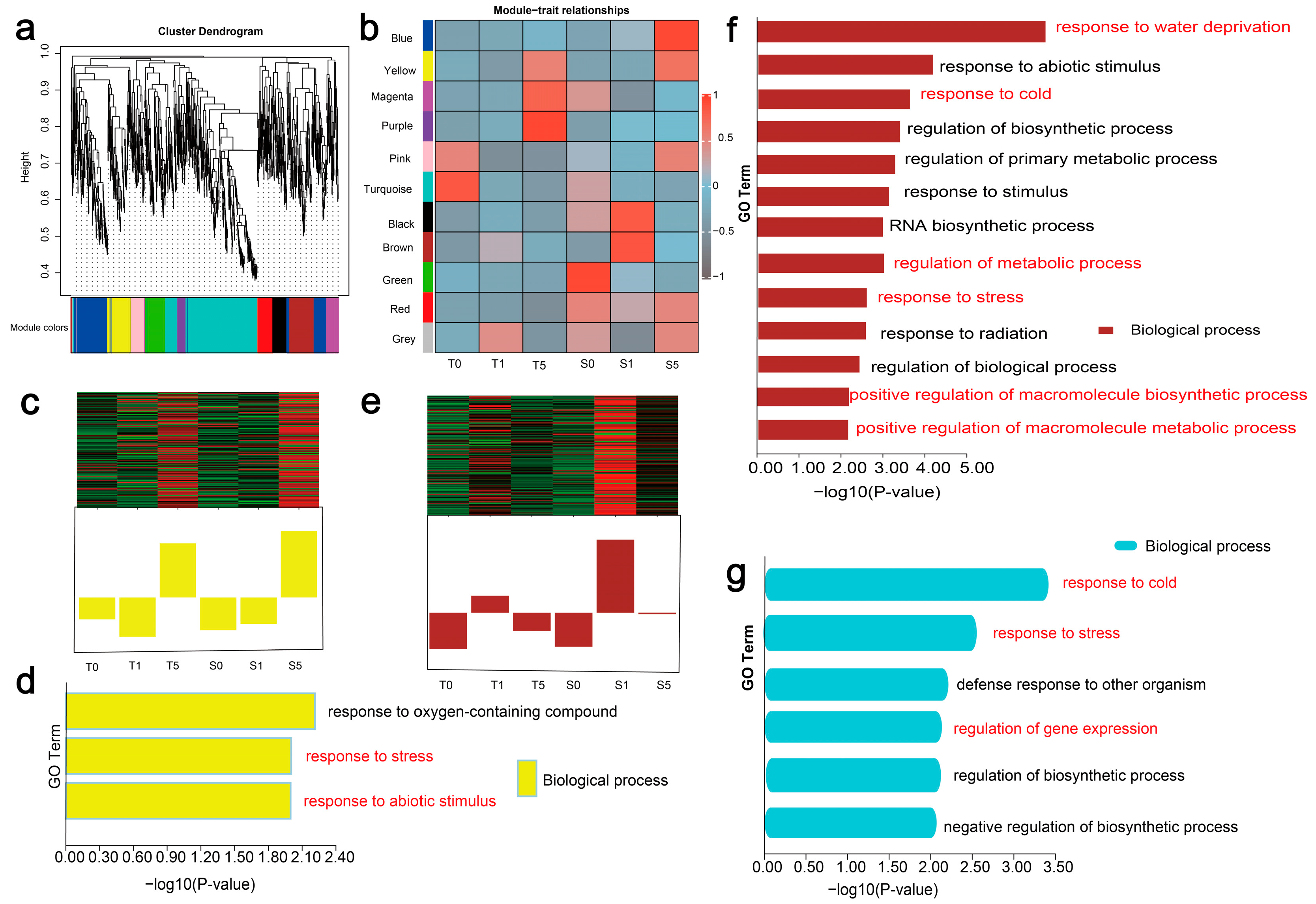
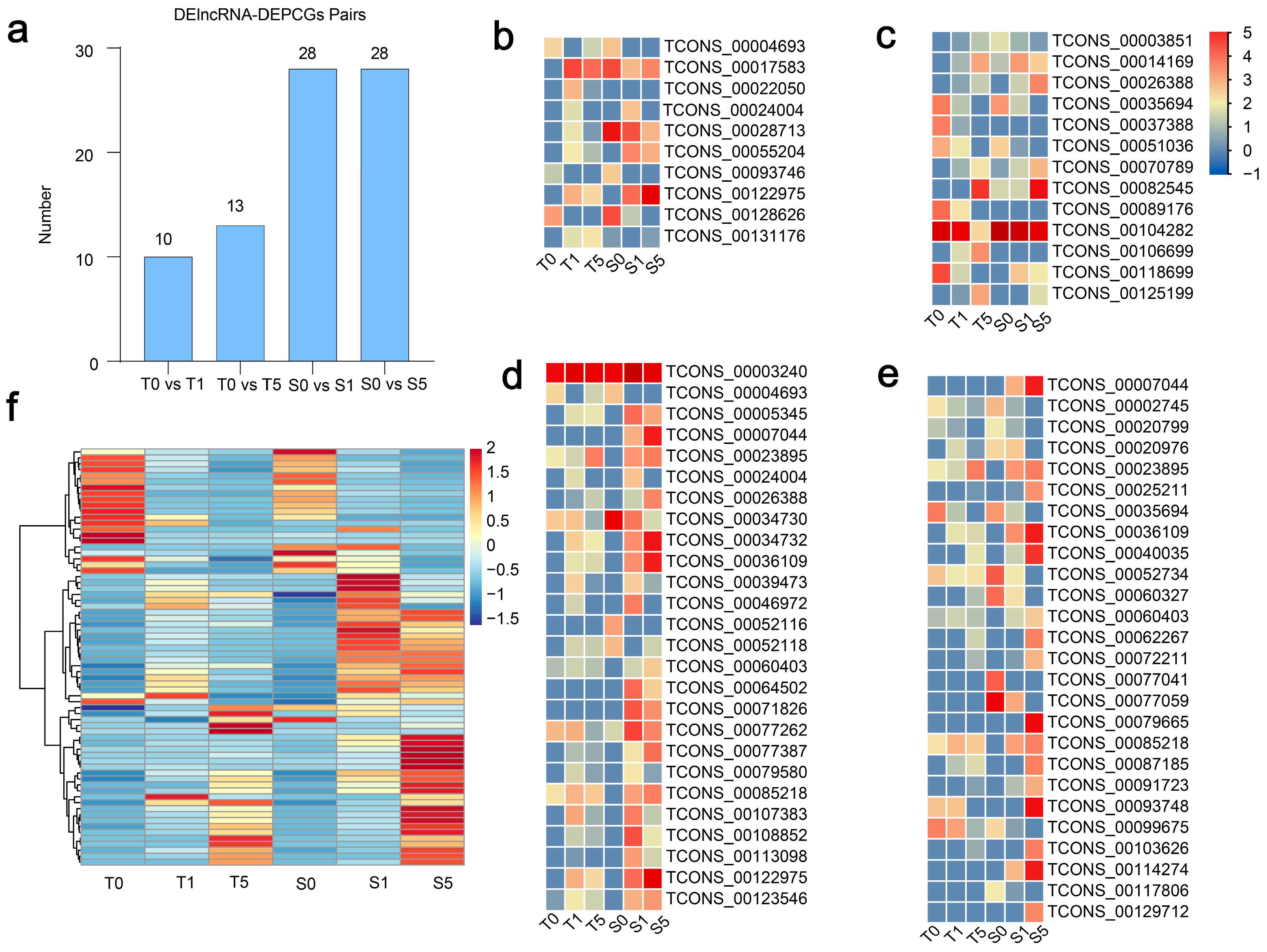
3.4. Expressional Dynamics of DElncRNAs in Two Cultivars Across Drought Stress Treatments
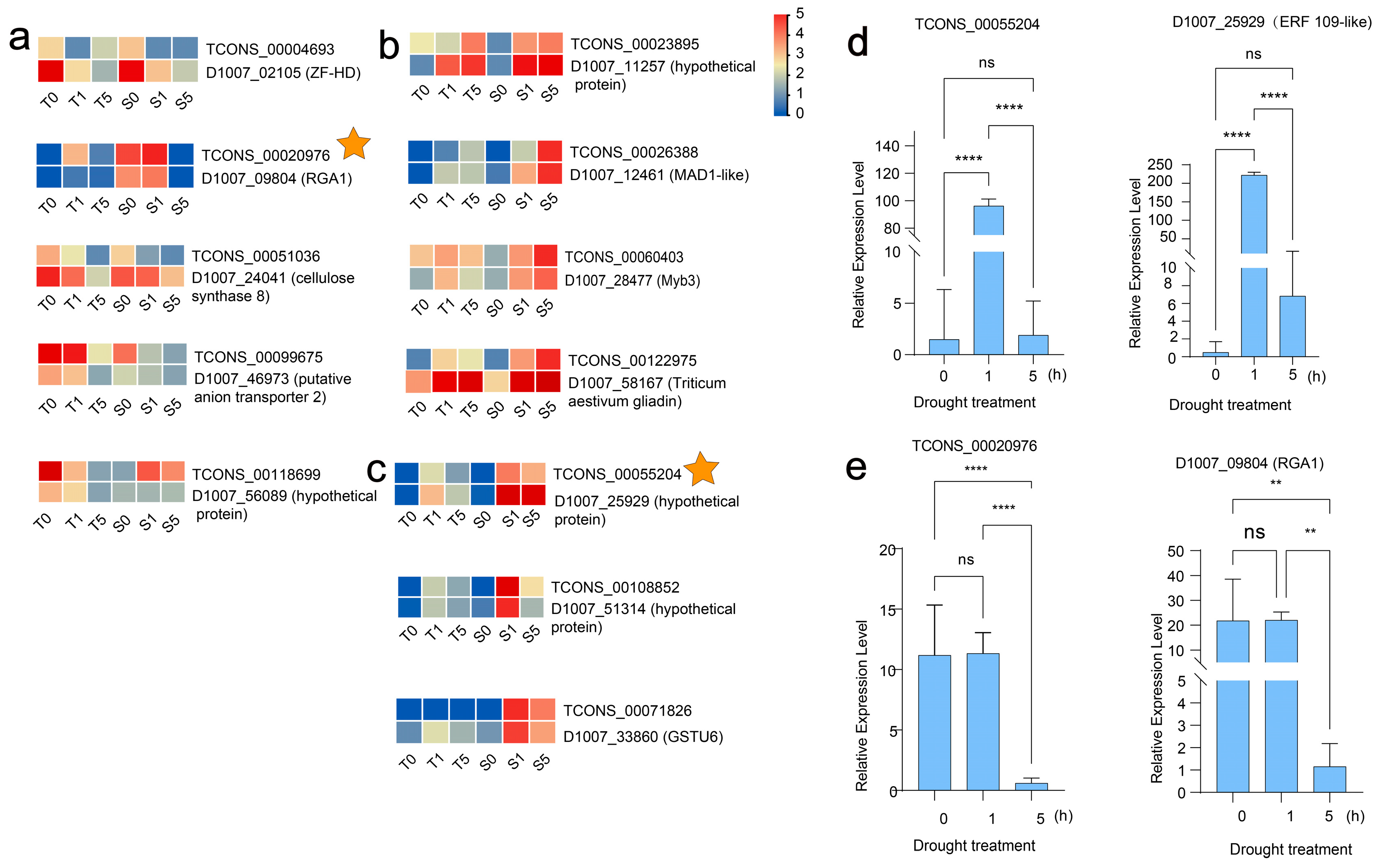
3.5. The Regulatory Diversity of DElncRNAs and Nearby Potential Target DEPCGs in Two Cultivars Across Drought Stress Treatments
3.6. In-Depth Functional Profiling of Putative LncRNA Target Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shafiq, S.; Li, J.; Sun, Q. Functions of plants long non-coding RNAs. Biochim. Et Biophys. Acta 2016, 1859, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Imaduwage, I.; Hewadikaram, M. Predicted roles of long non-coding RNAs in abiotic stress tolerance responses of plants. Mol. Hortic. 2024, 4, 20. [Google Scholar] [CrossRef]
- Palos, K.; Yu, L.; Railey, C.E.; Nelson Dittrich, A.C.; Nelson, A.D.L. Linking discoveries, mechanisms, and technologies to develop a clearer perspective on plant long noncoding RNAs. Plant Cell 2023, 35, 1762–1786. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.K.; Wang, H. Computational Analysis Predicts Hundreds of Coding lncRNAs in Zebrafish. Biology 2021, 10, 371. [Google Scholar] [CrossRef] [PubMed]
- Hung, T.; Chang, H.Y. Long noncoding RNA in genome regulation: Prospects and mechanisms. RNA Biol. 2010, 7, 582–585. [Google Scholar] [CrossRef] [PubMed]
- Sarropoulos, I.; Marin, R.; Cardoso-Moreira, M.; Kaessmann, H. Developmental dynamics of lncRNAs across mammalian organs and species. Nature 2019, 571, 510–514. [Google Scholar] [CrossRef]
- Liu, J.; Lao, L.; Chen, J.; Li, J.; Zeng, W.; Zhu, X.; Li, J.; Chen, X.; Yang, L.; Xing, Y.; et al. The IRENA lncRNA converts chemotherapy-polarized tumor-suppressing macrophages to tumor-promoting phenotypes in breast cancer. Nat. Cancer 2021, 2, 457–473. [Google Scholar] [CrossRef]
- Liang, W.W.; Müller, S.; Hart, S.K.; Wessels, H.H.; Méndez-Mancilla, A.; Sookdeo, A.; Choi, O.; Caragine, C.M.; Corman, A.; Lu, L.; et al. Transcriptome-scale RNA-targeting CRISPR screens reveal essential lncRNAs in human cells. Cell 2024, 187, 7637–7654.e29. [Google Scholar] [CrossRef]
- Yang, L.; Cheng, Y.; Yuan, C.; Zhou, Y.F.; Huang, Q.J.; Zhao, W.L.; He, R.R.; Jiang, J.; Qin, Y.C.; Chen, Z.T.; et al. The long non-coding RNA VIVIpary promotes seed dormancy release and pre-harvest sprouting through chromatin remodeling in rice. Mol. Plant, 2025; in press. [Google Scholar]
- Qin, T.; Zhao, H.; Cui, P.; Albesher, N.; Xiong, L. A Nucleus-Localized Long Non-Coding RNA Enhances Drought and Salt Stress Tolerance. Plant Physiol. 2017, 175, 1321–1336. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhu, P.; Hepworth, J.; Bloomer, R.; Antoniou-Kourounioti, R.L.; Doughty, J.; Heckmann, A.; Xu, C.; Yang, H.; Dean, C. Natural temperature fluctuations promote COOLAIR regulation of FLC. Genes Dev. 2021, 35, 888–898. [Google Scholar] [CrossRef]
- Liu, X.; Li, D.; Zhang, D.; Yin, D.; Zhao, Y.; Ji, C.; Zhao, X.; Li, X.; He, Q.; Chen, R.; et al. A novel antisense long noncoding RNA, TWISTED LEAF, maintains leaf blade flattening by regulating its associated sense R2R3-MYB gene in rice. New Phytol. 2018, 218, 774–788. [Google Scholar] [CrossRef]
- Gao, R.; Liu, P.; Irwanto, N.; Loh, R.; Wong, S.M. Upregulation of LINC-AP2 is negatively correlated with AP2 gene expression with Turnip crinkle virus infection in Arabidopsis thaliana. Plant Cell Rep. 2016, 35, 2257–2267. [Google Scholar] [CrossRef]
- Yang, T.; Ma, H.; Zhang, J.; Wu, T.; Song, T.; Tian, J.; Yao, Y. Systematic identification of long noncoding RNAs expressed during light-induced anthocyanin accumulation in apple fruit. Plant J. 2019, 100, 572–590. [Google Scholar] [CrossRef]
- Xu, K.; Zhao, Y.; Zhao, Y.; Feng, C.; Zhang, Y.; Wang, F.; Li, X.; Gao, H.; Liu, W.; Jing, Y.; et al. Soybean F-Box-Like Protein GmFBL144 Interacts With Small Heat Shock Protein and Negatively Regulates Plant Drought Stress Tolerance. Front. Plant Sci. 2022, 13, 823529. [Google Scholar] [CrossRef]
- El-Sappah, A.; Rather, S. Genomics Approaches to Study Abiotic Stress Tolerance in Plants. In Plant Abiotic Stress Physiology; Apple Academic Press: New York, NY, USA, 2021; pp. 25–46. [Google Scholar]
- Zeng, Y.; Muhammad, A.; Wan, L.; Gao, C.; Zhao, P.; El-Sappah, A.H. Epitranscriptomic modifications for enhancing abiotic stress resistance in plants. Front. Plant Sci. 2025, 16, 1538664. [Google Scholar] [CrossRef]
- Gupta, A.; Rico-Medina, A.; Caño-Delgado, A.I. The physiology of plant responses to drought. Science 2020, 368, 266–269. [Google Scholar] [CrossRef]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional genomics in plant abiotic stress responses and tolerance: From gene discovery to complex regulatory networks and their application in breeding. Proc. Jpn. Academy. Ser. B Phys. Biol. Sci. 2022, 98, 470–492. [Google Scholar] [CrossRef]
- Jin, X.; Wang, Z.; Li, X.; Ai, Q.; Wong, D.C.J.; Zhang, F.; Yang, J.; Zhang, N.; Si, H. Current perspectives of lncRNAs in abiotic and biotic stress tolerance in plants. Front. Plant Sci. 2023, 14, 1334620. [Google Scholar] [CrossRef]
- Li, J.; Zafar, S.; Javaid, A.; Perveen, S.; Hasnain, Z.; Ihtisham, M.; Abbas, A.; Usman, M.; El-Sappah, A.H.; Abbas, M. Zinc Nanoparticles (ZnNPs): High-Fidelity Amelioration in Turnip (Brassica rapa L.) Production under Drought Stress. Sustainability 2023, 15, 6512. [Google Scholar] [CrossRef]
- Mukherjee, A.; Dwivedi, S.; Bhagavatula, L.; Datta, S. Integration of light and ABA signaling pathways to combat drought stress in plants. Plant Cell Rep. 2023, 42, 829–841. [Google Scholar] [CrossRef]
- Muhammad Aslam, M.; Waseem, M.; Jakada, B.H.; Okal, E.J.; Lei, Z.; Saqib, H.S.A.; Yuan, W.; Xu, W.; Zhang, Q. Mechanisms of Abscisic Acid-Mediated Drought Stress Responses in Plants. Int. J. Mol. Sci. 2022, 23, 1084. [Google Scholar] [CrossRef]
- Morgil, H.; Tardu, M.; Cevahir, G.; Kavakli, İ.H. Comparative RNA-seq analysis of the drought-sensitive lentil (Lens culinaris) root and leaf under short- and long-term water deficits. Funct. Integr. Genom. 2019, 19, 715–727. [Google Scholar] [CrossRef]
- Sobreiro, M.B.; Collevatti, R.G.; Dos Santos, Y.L.A.; Bandeira, L.F.; Lopes, F.J.F.; Novaes, E. RNA-Seq reveals different responses to drought in Neotropical trees from savannas and seasonally dry forests. BMC Plant Biol. 2021, 21, 463. [Google Scholar] [CrossRef]
- Pang, J.; Zhang, X.; Ma, X.; Zhao, J. Spatio-Temporal Transcriptional Dynamics of Maize Long Non-Coding RNAs Responsive to Drought Stress. Genes 2019, 10, 138. [Google Scholar] [CrossRef]
- Rai, M.I.; Alam, M.; Lightfoot, D.A.; Gurha, P.; Afzal, A.J. Classification and experimental identification of plant long non-coding RNAs. Genomics 2019, 111, 997–1005. [Google Scholar] [CrossRef]
- Bohra, A.; Choudhary, M.; Bennett, D.; Joshi, R.; Mir, R.R.; Varshney, R.K. Drought-tolerant wheat for enhancing global food security. Funct. Integr. Genom. 2024, 24, 212. [Google Scholar] [CrossRef]
- Wang, Y.L.; Wei, Z.X.; Xu, Q.J.; Zeng, X.Q.; Yuan, H.J.; Tang, Y.W.; Tashi, N. The complete mitochondrial genome of Tibetan hulless barley. Mitochondrial DNA Part B Resour. 2016, 1, 430–431. [Google Scholar] [CrossRef]
- Zeng, X.; Long, H.; Wang, Z.; Zhao, S.; Tang, Y.; Huang, Z.; Wang, Y.; Xu, Q.; Mao, L.; Deng, G.; et al. The draft genome of Tibetan hulless barley reveals adaptive patterns to the high stressful Tibetan Plateau. Proc. Natl. Acad. Sci. USA 2015, 112, 1095–1100. [Google Scholar] [CrossRef]
- Yao, X.; Yao, Y.; An, L.; Li, X.; Bai, Y.; Cui, Y.; Wu, K. Accumulation and regulation of anthocyanins in white and purple Tibetan Hulless Barley (Hordeum vulgare L. var. nudum Hook. f.) revealed by combined de novo transcriptomics and metabolomics. BMC Plant Biol. 2022, 22, 391. [Google Scholar] [CrossRef]
- Wei, N.; Yue, X. Distribution of Core Root Microbiota of Tibetan Hulless Barley along an Altitudinal and Geographical Gradient in the Tibetan Plateau. Microorganisms 2022, 10, 1737. [Google Scholar] [CrossRef]
- Dou, X.; Zhou, Z.; Zhao, L. Identification and expression analysis of miRNAs in germination and seedling growth of Tibetan hulless barley. Genomics 2021, 113, 3735–3749. [Google Scholar] [CrossRef]
- Zheng, K.; Wu, X.; Xue, X.; Li, W.; Wang, Z.; Chen, J.; Zhang, Y.; Qiao, F.; Zhao, H.; Zhang, F.; et al. Transcriptome Screening of Long Noncoding RNAs and Their Target Protein-Coding Genes Unmasks a Dynamic Portrait of Seed Coat Coloration Associated with Anthocyanins in Tibetan Hulless Barley. Int. J. Mol. Sci. 2023, 24, 10587. [Google Scholar] [CrossRef]
- Liang, J.; Chen, X.; Deng, G.; Pan, Z.; Zhang, H.; Li, Q.; Yang, K.; Long, H.; Yu, M. Dehydration induced transcriptomic responses in two Tibetan hulless barley (Hordeum vulgare var. nudum) accessions distinguished by drought tolerance. BMC Genom. 2017, 18, 775. [Google Scholar] [CrossRef]
- Mayer, K.F.; Waugh, R.; Brown, J.W.; Schulman, A.; Langridge, P.; Platzer, M.; Fincher, G.B.; Muehlbauer, G.J.; Sato, K.; Close, T.J.; et al. A physical, genetic and functional sequence assembly of the barley genome. Nature 2012, 491, 711–716. [Google Scholar]
- Liang, J.; Deng, G.; Long, H.; Pan, Z.; Wang, C.; Cai, P.; Xu, D.; Nima, Z.-X.; Yu, M. Virus-induced silencing of genes encoding LEA protein in Tibetan hulless barley (Hordeum vulgare ssp. vulgare) and their relationship to drought tolerance. Mol. Breed. 2011, 30, 441–451. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Zeng, X.; Xu, T.; Ling, Z.; Wang, Y.; Li, X.; Xu, S.; Xu, Q.; Zha, S.; Qimei, W.; Basang, Y.; et al. An improved high-quality genome assembly and annotation of Tibetan hulless barley. Sci. Data 2020, 7, 139. [Google Scholar] [CrossRef]
- Langdon, W.B. Performance of genetic programming optimised Bowtie2 on genome comparison and analytic testing (GCAT) benchmarks. BioData Min. 2015, 8, 1. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef]
- Zandawala, M.; Bilal Amir, M.; Shin, J.; Yim, W.C.; Alfonso Yañez Guerra, L. Proteome-wide neuropeptide identification using NeuroPeptide-HMMer (NP-HMMer). Gen. Comp. Endocrinol. 2024, 357, 114597. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.J.; Yang, D.C.; Kong, L.; Hou, M.; Meng, Y.Q.; Wei, L.; Gao, G. CPC2: A fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017, 45, W12–W16. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, X.; Zhang, K.; Xiao, J.; Liu, F.; Yang, H.; Cai, Y.; Lai, F.; Fu, Q.; Wan, P. Transcriptomic Comparison of Rice lncRNAs in Response to Feeding by Brown Planthopper Populations with Different Virulence. Int. J. Mol. Sci. 2025, 26, 3486. [Google Scholar] [CrossRef]
- Holm, K.M.; Koepfli, K.P.; Pukazhenthi, B.S.; Ratan, A.; Fryxell, K.J.; Pham, M.; Weisz, D.; Dudchenko, O.; Aiden, E.L.; Lim, H.C. Chromosome-length genome assembly of the critically endangered Mountain bongo (Tragelaphus eurycerus isaaci): A resource for conservation and comparative genomics. G3 Genes|Genomes|Genet. 2025, jkaf109. [Google Scholar] [CrossRef]
- Copeland, C.S.; Marz, M.; Rose, D.; Hertel, J.; Brindley, P.J.; Santana, C.B.; Kehr, S.; Attolini, C.S.-O.; Stadler, P.F. Homology-based annotation of non-coding RNAs in the genomes of Schistosoma mansoni and Schistosoma japonicum. BMC Genom. 2009, 10, 464. [Google Scholar] [CrossRef]
- Engreitz, J.M.; Haines, J.E.; Perez, E.M.; Munson, G.; Chen, J.; Kane, M.; McDonel, P.E.; Guttman, M.; Lander, E.S. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 2016, 539, 452–455. [Google Scholar] [CrossRef]
- Alexander, R.D.; Wendelboe-Nelson, C.; Morris, P.C. The barley transcription factor HvMYB1 is a positive regulator of drought tolerance. Plant Physiol. Biochem. 2019, 142, 246–253. [Google Scholar] [CrossRef] [PubMed]
- Cheng, B.; Pei, W.; Wan, K.; Pan, R.; Zhang, W. LncRNA cis- and trans-regulation provides new insight into drought stress responses in wild barley. Physiol. Plant. 2024, 176, e14424. [Google Scholar] [CrossRef] [PubMed]
- Dawson, I.K.; Russell, J.; Powell, W.; Steffenson, B.; Thomas, W.T.B.; Waugh, R. Barley: A translational model for adaptation to climate change. New Phytol. 2015, 206, 913–931. [Google Scholar] [CrossRef] [PubMed]
- Singh, U.; Khemka, N.; Rajkumar, M.S.; Garg, R.; Jain, M. PLncPRO for prediction of long non-coding RNAs (lncRNAs) in plants and its application for discovery of abiotic stress-responsive lncRNAs in rice and chickpea. Nucleic Acids Res. 2017, 45, e183. [Google Scholar] [CrossRef]
- Cai, J.; Zhang, Y.; He, R.; Jiang, L.; Qu, Z.; Gu, J.; Yang, J.; Legascue, M.F.; Wang, Z.Y.; Ariel, F.; et al. LncRNA DANA1 promotes drought tolerance and histone deacetylation of drought responsive genes in Arabidopsis. EMBO Rep. 2024, 25, 796–812. [Google Scholar] [CrossRef]
- Pang, Y.; Zheng, K.; Min, Q.; Wang, Y.; Xue, X.; Li, W.; Zhao, H.; Qiao, F.; Han, S. Long Noncoding RNAs in Response to Hyperosmolarity Stress, but Not Salt Stress, Were Mainly Enriched in the Rice Roots. Int. J. Mol. Sci. 2024, 25, 6226. [Google Scholar] [CrossRef]
- Min, Q.; Zheng, K.; Liu, T.; Wang, Z.; Xue, X.; Li, W.; Liu, Y.; Zhang, Y.; Qiao, F.; Chen, J.; et al. Transcriptomic Profiles of Long Noncoding RNAs and Their Target Protein-Coding Genes Reveals Speciation Adaptation on the Qinghai-Xizang (Tibet) Plateau in Orinus. Biology 2024, 13, 349. [Google Scholar] [CrossRef]
- Deng, P.; Liu, S.; Nie, X.; Weining, S.; Wu, L. Conservation analysis of long non-coding RNAs in plants. Sci. China Life Sci. 2018, 61, 190–198. [Google Scholar] [CrossRef]
- Zhou, Y.F.; Zhang, Y.C.; Sun, Y.M.; Yu, Y.; Lei, M.Q.; Yang, Y.W.; Lian, J.P.; Feng, Y.Z.; Zhang, Z.; Yang, L.; et al. The parent-of-origin lncRNA MISSEN regulates rice endosperm development. Nat. Commun. 2021, 12, 6525. [Google Scholar] [CrossRef]
- Yang, X.; Liu, C.; Niu, X.; Wang, L.; Li, L.; Yuan, Q.; Pei, X. Research on lncRNA related to drought resistance of Shanlan upland rice. BMC Genom. 2022, 23, 336. [Google Scholar] [CrossRef] [PubMed]
- Bridges, M.C.; Daulagala, A.C.; Kourtidis, A. LNCcation: lncRNA localization and function. J. Cell Biol. 2021, 220, e202009045. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Fu, H.; Wu, Y.; Zheng, X. Function of lncRNAs and approaches to lncRNA-protein interactions. Sci. China Life Sci. 2013, 56, 876–885. [Google Scholar] [CrossRef] [PubMed]
- Feng, K.; Hou, X.L.; Xing, G.M.; Liu, J.X.; Duan, A.Q.; Xu, Z.S.; Li, M.Y.; Zhuang, J.; Xiong, A.S. Advances in AP2/ERF super-family transcription factors in plant. Crit. Rev. Biotechnol. 2020, 40, 750–776. [Google Scholar] [CrossRef]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Lee, Y.K.; Kumari, S.; Olson, A.; Hauser, F.; Ware, D. Role of a ZF-HD Transcription Factor in miR157-Mediated Feed-Forward Regulatory Module That Determines Plant Architecture in Arabidopsis. Int. J. Mol. Sci. 2022, 23, 8665. [Google Scholar] [CrossRef]
- Min, Q.; Zheng, K.; Pang, Y.; Fang, Y.; Zhang, Y.; Qiao, F.; Su, X.; Chen, J.; Han, S. Transcription factors in Orinus: Novel insights into transcription regulation for speciation adaptation on the Qinghai-Xizang (Tibet) Plateau. BMC Plant Biol. 2025, 25, 560. [Google Scholar] [CrossRef]
- Tian, J.; Yuan, P.; Gao, X.; Wang, H.; Wang, M.; Jiao, J.; Zhang, K.; Hao, P.; Song, C.; Zheng, X.; et al. The AP2/ERF transcription factor MhERF113-like positively regulates drought tolerance in transgenic tomato and apple. Plant Physiol. Biochem. 2025, 221, 109598. [Google Scholar] [CrossRef]
- Gao, L.; Lv, Q.; Wang, L.; Han, S.; Wang, J.; Chen, Y.; Zhu, W.; Zhang, X.; Bao, F.; Hu, Y.; et al. Abscisic acid-mediated autoregulation of the MYB41-BRAHMA module enhances drought tolerance in Arabidopsis. Plant Physiol. 2024, 196, 1608–1626. [Google Scholar] [CrossRef]
- Li, M.; Dong, H.; Li, J.; Dai, X.; Lin, J.; Li, S.; Zhou, C.; Chiang, V.L.; Li, W. PtrVCS2 Regulates Drought Resistance by Changing Vessel Morphology and Stomatal Closure in Populus trichocarpa. Int. J. Mol. Sci. 2023, 24, 4458. [Google Scholar] [CrossRef]
- Mráz, P.; Tarbush, E.; Müller-Schärer, H. Drought tolerance and plasticity in the invasive knapweed Centaurea stoebe s.l. (Asteraceae): Effect of populations stronger than those of cytotype and range. Ann. Bot. 2014, 114, 289–299. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Fang, Y.; Min, Q.; Zheng, K.; Pang, Y.; Chen, J.; Qiao, F.; Han, S. Transcriptomic Analysis Reveals the Role of Long Non-Coding RNAs in Response to Drought Stress in Tibetan Hulless Barley. Biology 2025, 14, 737. https://doi.org/10.3390/biology14070737
Wang Z, Fang Y, Min Q, Zheng K, Pang Y, Chen J, Qiao F, Han S. Transcriptomic Analysis Reveals the Role of Long Non-Coding RNAs in Response to Drought Stress in Tibetan Hulless Barley. Biology. 2025; 14(7):737. https://doi.org/10.3390/biology14070737
Chicago/Turabian StyleWang, Zitao, Yue Fang, Qinyue Min, Kaifeng Zheng, Yanrong Pang, Jinyuan Chen, Feng Qiao, and Shengcheng Han. 2025. "Transcriptomic Analysis Reveals the Role of Long Non-Coding RNAs in Response to Drought Stress in Tibetan Hulless Barley" Biology 14, no. 7: 737. https://doi.org/10.3390/biology14070737
APA StyleWang, Z., Fang, Y., Min, Q., Zheng, K., Pang, Y., Chen, J., Qiao, F., & Han, S. (2025). Transcriptomic Analysis Reveals the Role of Long Non-Coding RNAs in Response to Drought Stress in Tibetan Hulless Barley. Biology, 14(7), 737. https://doi.org/10.3390/biology14070737





