Genomic and Molecular Mechanisms of Goat Environmental Adaptation
Simple Summary
Abstract
1. Introduction
2. Overview of Goat Genomics Research
2.1. Construction and Refinement of the Goat Reference Genome
2.2. Features of the Goat Genome
3. Genetic Basis of Environmental Adaptation in Goats
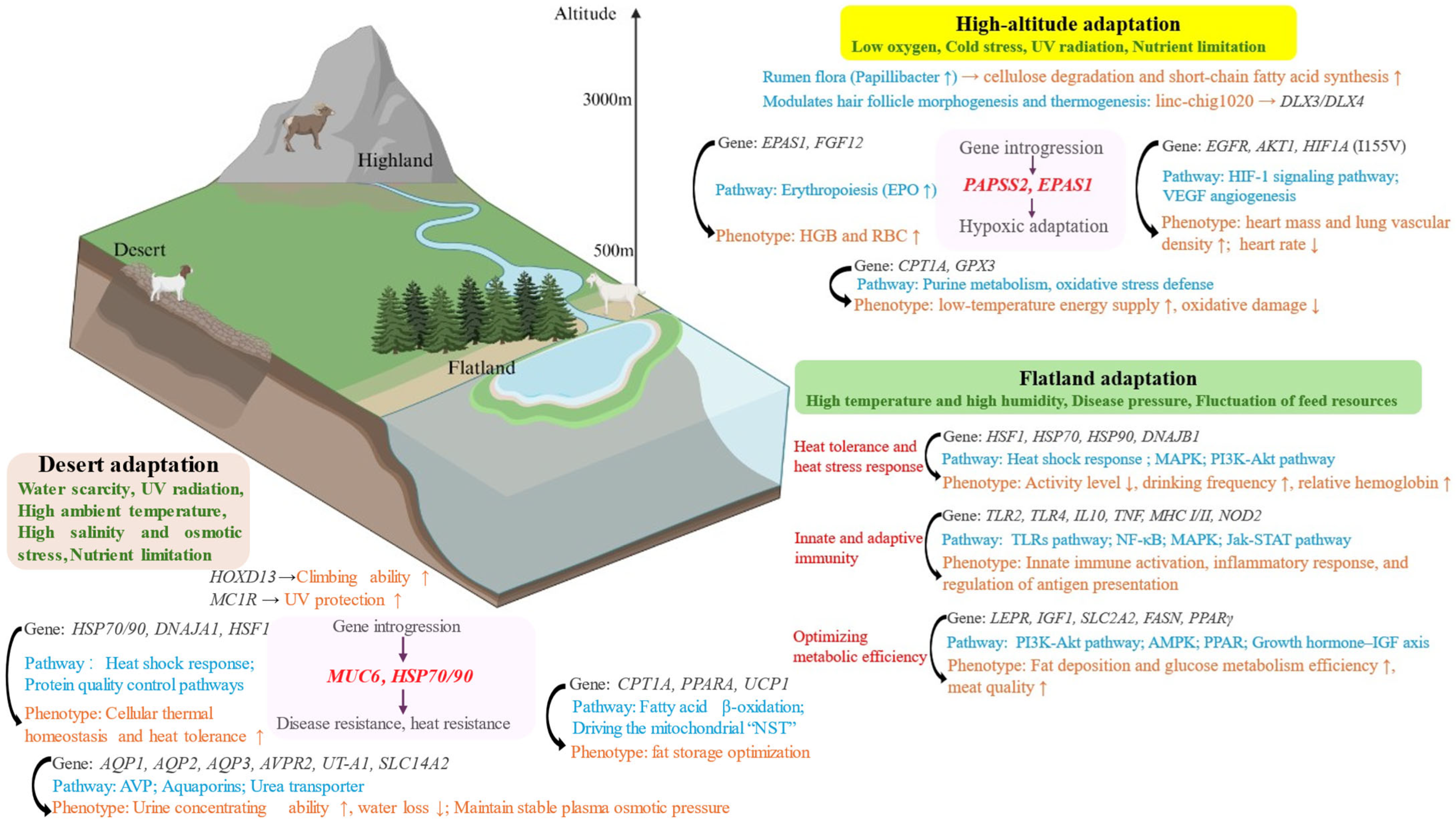
3.1. Evolution and Survival of Goats in Different Environments
3.1.1. Adaptation to Heat and Desert Environments
3.1.2. High-Altitude Adaptation
3.1.3. Cold Environment Adaptation
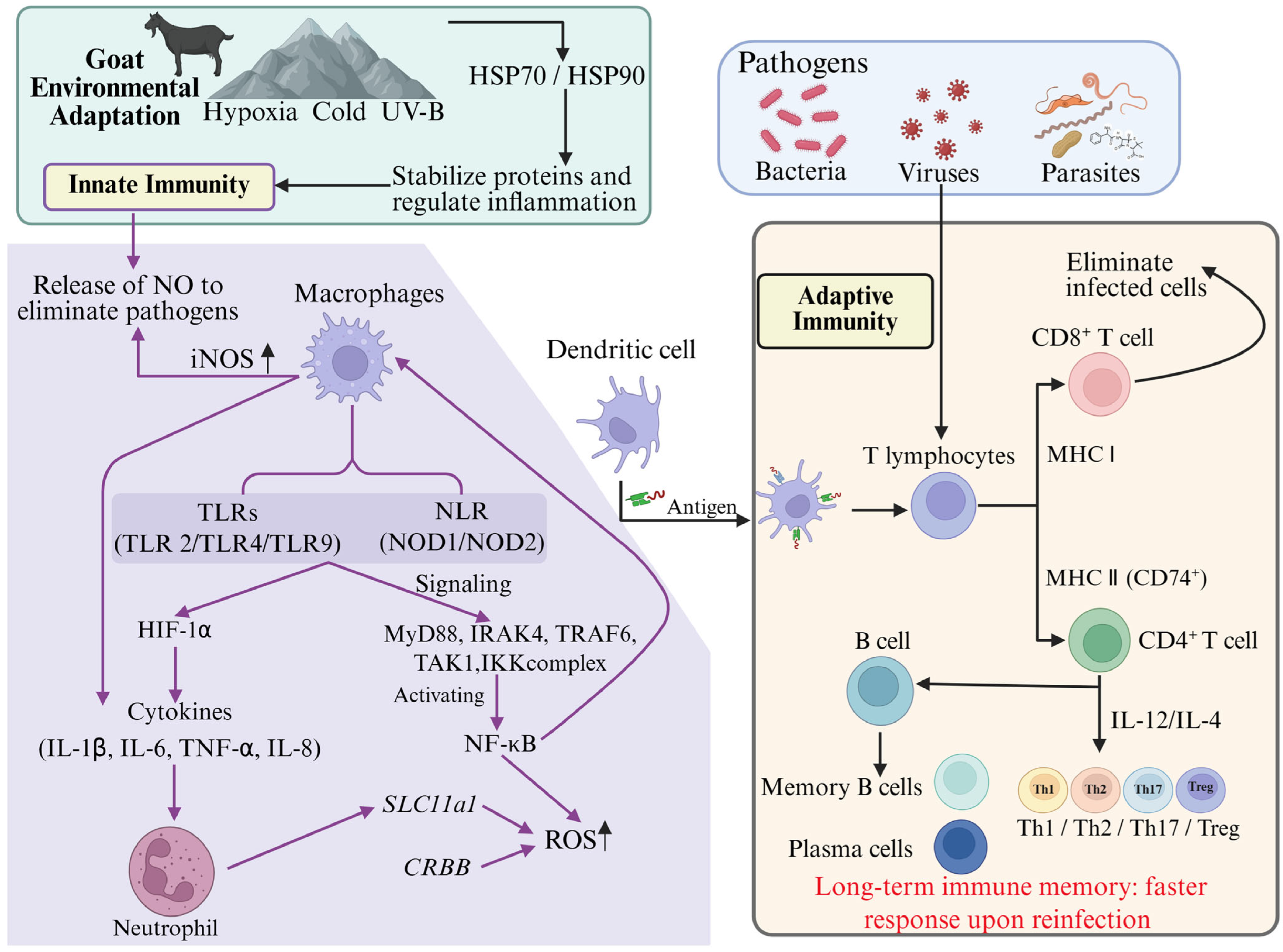
3.2. The Synergistic Mechanism of Immune Signaling Pathways and Environmental Adaptation
3.3. Identification of Key Adaptive Genes in Goats
4. Synergistic Action of Genomic and Epigenetic Regulation
5. Research Strategies for Environmental Adaptation from a Multi-Omics Perspective
5.1. Environmental Gradient–Phenotype Association Study
5.2. Multi-Omics Dynamic Monitoring and Functional Verification
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Daly, K.G.; Mattiangeli, V.; Hare, A.J.; Davoudi, H.; Fathi, H.; Doost, S.B.; Amiri, S.; Khazaeli, R.; Decruyenaere, D.; Nokandeh, J.; et al. Herded and Hunted Goat Genomes from the Dawn of Domestication in the Zagros Mountains. Proc. Natl. Acad. Sci. USA 2021, 118, e2100901118. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Wang, X.; Li, M.; Li, Y.; Yang, Z.; Wang, X.; Pan, X.; Gong, M.; Zhang, Y.; Guo, Y.; et al. The Origin of Domestication Genes in Goats. Sci. Adv. 2020, 6, eaaz5216. [Google Scholar] [CrossRef] [PubMed]
- Colli, L.; Milanesi, M.; Talenti, A.; Bertolini, F.; Chen, M.; Crisà, A.; Daly, K.G.; Del Corvo, M.; Guldbrandtsen, B.; Lenstra, J.A.; et al. Genome-Wide SNP Profiling of Worldwide Goat Populations Reveals Strong Partitioning of Diversity and Highlights Post-Domestication Migration Routes. Genet. Sel. Evol. 2018, 50, 58. [Google Scholar] [CrossRef] [PubMed]
- Qanbari, S.; Simianer, H. Mapping Signatures of Positive Selection in the Genome of Livestock. Livest. Sci. 2014, 166, 133–143. [Google Scholar] [CrossRef]
- Brito, L.F.; Kijas, J.W.; Ventura, R.V.; Sargolzaei, S.; Porto-Neto, L.R.; Cánovas, A.; Feng, Z.; Jafarikia, M.; Schenkel, F.S. Genetic Diversity and Signatures of Selection in Various Goat Breeds Revealed by Genome-Wide SNP Markers. BMC Genom. 2017, 18, 229. [Google Scholar] [CrossRef]
- Nanaei, H.A.; Cai, Y.; Alshawi, A.; Wen, J.; Hussain, T.; Fu, W.W.; Xu, N.Y.; Essa, A.; Alenstra, J.; Wang, X.; et al. Genomic Analysis of Indigenous Goats in Southwest Asia Reveals Evidence of Ancient Adaptive Introgression Related to Desert Climate. Zool. Res. 2023, 44, 20–29. [Google Scholar] [CrossRef]
- Zhang, H.; Duan, X.; Li, H.; Niu, L.; Wang, L.; Li, L.; Zhang, H.; Zhong, T. The Complete Mitochondrial Genome of Chinese Tibetan Goat (Capra hircus). Mitochondrial DNA A DNA Mapp. Seq. Anal. 2016, 27, 1161–1162. [Google Scholar] [CrossRef]
- Nair, M.R.R.; Sejian, V.; Silpa, M.V.; Fonsêca, V.F.C.; de Melo Costa, C.C.; Devaraj, C.; Krishnan, G.; Bagath, M.; Nameer, P.O.; Bhatta, R. Goat as the Ideal Climate-Resilient Animal Model in Tropical Environment: Revisiting Advantages over Other Livestock Species. Int. J. Biometeorol. 2021, 65, 2229–2240. [Google Scholar] [CrossRef]
- Bao, W.; Lei, C.; Wen, W. Genomic Insights into Ruminant Evolution: From Past to Future Prospects. Zool. Res. 2019, 40, 476–487. [Google Scholar] [CrossRef]
- Raoult, C.M.C.; Osthaus, B.; Hildebrand, A.C.G.; Mcelligott, A.G.; Nawroth, C. Goats Show Higher Behavioural Flexibility than Sheep in a Spatial Detour Task. R. Soc. Open Sci. 2021, 8, 201627. [Google Scholar] [CrossRef]
- Zieba, D.A.; Szczesna, M.; Klocek-Gorka, B.; Williams, G.L. Leptin as a Nutritional Signal Regulating Appetite and Reproductive Processes in Seasonally-Breeding Ruminants. J. Physiol. Pharmacol. 2008, 59 (Suppl. S9), 7–18. [Google Scholar] [PubMed]
- Lu, C.D. The Role of Goats in the World: Society, Science, and Sustainability. Small Rumin. Res. 2023, 227, 107056. [Google Scholar] [CrossRef]
- Gerber, P.J.; Henderson, B.; Makkar, H.; Hristov, A.; Oh, J.; Lee, C.-C.; Meinen, R.; Montes, F.; Ott, T.; Firkins, J.; et al. Mitigation of Greenhouse Gas Emissions in Livestock Production—A Review of Technical Options for Non-CO2 Emissions; Food and Agriculture Organization of the United Nations (FAO): Rome, Italy, 2013; Volume 177, ISBN 978-92-5-107658-3/978-92-5-107659-0. [Google Scholar]
- Dong, Y.; Xie, M.; Jiang, Y.; Xiao, N.; Du, X.; Zhang, W.; Tosser-Klopp, G.; Wang, J.; Yang, S.; Liang, J.; et al. Sequencing and Automated Whole-Genome Optical Mapping of the Genome of a Domestic Goat (Capra hircus). Nat. Biotechnol. 2013, 31, 135–141. [Google Scholar] [CrossRef]
- Hu, T.; Chitnis, N.; Monos, D.; Dinh, A. Next-Generation Sequencing Technologies: An Overview. Hum. Immunol. 2021, 82, 801–811. [Google Scholar] [CrossRef] [PubMed]
- Bickhart, D.M.; Rosen, B.D.; Koren, S.; Sayre, B.L.; Hastie, A.R.; Chan, S.; Lee, J.; Lam, E.T.; Liachko, I.; Sullivan, S.T.; et al. Single-Molecule Sequencing and Chromatin Conformation Capture Enable de Novo Reference Assembly of the Domestic Goat Genome. Nat. Genet. 2017, 49, 643–650. [Google Scholar] [CrossRef]
- Miga, K.H. Centromere Studies in the Era of “Telomere-to-Telomere” Genomics. Exp. Cell Res. 2020, 394, 112127. [Google Scholar] [CrossRef]
- Wu, H.; Luo, L.Y.; Zhang, Y.H.; Zhang, C.Y.; Huang, J.H.; Mo, D.X.; Zhao, L.M.; Wang, Z.X.; Wang, Y.C.; He-Hua, E.; et al. Telomere-to-Telomere Genome Assembly of a Male Goat Reveals Variants Associated with Cashmere Traits. Nat. Commun. 2024, 15, 10041. [Google Scholar] [CrossRef]
- Dong, Y.; Zhang, X.; Xie, M.; Arefnezhad, B.; Wang, Z.; Wang, W.; Feng, S.; Huang, G.; Guan, R.; Shen, W.; et al. Reference Genome of Wild Goat (Capra aegagrus) and Sequencing of Goat Breeds Provide Insight into Genic Basis of Goat Domestication. BMC Genom. 2015, 16, 431. [Google Scholar] [CrossRef]
- Siddiki, A.Z.; Baten, A.; Billah, M.; Alam, M.A.U.; Shawrob, K.S.M.; Saha, S.; Chowdhury, M.; Rahman, A.H.; Stear, M.; Miah, G. The Genome of the Black Bengal Goat (Capra hircus). BMC Res. Notes 2019, 12, 362. [Google Scholar] [CrossRef]
- Li, R.; Yang, P.; Dai, X.; Nanaei, H.A.; Fang, W.; Yang, Z.; Cai, Y.; Zheng, Z.; Wang, X.; Jiang, Y. A near Complete Genome for Goat Genetic and Genomic Research. Genet. Sel. Evol. 2021, 53, 74. [Google Scholar] [CrossRef]
- Peipei, B.; Jiaxin, L.; Shishuo, Z.; Xingquan, W.; Mian, G.; Xi, G.; Yudong, C.; Qimeng, Y.; Jiaqi, F.; Rongrong, L.; et al. A Graph-Based Goat Pangenome Reveals Structural Variations Involved in Domestication and Adaptation. Mol. Biol. Evol. 2024, 41, msae251. [Google Scholar] [CrossRef]
- Hiendleder, S.; Lewalski, H.; Wassmuth, R.; Janke, A.; Hiendleder, S.; Lewalski, H.; Wassmuth, R.; Janke, A. The Complete Mitochondrial DNA Sequence of the Domestic Sheep (Ovis aries) and Comparison with the Other Major Ovine Haplotype. J. Mol. Evol. 1998, 47, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Wang, M.; Zhang, Y.E.; Tan, S. The Power of “controllers”: Transposon-Mediated Duplicated Genes Evolve towards Neofunctionalization. J. Genet. Genom. 2023, 50, 462–472. [Google Scholar] [CrossRef]
- Khdhr, D.M.; Karim, K.J. Cytogenetic Study of Meriz Goat Breeds in Iraqi Kurdistan Region. Cell. Mol. Biol. 2024, 70, 155–160. [Google Scholar] [CrossRef] [PubMed]
- Evans, H.J.; Buckland, R.A.; Sumner, A.T. Chromosome Homology and Heterochromatin in Goat, Sheep and Ox Studied by Banding Techniques. Chromosoma 1973, 42, 383–402. [Google Scholar] [CrossRef]
- Fontanesi, L.; Beretti, F.; Martelli, P.L.; Colombo, M.; Dall’olio, S.; Occidente, M.; Portolano, B.; Casadio, R.; Matassino, D.; Russo, V. A First Comparative Map of Copy Number Variations in the Sheep Genome. Genomics 2011, 97, 158–165. [Google Scholar] [CrossRef]
- Yang, J.; Wang, D.F.; Huang, J.H.; Zhu, Q.H.; Luo, L.Y.; Lu, R.; Xie, X.L.; Salehian-Dehkordi, H.; Esmailizadeh, A.; Liu, G.E.; et al. Structural Variant Landscapes Reveal Convergent Signatures of Evolution in Sheep and Goats. Genome Biol. 2024, 25, 148. [Google Scholar] [CrossRef]
- Luo, L.Y.; Wu, H.; Zhao, L.M.; Zhang, Y.H.; Huang, J.H.; Liu, Q.Y.; Wang, H.T.; Mo, D.X.; Eer, H.H.; Zhang, L.Q.; et al. Telomere-to-Telomere Sheep Genome Assembly Identifies Variants Associated with Wool Fineness. Nat. Genet. 2025, 57, 218–230. [Google Scholar] [CrossRef]
- Jiang, Y.; Xie, M.; Chen, W.; Talbot, R.; Maddox, J.F.; Faraut, T.; Wu, C.; Muzny, D.M.; Li, Y.; Zhang, W.; et al. The Sheep Genome Illuminates Biology of the Rumen and Lipid Metabolism. Science 2014, 344, 1168–1173. [Google Scholar] [CrossRef]
- Hall, A.M.; Kou, K.; Chen, Z.; Pietka, T.A.; Kumar, M.; Korenblat, K.M.; Lee, K.; Ahn, K.; Fabbrini, E.; Klein, S.; et al. Evidence for Regulated Monoacylglycerol Acyltransferase Expression and Activity in Human Liver. J. Lipid Res. 2012, 53, 990–999. [Google Scholar] [CrossRef]
- Zhang, S.; Gao, X.; Jiang, Y.; Shen, Y.; Xie, H.; Pan, P.; Huang, Y.; Wei, Y.; Jiang, Q. Population Validation of Reproductive Gene Mutation Loci and Association with the Litter Size in Nubian Goat. Arch. Anim. Breed. 2021, 64, 375–386. [Google Scholar] [CrossRef]
- Shirazi, M.R.J.; Zamiri, M.J.; Salehi, M.S.; Moradi, S.; Tamadon, A.; Namavar, M.R.; Akhlaghi, A.; Tsutsui, K.; Caraty, A. Differential Expression of RFamide-Related Peptide, a Mammalian Gonadotrophin-Inhibitory Hormone Orthologue, and Kisspeptin in the Hypothalamus of Abadeh Ecotype Does during Breeding and Anoestrous Seasons. J. Neuroendocrinol. 2014, 26, 186–194. [Google Scholar] [CrossRef]
- Berisha, B.; Thaqi, G.; Schams, D.; Rodler, D.; Sinowatz, F.; Pfaffl, M.W. Effect of the Gonadotropin Surge on Steroid Receptor Regulation in Preovulatory Follicles and Newly Formed Corpora Lutea in the Cow. Domest. Anim. Endocrinol. 2024, 89, 106876. [Google Scholar] [CrossRef] [PubMed]
- Pramod, R.K.; Sharma, S.K.; Singhi, A.; Pan, S.; Mitra, A. Differential Ovarian Morphometry and Follicular Expression of BMP15, GDF9 and BMPR1B Influence the Prolificacy in Goat. Reprod. Domest. Anim. 2013, 48, 803–809. [Google Scholar] [CrossRef]
- Zhou, H.; Gong, H.; Li, S.; Luo, Y.; Hickford, J.G.H. A 57-Bp Deletion in the Ovine KAP6-1 Gene Affects Wool Fibre Diameter. J. Anim. Breed. Genet. 2015, 132, 301–307. [Google Scholar] [CrossRef]
- Plowman, J.E. Diversity of Trichocyte Keratins and Keratin Associated Proteins. Adv. Exp. Med. Biol. 2018, 1054, 21–32. [Google Scholar] [CrossRef]
- Shaobin, L.; Qiming, X.; Fangfang, Z.; Jiqing, W.; Zhaohua, H.; Jiang, H.; Xiu, L.; Yuzhu, L. Short Communication: A Highly Polymorphic Caprine Keratin-Associated Protein Gene Identified and Its Effect on Cashmere Traits. J. Anim. Sci. 2021, 99, skab233. [Google Scholar] [CrossRef]
- Zhao, M.; Zhou, H.; Hickford, J.G.H.; Gong, H.; Wang, J.; Hu, J.; Liu, X.; Li, S.; Hao, Z.; Luo, Y. Variation in the Caprine Keratin-Associated Protein 15-1 (KAP15-1) Gene Affects Cashmere Fibre Diameter. Arch. Anim. Breed. 2019, 62, 125–133. [Google Scholar] [CrossRef]
- Oudelaar, A.M.; Higgs, D.R. The Relationship between Genome Structure and Function. Genetics 2021, 22, 154–168. [Google Scholar] [CrossRef] [PubMed]
- Demir, E.; Ceccobelli, S.; Bilginer, U.; Pasquini, M.; Attard, G.; Karsli, T. Conservation and Selection of Genes Related to Environmental Adaptation in Native Small Ruminant Breeds: A Review. Ruminants 2022, 2, 255–270. [Google Scholar] [CrossRef]
- Costa, W.K.A.; Souza, E.L.; Beltrão-Filho, E.; Kelly, G.; Vasconcelos, S.G.; Santi-Gadelha, T.; de Almeida Gadelha, C.A.; Franco, O.L.; Magnani, M. Comparative Protein Composition Analysis of Goat Milk Produced by the Alpine and Saanen Breeds in Northeastern Brazil and Related Antibacterial Activities. PLoS ONE 2014, 9, e93361. [Google Scholar] [CrossRef] [PubMed]
- Hooper, H.B.; Silva, P.D.S.; de Oliveira, S.A.; Merighe, G.K.F.; Titto, C.G.; Negrão, J.A. Long-Term Heat Stress at Final Gestation: Physiological and Heat Shock Responses of Saanen Goats. Int. J. Biometeorol. 2021, 65, 2123–2135. [Google Scholar] [CrossRef] [PubMed]
- Suyasa, I.N.; Suardana, I.W.; Putra, I.G.A.A.; Suryani, N.N. Phenotype and Genotype of Boerka Goats Raised in Bali. Vet. World 2023, 16, 912–917. [Google Scholar] [CrossRef]
- Zhi, W.; Tang, K.; Yang, J.; Yang, T.; Chen, R.; Huang, J.; Tan, H.; Zhao, J.; Sheng, Z. Research on the Gut Microbiota of Hainan Black Goat. Animals 2022, 12, 3129. [Google Scholar] [CrossRef]
- Eom, J.S.; Choi, Y.; Lee, S.J.; Kim, H.S.; Jo, S.U.; Bae, D.; Lim, D.H.; Kim, E.T.; Kim, S.B.; Lee, S.S. Integrated Analysis of Rumen Metabolomics and Metataxonomics to Understand Changes in Metabolic and Microbial Community in Korean Native Goats under Heat Stress. Sci. Rep. 2024, 14, 31416. [Google Scholar] [CrossRef]
- Yao, Y.; Zhou, X.; Hadiatullah, H.; Li, C.; Wang, X.; Wang, S. Effects of Human, Caprine, and Bovine Milk Fat Globules on Microbiota Adhesion and Gut Microecology. J. Agric. Food Chem. 2021, 69, 9778–9787. [Google Scholar] [CrossRef]
- Choshniak, I.; Ben-Kohav, N.; Taylor, C.R.; Robertshaw, D.; Shkolnik, A. Metabolic Adaptations for Desert Survival in the Bedouin Goat. Am. J. Physiol. 1995, 268, R1101–R1110. [Google Scholar] [CrossRef]
- Wu, D.D.; Yang, C.P.; Wang, M.S.; Dong, K.Z.; Yan, D.W.; Hao, Z.Q.; Fan, S.Q.; Chu, S.Z.; Shen, Q.S.; Jiang, L.P.; et al. Convergent Genomic Signatures of High-Altitude Adaptation among Domestic Mammals. Natl. Sci. Rev. 2020, 7, 952–963. [Google Scholar] [CrossRef]
- Jin, M.; Lu, J.; Fei, X.; Lu, Z.; Quan, K.; Liu, Y.; Chu, M.; Di, R.; Wei, C.; Wang, H. Selection Signatures Analysis Reveals Genes Associated with High-Altitude Adaptation in Tibetan Goats from Nagqu, Tibet. Animals 2020, 10, 1599. [Google Scholar] [CrossRef] [PubMed]
- Laguë, S.L.; Ivy, C.M.; York, J.M.; Dawson, N.J.; Chua, B.A.; Alza, L.; Scott, G.R.; McCracken, K.G.; Milsom, W.K. Gas Exchange, Oxygen Transport and Metabolism in High-Altitude Waterfowl. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2025, 380, 20230424. [Google Scholar] [CrossRef]
- Renzheng, J.; Ciren, B. Comparison on Several Hematologic Value of Goat in Tibet Plateau at Different Altitude, Southwest China. Southwest China J. Agr. Sci. 1992, 1, 014. [Google Scholar]
- Zhao, B.; Wu, C.; Sammad, A.; Ma, Z.; Suo, L.; Wu, Y.; Fu, X. The Fiber Diameter Traits of Tibetan Cashmere Goats Are Governed by the Inherent Differences in Stress, Hypoxic, and Metabolic Adaptations: An Integrative Study of Proteome and Transcriptome. BMC Genom. 2022, 23, 191. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Suo, L.; Wu, Y.; Chen, T.; Tulafu, H.; Lu, Q.; Liu, W.; Sammad, A.; Wu, C.; Fu, X. Stress Adaptation in Tibetan Cashmere Goats Is Governed by Inherent Metabolic Differences and Manifested through Variable Cashmere Phenotypes. Genomics 2024, 116, 110801. [Google Scholar] [CrossRef]
- Kumar, P.; Bharti, V.K.; Kumar, K. Effect of Short-term Exposure to High-altitude Hypoxic Climate on Feed-intake, Blood Glucose Level and Physiological Responses. Int. J. Biometeorol. 2024, 68, 795–806. [Google Scholar] [CrossRef]
- do Prado Paim, T.; Borges, B.O.; de Mello Tavares Lima, P.; Gomes, E.F.; Dallago, B.S.L.; Fadel, R.; de Menezes, A.M.; Louvandini, H.; Canozzi, M.E.A.; Barcellos, J.O.J.; et al. Thermographic Evaluation of Climatic Conditions on Lambs from Different Genetic Groups. Int. J. Biometeorol. 2013, 57, 59–66. [Google Scholar] [CrossRef]
- Wang, F.H.; Zhang, L.; Gong, G.; Yan, X.C.; Zhang, L.T.; Zhang, F.T.; Liu, H.F.; Lv, Q.; Wang, Z.Y.; Wang, R.J.; et al. Genome-Wide Association Study of Fleece Traits in Inner Mongolia Cashmere Goats. Anim. Genet. 2021, 52, 375–379. [Google Scholar] [CrossRef]
- Zhao, L.; Li, M.; Xiao, M.; Chen, M.; Yang, H.; Zhao, Y. A Role for miRNAs in the Regulation of Brown Adipose Tissue Whitening in Goats (Capra hircus). J. Anim. Sci. 2024, 102, skae124. [Google Scholar] [CrossRef]
- Jiang, T.; Su, D.; Liu, X.; Wang, Y.; Wang, L. Transcriptomic Analysis Reveals Fibroblast Growth Factor 11 (FGF11) Role in Brown Adipocytes in Thermogenic Regulation of Goats. Int. J. Mol. Sci. 2023, 24, 10838. [Google Scholar] [CrossRef]
- Liu, X.; Tang, J.; Zhang, R.; Zhan, S.; Zhong, T.; Guo, J.; Wang, Y.; Cao, J.; Li, L.; Zhang, H. Cold Exposure Induces Lipid Dynamics and Thermogenesis in Brown Adipose Tissue of Goats. BMC Genom. 2022, 23, 528. [Google Scholar] [CrossRef]
- Su, D.; Song, Y.; Li, D.; Yang, S.; Zhan, S.; Zhong, T.; Guo, J.; Cao, J.; Li, L.; Zhang, H. Cold Exposure Affects Glucose Metabolism, Lipid Droplet Deposition and Mitophagy in Skeletal Muscle of Newborn Goats. Domest. Anim. Endocrinol. 2024, 88, 106847. [Google Scholar] [CrossRef] [PubMed]
- Coloma-García, W.; Mehaba, N.; Such, X.; Caja, G.; Salama, A.A.K. Effects of Cold Exposure on Some Physiological, Productive, and Metabolic Variables in Lactating Dairy Goats. Animals 2020, 10, 2383. [Google Scholar] [CrossRef] [PubMed]
- Be, K.E.; Ehrlenbruch, R. Thermoregulatory Behavior of Dairy Goats at Low Temperatures and the Use of Outdoor Yards. Can. J. Anim. Sci. 2013, 93, 35–41. [Google Scholar] [CrossRef]
- Heyman, B. Antibody Feedback Regulation. Immunol. Rev. 2024, 328, 126–142. [Google Scholar] [CrossRef]
- Bhattacharyya, N.D.; Feng, C.G. Regulation of T Helper Cell Fate by TCR Signal Strength. Front. Immunol. 2020, 11, 624. [Google Scholar] [CrossRef]
- Adebayo, M.; Singh, S.; Singh, A.; Dasgupta, S. Mitochondrial Fusion and Fission: The Fine-tune Balance for Cellular Homeostasis. FASEB J. 2021, 35, e21620. [Google Scholar] [CrossRef]
- Wen, F.; Gao, J.; Zhang, G.; Guo, S.; Zhang, X.; Han, S.; Feng, X.; Chen, X.; Hu, J. ROS-DRP1-Mediated Excessive Mitochondrial Fission and Autophagic Flux Inhibition Contribute to Heat Stress-Induced Apoptosis in Goat Sertoli Cells. J. Anim. Sci. Biotechnol. 2025, 16, 58. [Google Scholar] [CrossRef]
- Feng, R.; Zhao, J.; Zhang, Q.; Zhu, Z.; Zhang, J.; Liu, C.; Zheng, X.; Wang, F.; Su, J.; Ma, X. Generation of Anti-Mastitis Gene-Edited Dairy Goats with Enhancing Lysozyme Expression by Inflammatory Regulatory Sequence Using ISDra2-TnpB System. Adv. Sci. 2024, 11, e2404408. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Qi, Y.; Cao, Y.; Zhang, X.; Wang, Y.; Liu, Q.; Zhang, J.; Zhou, G.; Ai, Y.; Wei, S.; et al. Domain Fusion TLR2-4 Enhances the Autophagy-Dependent Clearance of Staphylococcus Aureus in the Genetic Engineering Goat. eLife 2022, 11, e78044. [Google Scholar] [CrossRef]
- Zhan, B.; Fadista, J.; Thomsen, B.; Hedegaard, J.; Panitz, F.; Bendixen, C. Global Assessment of Genomic Variation in Cattle by Genome Resequencing and High-Throughput Genotyping. BMC Genom. 2011, 12, 557. [Google Scholar] [CrossRef] [PubMed]
- Cozzi, P.; Manunza, A.; Ramirez-Diaz, J.; Tsartsianidou, V.; Gkagkavouzis, K.; Peraza, P.; Johansson, A.M.; Arranz, J.J.; Freire, F.; Kusza, S.; et al. SMARTER-Database: A Tool to Integrate SNP Array Datasets for Sheep and Goat Breeds. GigaByte 2024, 2024, gigabyte139. [Google Scholar] [CrossRef]
- Peng, W.; Zhang, Y.; Gao, L.; Wang, S.; Liu, M.; Sun, E.; Lu, K.; Zhang, Y.; Li, B.; Li, G. Investigation of Selection Signatures of Dairy Goats Using Whole-Genome Sequencing Data. BMC Genom. 2025, 26, 234. [Google Scholar] [CrossRef] [PubMed]
- Joost, S.; Kalbermatten, M.; Bonin, A. Spatial Analysis Method (SAM): A Software Tool Combining Molecular and Environmental Data to Identify Candidate Loci for Selection. Mol. Ecol. Resour. 2010, 8, 957–960. [Google Scholar] [CrossRef]
- Frichot, E.; Schoville, S.D.; Bouchard, G.; Olivier, F. Testing for Associations between Loci and Environmental Gradients Using Latent Factor Mixed Models. Mol. Biol. Evol. 2013, 30, 1687–1699. [Google Scholar] [CrossRef]
- Racimo, F. Testing for Ancient Selection Using Cross-Population Allele Frequency Differentiation. Genetics 2016, 202, 733–750. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Zhang, W.; Zhang, H.; Yang, X.; Ceccobelli, S.; Zhao, Y.; E, G. Identification of Goat Supernumerary Teat Phenotype Using Wide-Genomic Copy Number Variants. Animals 2024, 14, 3252. [Google Scholar] [CrossRef]
- Burak, Y.; Davide, M.; Ludovica, M.; Rodrigo, F.; Mayukh, M.; Luca, P. Improving Selection Detection with Population Branch Statistic on Admixed Populations. Genome Biol. Evol. 2021, 13, evab039. [Google Scholar] [CrossRef]
- Pegolo, S.; Bisutti, V.; Lfm, M.; Cecchinato, A.; Amalfitano, N.; Dettori, M.L.; Pazzola, M.; Vacca, G.M.; Bittante, G. Genome-Wide Landscape of Genetic Diversity, Runs of Homozygosity, and Runs of Heterozygosity in Five Alpine and Mediterranean Goat Breeds. J. Anim. Sci. Biotechnol. 2025, 16, 33. [Google Scholar] [CrossRef] [PubMed]
- Sheidai, M.; Malekmohammadi, L.; Ghahremaninejad, F.; Danehkar, A.; Koohdar, F. A New Computational Method to Estimate Adaptation Time in Avicennia by Using Divergence Time. Sci. Rep. 2024, 14, 24158. [Google Scholar] [CrossRef]
- Berthouly, C.; Ngoc, D.D.; Thevenon, S.; Bouchel, D.; Maillard, J.C. How Does Farmer Connectivity Influence Livestock Genetic Structure? A Case-Study in a Vietnamese Goat Population. Mol. Ecol. 2010, 18, 3980–3991. [Google Scholar] [CrossRef]
- Chen, Y.; Li, R.; Sun, J.; Li, C.; Xiao, H.; Chen, S. Genome-Wide Population Structure and Selection Signatures of Yunling Goat Based on RAD-Seq. Animals 2022, 12, 2401. [Google Scholar] [CrossRef]
- Peng, W.; Zhang, Y.; Gao, L.; Shi, W.; Liu, Z.; Guo, X.; Zhang, Y.; Li, B.; Li, G.; Cao, J. Selection Signatures and Landscape Genomics Analysis to Reveal Climate Adaptation of Goat Breeds. BMC Genom. 2024, 25, 420. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.X.; Asadollahpour Nanaei, H.; Jafarpour Negari, N.; Amiri Roudbar, M.; Amiri Ghanatsaman, Z.; Niyazbekova, Z.; Yang, X.J. Genomic Analysis Uncovers Novel Candidate Genes Related to Adaptation to Tropical Climates and Milk Production Traits in Native Goats. BMC Genom. 2024, 25, 477. [Google Scholar] [CrossRef]
- Yadav, V.P.; Dangi, S.S.; Chouhan, V.S.; Gupta, M.; Dangi, S.K.; Singh, G.; Maurya, V.P.; Kumar, P.; Sarkar, M. Expression Analysis of NOS Family and HSP Genes during Thermal Stress in Goat (Capra hircus). Int. J. Biometeorol. 2016, 60, 381–389. [Google Scholar] [CrossRef] [PubMed]
- Chebii, V.J.; Oyola, S.O.; Kotze, A.; Entfellner, J.B.D.; Mutuku, J.M.; Agaba, M. Genome-Wide Analysis of Nubian Ibex Reveals Candidate Positively Selected Genes That Contribute to Its Adaptation to the Desert Environment. Animals 2020, 10, 2181. [Google Scholar] [CrossRef]
- Tian, D.; Zhou, B.; Han, B.; Liu, S.; Tian, F.; Qi, D.; Zhao, K. Deep Genome Sequencing Provides Potential Novel Insights into Plateau Adaptations in Domestication of Goats to Extreme Environments. Res. Sq. 2021. [Google Scholar] [CrossRef]
- Kim, E.S.; Elbeltagy, A.R.; Aboul-Naga, A.M.; Rischkowsky, B.; Sayre, B.; Mwacharo, J.M.; Rothschild, M.F. Multiple Genomic Signatures of Selection in Goats and Sheep Indigenous to a Hot Arid Environment. Heredity 2016, 116, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Onzima, R.B.; Upadhyay, M.R.; Doekes, H.P.; Brito, L.F.; Bosse, M.; Kanis, E.; Groenen, M.A.M.; Crooijmans, R.P.M.A. Genome-Wide Characterization of Selection Signatures and Runs of Homozygosity in Ugandan Goat Breeds. Front. Genet. 2018, 9, 318. [Google Scholar] [CrossRef]
- Song, S.; Yao, N.; Yang, M.; Liu, X.; Dong, K.; Zhao, Q.; Pu, Y.; He, X.; Guan, W.; Yang, N.; et al. Exome Sequencing Reveals Genetic Differentiation Due to High-Altitude Adaptation in the Tibetan Cashmere Goat (Capra hircus). BMC Genom. 2016, 17, 122. [Google Scholar] [CrossRef]
- Zargar, R.; Urwat, U.; Malik, F.; Shah, R.A.; Bhat, M.H.; Naykoo, N.A.; Khan, F.; Khan, H.M.; Ahmed, S.M.; Vijh, R.K.; et al. Molecular Characterization of RNA Binding Motif Protein 3 (RBM3) Gene from Pashmina Goat. Res. Vet. Sci. 2015, 98, 51–58. [Google Scholar] [CrossRef]
- Zhao, L.; Yang, H.; Li, M.; Xiao, M.; Li, X.; Cheng, L.; Cheng, W.; Chen, M.; Zhao, Y. Global Gene Expression Profiling of Perirenal Brown Adipose Tissue Whitening in Goat Kids Reveals Novel Genes Linked to Adipose Remodeling. J. Anim. Sci. Biotechnol. 2024, 15, 47. [Google Scholar] [CrossRef]
- Wang, X.; Liu, J.; Zhou, G.; Guo, J.; Yan, H.; Niu, Y.; Li, Y.; Yuan, C.; Geng, R.; Lan, X. Whole-Genome Sequencing of Eight Goat Populations for the Detection of Selection Signatures Underlying Production and Adaptive Traits. Sci. Rep. 2016, 6, 38932. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yang, L.; Lin, X.; Peng, P.; Shen, W.; Tang, S.; Lan, X.; Wan, F.; Yin, Y.; Liu, M. Effects of Genetic Variation of the Sorting Nexin 29 (SNX29) Gene on Growth Traits of Xiangdong Black Goat. Animals 2022, 12, 3461. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Wang, Z.; Sun, J.; Huang, Y.; Lei, C. Identification of Genomic Characteristics and Selective Signals in a Du’an Goat Flock. Animals 2020, 10, 994. [Google Scholar] [CrossRef]
- Sasazaki, S.; Tomita, K.; Nomura, Y.; Kawaguchi, F.; Kunieda, T.; Shah, M.K.; Mannen, H. FGF5 and EPAS1 Gene Polymorphisms Are Associated with High-Altitude Adaptation in Nepalese Goat Breeds. Anim. Sci. J. 2021, 92, e13640. [Google Scholar] [CrossRef] [PubMed]
- Kumar, C.; Song, S.; Jiang, L.; He, X.; Zhao, Q.; Pu, Y.; Malhi, K.K.; Kamboh, A.A.; Ma, Y. Sequence Characterization of DSG3 Gene to Know Its Role in High-Altitude Hypoxia Adaptation in the Chinese Cashmere Goat. Front. Genet. 2018, 9, 553. [Google Scholar] [CrossRef]
- Kalds, P.; Crispo, M.; Li, C.; Tesson, L.; Anegón, I.; Chen, Y.; Wang, X.; Menchaca, A. Generation of Double-Muscled Sheep and Goats by CRISPR/Cas9-Mediated Knockout of the Myostatin Gene. Methods Mol. Biol. 2022, 2495, 295–323. [Google Scholar] [CrossRef]
- Donkin, I.; Barrès, R. Sperm Epigenetics and Influence of Environmental Factors. Mol. Metab. 2018, 14, 1–11. [Google Scholar] [CrossRef]
- Berger, S.L.; Kouzarides, T.; Shiekhattar, R.; Shilatifard, A. An Operational Definition of Epigenetics. Genes Dev. 2009, 23, 781–783. [Google Scholar] [CrossRef]
- Gamal, L.; Noshy, M.M.; Aboul-Naga, A.M.; Sabit, H.; El-Shorbagy, H.M. DNA Methylation of GDF-9 and GHR Genes as Epigenetic Regulator of Milk Production in Egyptian Zaraibi Goat. Genes Genom. 2024, 46, 135–148. [Google Scholar] [CrossRef]
- Yang, C.; He, J.; Mao, J.; Ren, Y.; Liu, G.; Wei, C.; Zhang, G.; Tian, K.; Huang, X. Genome-Wide DNA Methylation Analysis and Functional Validation of Litter Size Traits in Jining Grey Goats. Genes 2024, 15, 353. [Google Scholar] [CrossRef]
- Yun, M.; Wu, J.; Workman, J.L.; Li, B. Readers of Histone Modifications. Cell Res. 2011, 21, 564–578. [Google Scholar] [CrossRef] [PubMed]
- Pei, X.; Li, H.; Yu, H.; Wang, W.; Mao, D. APN/AdipoRon Regulates Luteal Steroidogenesis through AMPK/EZH2/H3K27me3 in Goats. J. Steroid Biochem. Mol. Biol. 2025, 247, 106653. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Mattick, J.S. Structure and Function of Long Noncoding RNAs in Epigenetic Regulation. Nat. Struct. Mol. Biol. 2013, 20, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Chao, T.; Wang, Y.; Xuan, R.; Guo, Y.; He, P.; Zhang, L.; Wang, J. Transcriptome Analysis Revealed the Characteristics and Functions of Long Non-Coding RNAs in the Hypothalamus during Sexual Maturation in Goats. Front. Vet. Sci. 2024, 11, 1404681. [Google Scholar] [CrossRef]
- Mao, S.; Dong, S.; Hou, B.; Li, Y.; Sun, B.; Guo, Y.; Deng, M.; Liu, D.; Liu, G. Transcriptome Analysis Reveals Pituitary lncRNA, circRNA and mRNA Affecting Fertility in High- and Low-Yielding Goats. Front. Genet. 2023, 14, 1303031. [Google Scholar] [CrossRef] [PubMed]
- Ainash, C.; Goodrich, J.M.; Fabiola, L.V.; Maria, R.C.; Melisa, K.; Brutsaert, T.D.; Dolinoy, D.C.; Bigham, A.W. Genome-Wide Epigenetic Signatures of Adaptive Developmental Plasticity in the Andes. Genome Biol. Evol. 2021, 13, evaa239. [Google Scholar] [CrossRef]
- Yan, Z.; Yang, J.; Wei, W.T.; Zhou, M.L.; Mo, D.X.; Wan, X.; Ma, R.; Wu, M.M.; Huang, J.H.; Liu, Y.J. A Time-Resolved Multi-Omics Atlas of Transcriptional Regulation in Response to High-Altitude Hypoxia across Whole-Body Tissues. Nat. Commun. 2024, 15, 3970. [Google Scholar] [CrossRef]
- Dan, H.; Zhong, H.; Akhatayeva, Z.; Lin, K.; Xu, S. Whole-Genome Selective Scans Detect Genes Associated with Cashmere Traits and Climatic Adaptation in Cashmere Goats (Capra hircus) in China. Genes 2025, 16, 292. [Google Scholar] [CrossRef]
- Laquatra, C.; Sanchez-Martin, C.; Minervini, G.; Moroni, E.; Rasola, A. HIF1α-Dependent Induction of the Mitochondrial Chaperone TRAP1 Regulates Bioenergetic Adaptations to Hypoxia. Cell Death Dis. 2021, 12, 434. [Google Scholar] [CrossRef]
- Benjelloun, B.; Leempoel, K.; Boyer, F.; Stucki, S.; Streeter, I.; Orozco-terWengel, P.; Alberto, F.J.; Servin, B.; Biscarini, F.; Alberti, A.; et al. Multiple Genomic Solutions for Local Adaptation in Two Closely Related Species (Sheep and Goats) Facing the Same Climatic Constraints. Mol. Ecol. 2024, 33, e17257. [Google Scholar] [CrossRef]
- Li, C.; Wu, Y.; Chen, B.; Cai, Y.; Guo, J.; Leonard, A.S.; Kalds, P.; Zhou, S.; Zhang, J.; Zhou, P.; et al. Markhor-Derived Introgression of a Genomic Region Encompassing PAPSS2 Confers High-Altitude Adaptability in Tibetan Goats. Mol. Biol. Evol. 2022, 39, msac253. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Ma, J.; Long, K.; Zhang, J.; Qiu, W.; Li, Y.; Jin, L.; Wang, X.; Jiang, A.; Liu, L.; et al. Comparative microRNA Transcriptomes in Domestic Goats Reveal Acclimatization to High Altitude. Front. Genet. 2020, 11, 809. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhang, X.; Zhaxi, Y.; Pan, C.; Zhang, Z.; Pan, J.; Shahzad, K.; Sun, F.; Zhen, Y.; Jinmei, J.; et al. Integrative Multi-Omics Analysis Reveals Liver-Gut Axis Adaptation in High-Altitude Goats. Comp. Biochem. Physiol. Part D Genom. Proteom. 2025, 54, 101422. [Google Scholar] [CrossRef] [PubMed]
- Velayudhan, S.M.; Sejian, V.; Devaraj, C.; Manjunathareddy, G.B.; Ruban, W.; Kadam, V.; König, S.; Bhatta, R. Novel Insights to Assess Climate Resilience in Goats Using a Holistic Approach of Skin-Based Advanced NGS Technologies. Int. J. Mol. Sci. 2023, 24, 10319. [Google Scholar] [CrossRef]
| Breed | Genome Versions | Sequencing Technology | Genome Size/Gb | Assembling Evaluation Metrics | Time | Reference | ||
|---|---|---|---|---|---|---|---|---|
| Contig N50/Mb | BUSCO | Number of Gaps | ||||||
| Yunnan black goat | CHIR_1.0 | Illumina | 2.8 | 0.15 | 95% | 256,764 | 2013 | [14] |
| Capra aegagrus | CapAeg_1.0 | Illumina | 2.8 | 0.15 | 90.7% | 279,195 | 2015 | [19] |
| Black Bengal goat | CVASU_BBG_1.0 | Illumina | 3.0 | 26.2 | 82.5% | 3943 | 2019 | [20] |
| Saanen dairy goat | Saanen_v1 | Illumina, PacBio, Hi-C | 2.7 | 46.2 | 98.3% | 169 | 2020 | [21] |
| San Clemente Island goat | ARS1 | Illumina, PacBio, Hi-C | 2.9 | 18.70 | 96% | 649 | 2022 | [16] |
| Bore Goat | ASM4458790v1 | Illumina, PacBio HiFi, Hi-C | 2.9 | 34.22 | 95.6% | 165 | 2024 | [22] |
| Nubian Goat | ASM4458793v1 | PacBio HiFi | 2.9 | 72.12 | 95.6% | 119 | 2024 | [22] |
| Inner Mongolia cashmere goat | T2T-goat1.0 | PacBio HiFi, Ultra-long ONT, Bionano and Hi-C | 2.9 | 100.8 | 98.0% | 0 | 2024 | [18] |
| Tibetan goat | ASM4646407v1 | Illumina, PacBio, Hi-C | 2.9 | 97.6 | 96.4% | 243 | 2025 | [7] |
| Category | Statistical Methods | Detection Signal | Underlying Model | Individual/ Population Data | Reference |
|---|---|---|---|---|---|
| Population Differentiation | XP-CLR | Selective sweeps with allele frequency shifts | Composite Likelihood Model | population | [75] |
| Fst | Allele frequency divergence between populations | Wright’s Island Model | population | [76] | |
| Haplotype and LD-based Methods | XP-EHH | Completed selective sweeps | Extended Haplotype Homozygosity (EHH) Model | individual | [77] |
| ROH | Artificial selection/demographic bottlenecks | Homozygosity Segment-based Analysis | individual | [78] | |
| Environment Association Analysis | LFMM | Correlation between allele frequency and environment | Latent Factor Mixed Model (LFMM) | individual | [79] |
| SAM | Genetic structure coupled with geographic space | Spatial Statistics Models (sPCA) | individual | [80] |
| Adaptation Conditions | Species | Methods | Gene/Protein | Function | References |
|---|---|---|---|---|---|
| Tropical | Native Goats of Pakistan | SAM, Fst, θπ, | KITLG, HSPB9, HSP70, HSPA12B, NBEA | Thermotolerance | [83] |
| Barbari goat | RT-PCR, Western Blot, Immunocytochemistry | iNOS, eNOS, cNOS, HSP70, HSP90 | Improves heat stress and maintains cellular integrity and homeostasis in goats | [84] | |
| 130 domesticated species (Abergelle, Garganica, etc.) | Fst, XPEHH, LFMM, SAM | WDR75, SCN7A, PLCB1 | Thermotolerance | [82] | |
| Desert | Iraq goat and Pakistan goat | Fst, θπ and Tajima’s D statistics | KITLG | Fur color change | [6] |
| Capra nubiana | WGS, dN/dS ratio analysis, Fst | ABCA12, ASCL4, UVSSA | Participates in skin barrier protection | [85] | |
| Qaidam cashmere goats | WGS, Fst, Tajima’s D statistics | CNGA4, Camk2b | Enhance immune system function and resist adverse external factors | [86] | |
| Barki goats | Fst, iHS | FGF2, GNAI3, PLCB1 | Heat resistance and melanin production | [87] | |
| Ugandan goat | Fst, hapFLK, ROH | MTOR, MAPK3, HOXC12, IGF1, KPNA4, PPP1R36 | Involvement in the FAS pathway and regulation of HSP stress-induced heat tolerance | [88] | |
| High altitude | Tibetan goat | Exome sequencing, SAM, Fst | EPAS1, SIRT1, ICAM1, EDNRA, YES1 | Regulation of O2 utilization, inflammatory response, hemodynamics and cellular signaling | [89] |
| Pashmina goat, Bakerwal goat | qRT-PCR, Whole-length amplification | RBM3 | Activation of expression under low-temperature stress conditions | [90] | |
| Chinese indigenous goats | SNP Detection | DSG3 | Mutation site polymorphism is strongly associated with hypoxia adaptation | [50] | |
| Dazu black goat | Thermographic evaluation, RNA-seq | UCP1, CIDEA, PPARGC1a | Regulation of BAT thermogenesis in goats | [91] | |
| Tibetan goat | iHS, ZHp, di | CDK2, SOCS2, NOXA1, ENPEP | Hypoxia adaptation | [92] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, Y.; Ma, R.; Li, D.; Gao, Y.; Sheng, Z.; Shi, J.; Peng, Y.; Gao, Z.; Deng, W.; He, X. Genomic and Molecular Mechanisms of Goat Environmental Adaptation. Biology 2025, 14, 654. https://doi.org/10.3390/biology14060654
Lu Y, Ma R, Li D, Gao Y, Sheng Z, Shi J, Peng Y, Gao Z, Deng W, He X. Genomic and Molecular Mechanisms of Goat Environmental Adaptation. Biology. 2025; 14(6):654. https://doi.org/10.3390/biology14060654
Chicago/Turabian StyleLu, Ying, Ruoshan Ma, Dongfang Li, Yuyang Gao, Zhengmei Sheng, Jinpeng Shi, Yilong Peng, Zhengdong Gao, Weidong Deng, and Xiaoming He. 2025. "Genomic and Molecular Mechanisms of Goat Environmental Adaptation" Biology 14, no. 6: 654. https://doi.org/10.3390/biology14060654
APA StyleLu, Y., Ma, R., Li, D., Gao, Y., Sheng, Z., Shi, J., Peng, Y., Gao, Z., Deng, W., & He, X. (2025). Genomic and Molecular Mechanisms of Goat Environmental Adaptation. Biology, 14(6), 654. https://doi.org/10.3390/biology14060654





