Morphological and Phylogenetic Analysis of a New Jellyfish of Phyllorhiza (Scyphozoa, Mastigiidae) from the East China Sea
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
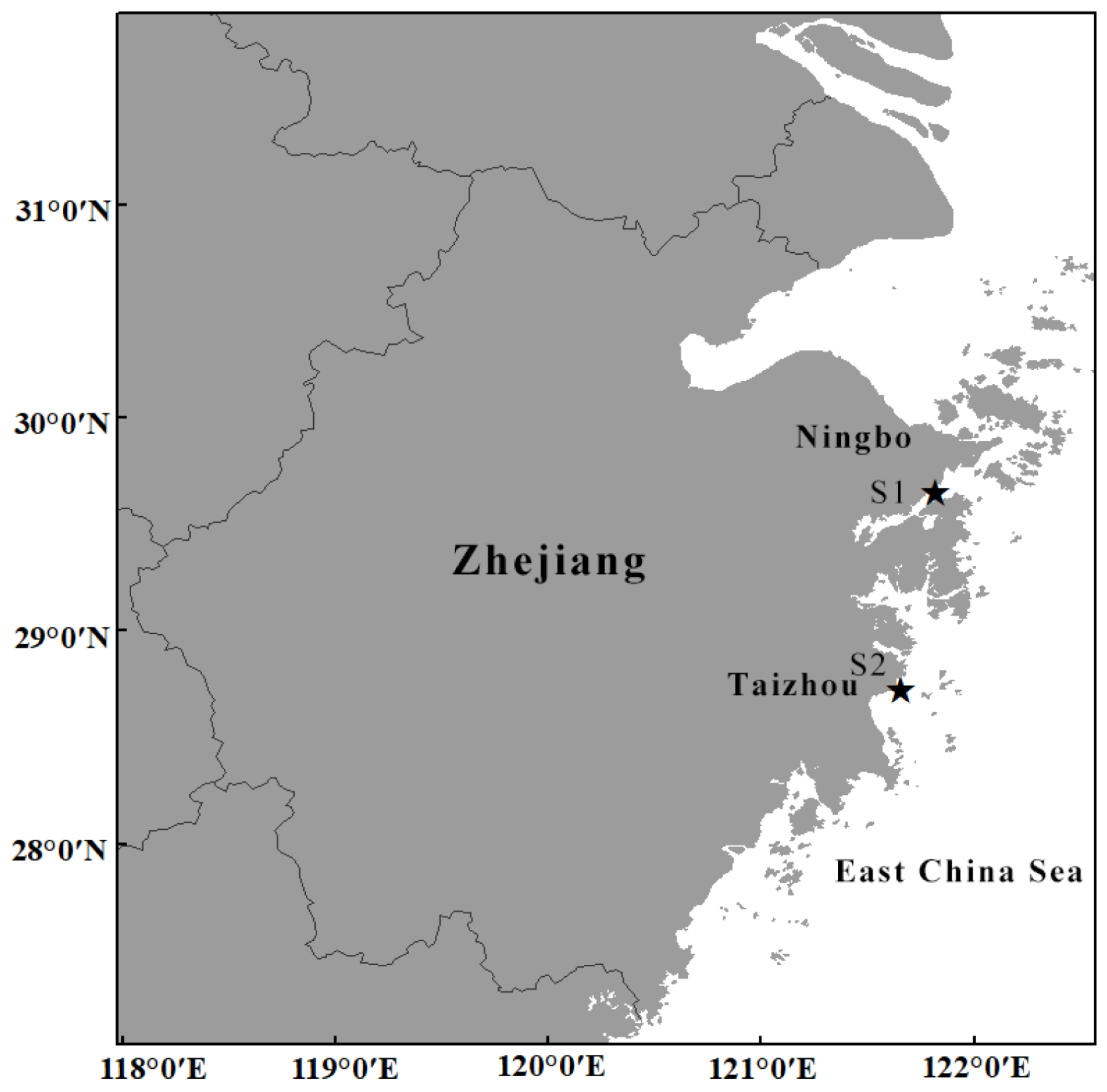
2.1. Sample Collection and Morphological Examination
2.2. DNA Extraction, Amplification and Sequencing
2.3. Genetic Data Analysis
| Markers | Primer | Sequense | PCR Conditions | Reference | Length (bp) | Model (BI) | Model (ML) |
|---|---|---|---|---|---|---|---|
| 16S | 16S-CB1 | 5′-TCGACTGTTTACCAAAAACATAGC-3′ | 33–35 cycles (94 °C for 45 s, 50–52 °C for 45 s, and 72 °C for 60 s) and 72 °C for 300 s | [24] | ≈640 | GTR+F+G4 | TIM2+F+G4. |
| 16S-CB2 | 5′-ACGGAATGAACTCAAATCATGTAAG-3′ | ||||||
| COI | LCO1490 | 5′-GGTCAACAAATCATAAAGATATTGG-3′ | 33–35 cycles (94 °C for 45 s, 50–55 °C for 45–60 s, and 72 °C for 60 s) and 72 °C for 600 s | [25] | ≈700 | GTR+F+G4 | TIM2+F+G4 |
| HCO2198 | 5′-TAAACTTCAGGGTGACCAAAAAATCA-3′ | ||||||
| COXIF | 5′-GTATTTTCTCTGGCGTACTAGGTGC-3′ | present study | ≈640 | ||||
| COXIR | 5′-ATAAATGCTGATATAAGATGGGGTC-3′ | ||||||
| 28S | Cassiopea 28S F | 5’-GRCGGCGAATTGTAGTCTCGA-3’ | 38 cycles (94 °C for 45 s, 47–55 °C for 60–90 s, and 72 °C for 70–90 s) and 72 °C for 600 s | [13] | ≈1010 | GTR+F+G4 | TN+F+G4 |
| Aa H28S 1039 | 5’-GTCTTTCGCCCCTATACCCA-3’ | ||||||
| 18S | Aa L18S 12 | 5’-TCCTGCCAGTAGTCATATGCTTG-3’ | 38 cycles (94 °C for 45–50 s, 47–54 °C for 70 s, and 72 °C for 70–90 s) and 72 °C for 600 s | [26] | ≈1770 | HKY+F+I | HKY+F+I |
| Aa H 18S 1798 | 5′-CCTACGGAAACCTTGTTACGA-3′ | ||||||
| Aa H18S 1318 FC | 5′-CAGACAAATCACTCCACCAAC-3′ | ||||||
| Aa H18S 1318 RC | 5′-GTTGGTGGAGTGATTTGTCTG-3′ |
| GenBank Accession No. | |||||||
|---|---|---|---|---|---|---|---|
| Species | Voucher/Isolate No. | Location | COI | 16S | 18S | 28S | Reference |
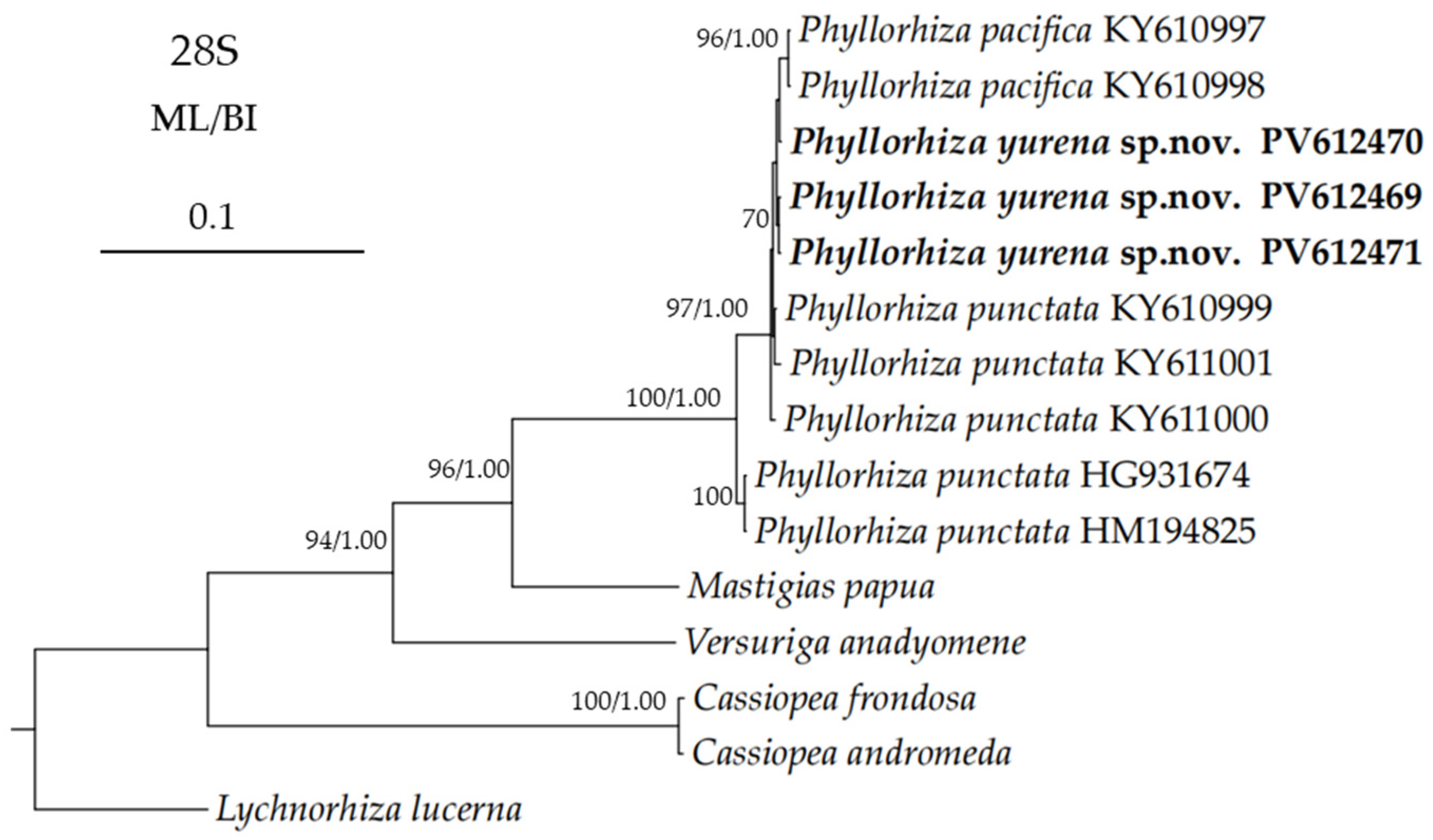
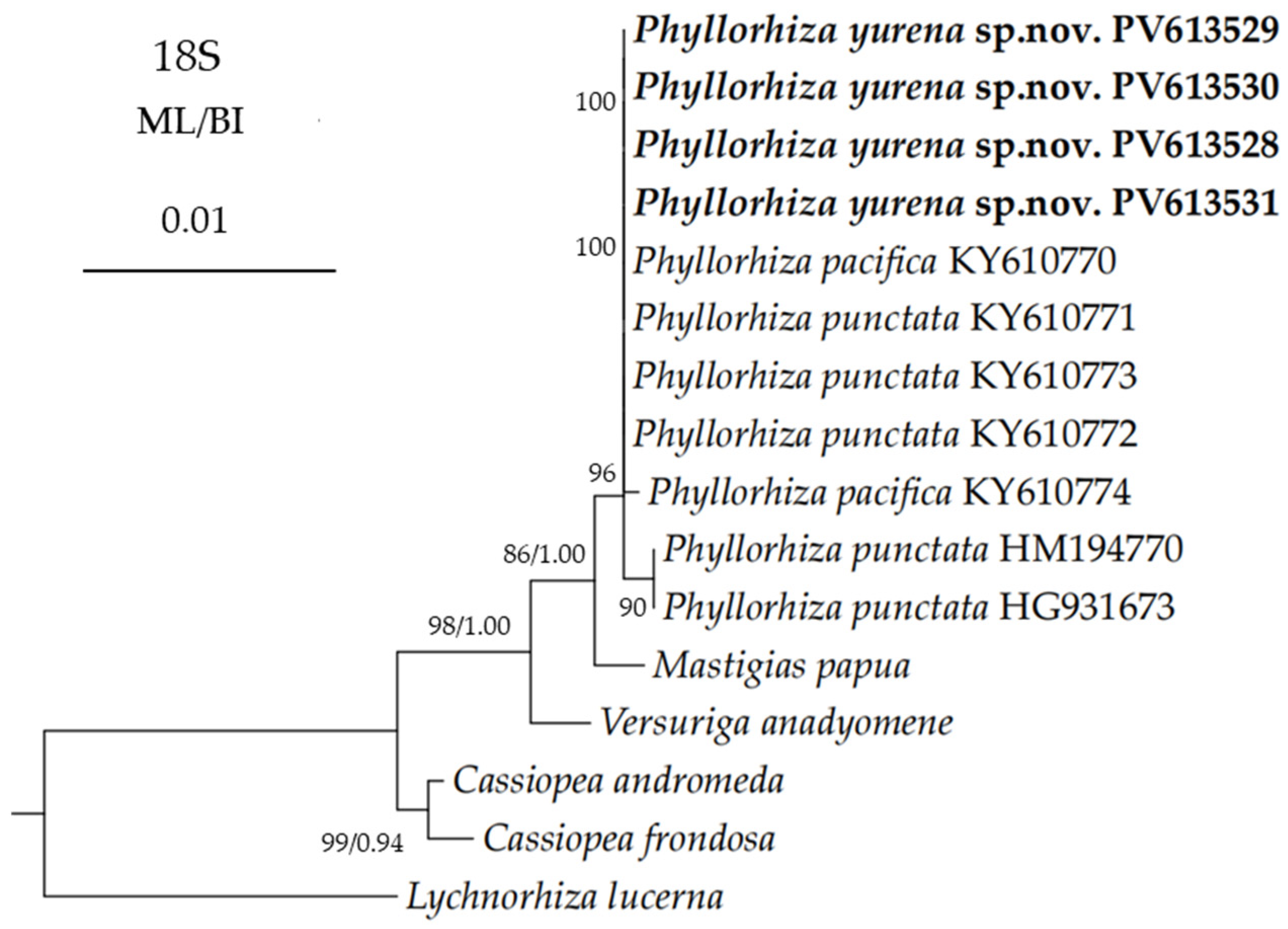
| Phyllorhiza yurena sp. nov. | TIO-SRMP001 | East Sea of China | PV366411 | PV367414 | PV613531 | PV612471 | present study |
| Phyllorhiza yurena sp. nov. | TIO-SRMP002 | East Sea of China | PV366405 | PV367408 | PV613528 | PV612469 | present study |
| Phyllorhiza yurena sp. nov. | NB-2023003 | East Sea of China | PV366406 | PV367409 | – | – | present study |
| Phyllorhiza yurena sp. nov. | NB-2023004 | East Sea of China | PV366408 | PV367410 | PV613529 | PV612470 | present study |
| Phyllorhiza yurena sp. nov. | NB-2023005 | East Sea of China | PV366409 | PV367411 | – | – | present study |
| Phyllorhiza yurena sp. nov. | NB-2023006 | East Sea of China | PV366407 | PV367412 | PV613530 | – | present study |
| Phyllorhiza yurena sp. nov. | NB-2023007 | East Sea of China | PV366410 | PV367413 | – | – | present study |
| Phyllorhiza pacifica | 12198 | Bangladesh: Teknaf | PP945789 | – | – | – | Khanam et al., unpublished |
| Phyllorhiza pacifica | M0D022675C_THKRKOB | Thailand | – | KY610622 | – | – | [13] |
| Phyllorhiza pacifica | M0D022673A_THKRKOP | Thailand | – | KY610623 | KY610770 | KY610998 | [13] |
| Phyllorhiza pacifica | M0D022675C_THKRKOB | Thailand | – | – | KY610774 | KY610997 | [13] |
| Phyllorhiza cf. pacifica | M0D21426B_IDJISUY | Indonesia | MN395673 | – | – | – | [27] |
| Phyllorhiza punctata | – | Israel | – | HG931681 | – | – | [28] |
| Phyllorhiza punctata | NO.5-16S | Thailand | – | KT982716 | – | – | [29] |
| Phyllorhiza punctata | PAZ072019_1 | Mexico | MT899235 | MT902932 | – | – | [12] |
| Phyllorhiza punctata | PAZ072019_2 | Mexico | MT904380 | MT902935 | – | – | [12] |
| Phyllorhiza punctata | M0D00662L | Australia | – | – | HM194770 | – | [26] |
| Phyllorhiza punctata | M0D014781M_MXBSAGO | Mexico | KY611062 | KY610625 | KY610773 | KY610999 | [27] |
| Phyllorhiza punctata | M0D0147830_MXBSCPC | Mexico | KY611060 | KY610626 | KY610771 | KY611000 | [27] |
| Phyllorhiza punctata | M0D014780L_MXBSMAG | Mexico | KY611061 | KY610627 | KY610772 | KY611001 | [27] |
| Phyllorhiza punctata | M0D00662L | Australia | – | KY610624 | – | HM194825 | [26,27] |
| Phyllorhiza punctata | Sc18.1.1 | Gulf of Mexico | GQ120101 | – | – | – | [30] |
| Phyllorhiza punctata | 2_S8GB | Australia | EU363342 | – | – | – | [31] |
| Phyllorhiza punctata | 1_S8GB | Australia | EU363341 | – | – | – | [31] |
| Phyllorhiza punctata | PPMJ4 | Malaysia | JN203010 | JN202945 | – | – | [32] |
| Phyllorhiza punctata | Phy | Malaysia | – | JN202946 | – | – | [32] |
| Phyllorhiza punctata | PPKS0612 | Malaysia | JN203009 | – | – | – | [32] |
| Phyllorhiza punctata | PPKS0912 | Malaysia | JN203008 | – | – | – | [32] |
| Phyllorhiza punctata | PPKS1012 | Malaysia | JN203007 | – | – | – | [32] |
| Phyllorhiza punctata | PPKS0712 | Malaysia | JN203006 | – | – | – | [32] |
| Phyllorhiza punctata | PPKS0512 | Malaysia | JN203005 | – | – | – | [32] |
| Phyllorhiza punctata | PPKS0412 | Malaysia | JN203004 | – | – | – | [32] |
| Phyllorhiza punctata | PPKS0112 | Malaysia | JN203003 | – | – | – | [32] |
| Phyllorhiza punctata | PPPP04 | Malaysia | JN203002 | – | – | – | [32] |
| Phyllorhiza punctata | PPPP03 | Malaysia | JN203001 | – | – | – | [32] |
| Phyllorhiza punctata | PPPP01 | Malaysia | JN203000 | – | – | – | [32] |
| Phyllorhiza punctata | PPKS0110 | Malaysia | JN202999 | – | – | – | [32] |
| Phyllorhiza punctata | PPKS0310 | Malaysia | JN202998 | – | – | – | [32] |
| Phyllorhiza punctata | M0D013181Y | Australia | KU900939 | KU901025 | – | – | [33] |
| Phyllorhiza punctata | M0D013180X | Australia | KU900938 | KU901024 | – | – | [33] |
| Phyllorhiza punctata | – | Eastern Mediterranean | – | – | HG931673 | HG931674 | [28] |
| Phyllorhiza punctata | – | Gulf of Mexico | – | JX393272 | – | – | [34] |
| Phyllorhiza punctata | – | Singapore | OR400205 | OR400205 | – | – | [35] |
| Phyllorhiza punctata | – | Singapore | OR400201 | OR400201 | – | – | [35] |
| Phyllorhiza punctata | – | Singapore | NC_084193 | NC_084193 | – | – | [35] |
| Lychnorhiza lucerna | M0D016088T_NIANGBW | Nicaragua | KY611034 | KY610591 | KY610785 | KY610906 | [13] |
| Cassiopea frondosa | M0D021382J_PABTBDE | Panama | KY610557 | KY610615 | KY610767 | KY611004 | [13] |
| Cassiopea andromeda | M0D006024R_MXBCISJ | Mexico | KY610551 | KY610609 | KY610763 | KY611005 | [13] |
| Mastigias papua | M0D06000T | Palau | – | – | HM194796 | HM194849 | [33] |
| Mastigias papua | M0D005915M | Palau | KU901397 | KU901021 | – | – | [33] |
| Versuriga anadyomene | M0D00095Q | Palau | – | – | HM194768 | HM194823 | [33] |
| Versuriga anadyomene | - | South China Sea | KX904853 | KX904852 | – | – | [36] |
3. Results
3.1. Systematics
3.2. Genetic Distance and Phylogenetic Analyses
4. Discussion
4.1. Morphological Differences Between the New Species and Congeners
4.2. DNA Barcoding Analysis of Phyllorhiza
4.3. Key to the Species of the Genus Phyllorhiza
- 1.
- Umbrella broad and flat, mouth arms slender, strongly compressed ……………………………………………………………………………………………………………‥Phyllorhiza luzoni Mayer, 1915
- -
- Umbrella hemispherical ……………………………………………………………………………………………………………………………………………………………………………………………………2
- 2.
- Marginal lappets marked darker streaks, the disk is papillous or tuberculated, and the tubercles are larger toward the summit of the umbrella ………………………………………………………………………………………………………………………………………………………………………‥‥‥.Phyllorhiza chinensis L. Agassiz, 1862
- -
- Marginal lappets without darker streaks …………………………………………………………………………………………………………………………………………………………………………………3
- 3.
- Marginal lappets with 24 very long filaments …………………………………………………………………………………………………………………………………‥Phyllorhiza trifolium Haeckel, 1880
- -
- Marginal lappets without filaments ………………………………………………………………………………………………………………………………………………………………………………………4
- 4.
- Terminal appendages with a distal swelling ……………………………………………………………………………………………………………………………………………………………………………5
- -
- Terminal appendages without a distal swelling, umbrella flat-hemispherical and up to 400 mm wide, exumbrella with numerous minute brown spots, terminal appendages nearly as long as the mouth arms; predominantly purple ………………………………………………………………………………………………………………………………………………Phyllorhiza pacifica (Light, 1921)
- 5.
- Umbrella up to 600 mm wide, jelly very thick, exumbrella with finely granular surface; marginal lappets some broad and double, others simple, altogether up to 14 in each octant; arm-disk with numerous filaments; lower parts of mouth arms with a terminal white club-shaped appendage, some of which up to two-thirds as long as the mouth arms themselves …………………………………………………………………………………………………………………………………………………………………‥Phyllorhiza punctata von Lendenfeld, 1884
- -
- Umbrella up to 450 mm wide, exumbrella with white spots and warts; with numerous short, club-shaped filamentous appendages on the lower parts of each mouth arm; mouth arms three-winged, and mouthlets occupy two-thirds the length of mouth arms ……………………………………………………………………………………………………………………………………………………………………….Phyllorhiza yurena Chen, Hu & Xing sp. nov.
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Richardson, A.J.; Bakun, A.; Hays, G.C.; Gibbons, M.J. The jellyfish joyride: Causes, consequences and management responses to a more gelatinous future. Trends Ecol. Evol. 2009, 24, 312–322. [Google Scholar] [CrossRef] [PubMed]
- Chi, X.; Dierking, J.; Hoving, H.J.; Lüskow, F.; Denda, A.; Christiansen, B.; Sommer, U.; Hansen, T.; Javidpour, J. Tackling the jelly web: Trophic ecology of gelatinous zooplankton in oceanic food webs of the eastern tropical Atlantic assessed by stable isotope analysis. Limnol. Oceanogr. 2021, 66, 289–305. [Google Scholar] [CrossRef]
- Sigurdsson, G.M.; Lüskow, F.; Gislason, A.; Svavarsson, J. Detached tentacles of lion’s mane jellyfish Cyanea capillata can injure aquaculture fish. Aquac. Environ. Interact. 2024, 16, 263–266. [Google Scholar] [CrossRef]
- Condon, R.H.; Lucas, C.H.; Pitt, K.A.; Uye, S.-i. Jellyfish blooms and ecological interactions. Mar. Ecol. Prog. Ser. 2014, 510, 109–110. [Google Scholar] [CrossRef]
- Lucas, C.H.; Jones, D.O.; Hollyhead, C.J.; Condon, R.H.; Duarte, C.M.; Graham, W.M.; Robinson, K.L.; Pitt, K.A.; Schildhauer, M.; Regetz, J. Gelatinous zooplankton biomass in the global oceans: Geographic variation and environmental drivers. Glob. Ecol. Biogeogr. 2014, 23, 701–714. [Google Scholar] [CrossRef]
- Holst, S.; Laakmann, S. Morphological and molecular discrimination of two closely related jellyfish species, Cyanea capillata and C. lamarckii (Cnidaria, Scyphozoa), from the northeast Atlantic. J. Plankton Res. 2014, 36, 48–63. [Google Scholar] [CrossRef]
- Lüskow, F.; Bezio, N.; Caputo, L.; Chi, X.; Dumont, H.J.; Karunarathne, K.D.; López-González, P.J.; Mańko, M.K.; Marchessaux, G.; Suzuki, K.S. Hidden gems: Scattered knowledge hampered freshwater jellyfish research over the past one-and-a-half centuries. Ecol. Evol. 2024, 14, e70350. [Google Scholar] [CrossRef]
- Dong, Z.; Morandini, A.C.; Schiariti, A.; Wang, L.; Sun, T. First record of Phyllorhiza sp.(Cnidaria: Scyphozoa) in a Chinese coastal aquaculture pond. PeerJ 2019, 7, e6191. [Google Scholar] [CrossRef]
- Bayha, K.M.; Graham, W.M. First confirmed reports of the rhizostome jellyfish Mastigias(Cnidaria: Rhizostomeae) in the Atlantic basin. Aquat. Invasions 2011, 6, 361–366. [Google Scholar] [CrossRef]
- Goldstein, J.; Steiner, U.K. Ecological drivers of jellyfish blooms–The complex life history of a ‘well-known’medusa (Aurelia aurita). J. Anim. Ecol. 2020, 89, 910–920. [Google Scholar] [CrossRef]
- Lucas, C.H.; Graham, W.M.; Widmer, C. Jellyfish life histories: Role of polyps in forming and maintaining scyphomedusa populations. Adv. Mar. Biol. 2012, 63, 133–196. [Google Scholar] [CrossRef] [PubMed]
- Rosales-Catalán, L.; Estrada-González, M.C.; Rivera-Pérez, C.; Sanchez, M.A.R.; Gamero-Mora, E.; Morandini, A.C.; Mendoza-Becerril, M.A. Genetic and morphological evidence of the presence of Phyllorhiza punctata in the southwestern Gulf of California (NE Pacific Ocean). Aquat. Invasions 2021, 16, 637–652. [Google Scholar] [CrossRef]
- Daglio, L.G.; Dawson, M.N. Species richness of jellyfishes (Scyphozoa: Discomedusae) in the Tropical Eastern Pacific: Missed taxa, molecules, and morphology match in a biodiversity hotspot. Invertebr. Syst. 2017, 31, 635–663. [Google Scholar] [CrossRef]
- Gambill, M.; Jarms, G. Can Aurelia (Cnidaria, Scyphozoa) species be differentiated by comparing their scyphistomae and ephyrae? Eur. J. Taxon. 2014, 107, 1–23. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.; MrBayes, J.H. 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Xiang, C.Y.; Gao, F.; Jakovlić, I.; Lei, H.P.; Hu, Y.; Zhang, H.; Zou, H.; Wang, G.T.; Zhang, D. Using PhyloSuite for molecular phylogeny and tree-based analyses. Imeta 2023, 2, e87. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Hillis, D.M.; Bull, J.J. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 1993, 42, 182–192. [Google Scholar] [CrossRef]
- Alfaro, M.E.; Zoller, S.; Lutzoni, F. Bayes or bootstrap? A simulation study comparing the performance of Bayesian Markov chain Monte Carlo sampling and bootstrapping in assessing phylogenetic confidence. Mol. Biol. Evol. 2003, 20, 255–266. [Google Scholar] [CrossRef]
- Collins, A.G.; Bentlage, B.; Lindner, A.; Lindsay, D.; Haddock, S.H.D.; Jarms, G.; Norenburg, J.L.; Jankowski, T.; Cartwright, P. Phylogenetics of Trachylina (Cnidaria: Hydrozoa) with new insights on the evolution of some problematical taxa. J. Mar. Biol. Assoc. UK 2008, 88, 1673–1685. [Google Scholar] [CrossRef]
- Folmer, R.; Nilges, M.; Folkers, P.; Konings, R.; Hilbers, C. A model of the complex between single-stranded DNA and the single-stranded DNA binding protein encoded by gene V of filamentous bacteriophage M13. J. Mol. Biol. 1994, 240, 341–357. [Google Scholar] [CrossRef]
- Bayha, K.M.; Dawson, M.N. New family of allomorphic jellyfishes, Drymonematidae (Scyphozoa, Discomedusae), emphasizes evolution in the functional morphology and trophic ecology of gelatinous zooplankton. Biol. Bull. 2010, 219, 249–267. [Google Scholar] [CrossRef]
- Gómez Daglio, L.; Hayati, R.; Coleman, T.; Han, Y.-M.; Muzaki, F.; Aunurohim; de Bellard, M.E.; Saptarini, D. Species composition of Discomedusae jellyfish (Scyphozoa) in the coastal waters of Eastern Surabaya, East Java. Mar. Biodivers. 2022, 52, 23. [Google Scholar] [CrossRef]
- Mizrahi, G. Phylogenetic Analysis of Gelatinous Marine Fauna in the Eastern Mediterranean Basin: An Ecosystem Under Anthropogenic Stress; University of Haifa: Haifa, Israel, 2014. [Google Scholar]
- Ruijuan, L.; Jie, X.; Xuelei, Z.; Aungtonya, C. Genetic analysis of common venomous Cubozoa and Scyphozoa in Thailand waters. Acta Oceanol. Sin. 2016, 38, 51–61. (In Chinese) [Google Scholar]
- Ortman, B.D.; Bucklin, A.; Pagès, F.; Youngbluth, M. DNA barcoding the Medusozoa using mtCOI. Deep Sea Res. Part II Top. Stud. Oceanogr. 2010, 57, 2148–2156. [Google Scholar] [CrossRef]
- Daryanabard, R.; Dawson, M.N. Jellyfish blooms: Crambionella orsini (Scyphozoa: Rhizostomeae) in the gulf of Oman, Iran, 2002–2003. J. Mar. Biol. Assoc. UK 2008, 88, 477–483. [Google Scholar] [CrossRef]
- Rizman-Idid, M.; Farrah-Azwa, A.B.; Chong, V.C. Preliminary taxonomic survey and molecular documentation of jellyfish species (Cnidaria: Scyphozoa and Cubozoa) in Malaysia. Zool. Stud. 2016, 55, e35. [Google Scholar] [PubMed]
- Swift, H.F.; Daglio, L.G.; Dawson, M. Three routes to crypsis: Stasis, convergence, and parallelism in the Mastigias species complex (Scyphozoa, Rhizostomeae). Mol. Phylogenet. Evol. 2016, 99, 103–115. [Google Scholar] [CrossRef]
- Sparmann, S.F. Contributions to the Molecular Phylogeny, Phylogeography, and Taxonomy of Scyphozoan Jellyfish. Master’s Thesis, University of British Columbia, Vancouver, BC, USA, 2012. [Google Scholar]
- Ling, M.K.; Yap, N.W.L.; Iesa, I.B.; Yip, Z.T.; Huang, D.; Quek, Z.B.R. Revisiting mitogenome evolution in Medusozoa with eight new mitochondrial genomes. Iscience 2023, 26, 108252. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Dong, Z.; Li, Y. Versuriga anadyomene, a newly recorded scyphozoan jellyfish (Scyphozoa: Rhizostomae) in Chinese waters. J. Oceanol. Limnol. 2019, 37, 266–272. [Google Scholar] [CrossRef]
- Collins, A.G.; Morandini, A.C.; Cartwright, P. World List of Scyphozoa. Phyllorhiza Agassiz. 1862. Accessed Through: World Register of Marine Species. Available online: https://www.marinespecies.org/aphia.php?p=taxdetails&id=135256 (accessed on 6 March 2025).
- Light, S. Further notes on Philippine scyphomedusan jellyfishes. Philipp. J. Sci. 1921, 18, 25–47. [Google Scholar]
- Martellos, S.; Ukosich, L.; Avian, M. JellyWeb: An interactive information system on Scyphozoa, Cubozoa and Staurozoa. ZooKeys 2016, 554, 1–25. [Google Scholar] [CrossRef]
- Mayor, A.G. Medusae of the Philippines and of Torres Straits; Carnegie Institution of Washington: Washington, DC, USA, 1915. [Google Scholar]
- Jarms, G.; Morandini, A.; Schmidt-Rhaesa, A.; Giere, O.; Straehler-Pohl, I. World atlas of jellyfish. Bhandlungen des Naturwissenschaftlichen Vereins in Hamburg; Dölling und Galitz Verlag: Hamburg, Germany, 2019. [Google Scholar]
- Mayer, A.G. The Scyphomedusae; Carnegie Institution of Washington: Washington, DC, USA, 1910; Volume 3, pp. 56–76. [Google Scholar]
- Arsiranant, I.; Xiao, J.; Sriboonyuang, P.; Jamsawang, N.; Pengchumrus, W.; Thongbai, N.; Aungtonya, C.; Detsri, U.; Kootsomboon, P.; Zhang, X. The Genus Phyllorhiza (Rhizostomeae: Mastigiidae) in Coastal Waters of the Andaman Sea and the Gulf of Thailand. Phuket Mar. Biol. Cent. Res. Bull. 2020, 77, 61–76. [Google Scholar] [CrossRef]
- Agassiz, L. Contributions to the Natural History of the United States of America; Little, Brown: Boston, MA, USA, 1857; Volume 1. [Google Scholar]
- Haeckel, E. Das System der Medusen: Erster Theil Einer Monographie der Medusen. Atlas. System der Acraspeden. Zweite Hälfte der ersten Theils; G. Fischer: Schaffhausen, Switzerland, 1880. [Google Scholar]
- Lendenfeld, R. The Scyphomedusae of the Southern Hemisphere; Sydney Linnean Society of New South Wales: Manly, Australia, 1884. [Google Scholar]







| Body Proportions | Results |
|---|---|
| CDD/TBD | 49.4~55.9% |
| TMLL/TBD | 20.9~26.7% |
| RLL/TMLL | 32.3~42.8% |
| LStL/TMLL | 57.2~67.7% |
| ML/CDD | 44.1~48.5% |
| Species/Population | Location | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Phyllorhiza yurena sp. nov. PV366405-7 | East China Sea | 0 | |||||||||||||||||||||
| 2 | P. yurenasp. nov. PV366408-11 | East China Sea | 0.20% | 0 | ||||||||||||||||||||
| 3 | Phyllorhiza cf. pacifica MN395673 | Indonesia | 2.24% | 2.03% | - | |||||||||||||||||||
| 4 | P. punctata JN203007 | Malaysia | 2.03% | 1.82% | 0.60% | - | P. punctata Group I | |||||||||||||||||
| 5 | P. punctata JN203004 | Malaysia | 2.03% | 1.82% | 0.20% | 0.40% | - | |||||||||||||||||
| 6 | P. punctata JN203010, OR400205 | Malaysia, Singapore | 2.24% | 2.03% | 0.80% | 0.20% | 0.60% | 0 | ||||||||||||||||
| 7 | P. punctata JN203001-3,6,8,9 | Malaysia | 2.24% | 2.03% | 0.80% | 1.00% | 0.60% | 1.21% | 0 | |||||||||||||||
| 8 | P. punctata JN202999 | Malaysia | 1.82% | 1.62% | 0.40% | 0.20% | 0.20% | 0.40% | 0.80% | - | ||||||||||||||
| 9 | P. punctata JN203000 | Malaysia | 2.44% | 2.24% | 1.00% | 1.21% | 0.80% | 1.41% | 0.60% | 1.00% | - | |||||||||||||
| 10 | P. punctata JN203005 | Malaysia | 2.24% | 2.03% | 1.21% | 1.41% | 1.00% | 1.62% | 0.80% | 1.21% | 0.20% | - | ||||||||||||
| 11 | P. punctata OR400201, NC_084193 | Singapore | 10.00% | 10.24% | 11.21% | 10.97% | 10.97% | 10.97% | 10.72% | 10.72% | 11.46% | 11.21% | - | Group II | ||||||||||
| 12 | P. punctata KY611060-2, MT899235, MT904380 | Mexico | 9.74% | 9.98% | 10.95% | 10.70% | 10.70% | 10.70% | 10.46% | 10.46% | 11.19% | 10.95% | 1.00% | 0 | ||||||||||
| 13 | P. punctata GQ120101 | Mexico | 9.52% | 9.76% | 10.72% | 10.48% | 10.48% | 10.48% | 10.24% | 10.24% | 10.97% | 10.72% | 0.80% | 0.20% | - | |||||||||
| 14 | P. punctata EU363342 | Australia | 9.01% | 9.25% | 10.20% | 9.96% | 9.96% | 9.96% | 10.20% | 9.72% | 10.44% | 10.20% | 4.99% | 4.77% | 4.56% | - | Group III | |||||||
| 15 | P. punctata EU363341 | Australia | 8.78% | 9.01% | 9.96% | 9.72% | 9.72% | 9.72% | 9.96% | 9.48% | 10.20% | 9.96% | 4.78% | 4.55% | 4.34% | 0.20% | - | |||||||
| 16 | P. punctata KU900938, KU900939 | Australia | 9.01% | 9.25% | 10.20% | 9.96% | 9.96% | 9.96% | 10.20% | 9.72% | 10.44% | 10.20% | 4.99% | 4.77% | 4.56% | 0.40% | 0.20% | 0 | ||||||
| 17 | P. pacifica PP945789 | Bangladesh | 10.46% | 10.22% | 11.19% | 11.94% | 11.44% | 11.69% | 11.69% | 11.69% | 11.94% | 11.69% | 11.91% | 11.64% | 11.42% | 11.38% | 11.13% | 10.89% | - | |||||
| 18 | Mastigias papua | Palau | 21.71% | 21.71% | 22.27% | 22.27% | 21.99% | 22.27% | 22.84% | 21.99% | 21.99% | 21.71% | 23.56% | 23.24% | 22.98% | 22.40% | 22.11% | 20.64% | 23.82% | - | ||||
| 19 | Versuriga anadyomene | Palau | 19.48% | 19.21% | 20.28% | 20.55% | 20.28% | 20.55% | 20.55% | 20.28% | 20.83% | 20.55% | 21.19% | 20.62% | 20.37% | 20.64% | 20.37% | 22.40% | 18.18% | 18.73% | - | |||
| 20 | Cassiopea andromeda | Mexico | 22.14% | 22.14% | 22.14% | 22.70% | 22.42% | 22.98% | 21.86% | 22.42% | 22.42% | 22.70% | 22.96% | 23.51% | 23.25% | 23.86% | 23.57% | 23.86% | 26.14% | 27.97% | 23.50% | - | ||
| 21 | Cassiopea frondosa | Panama | 23.53% | 23.53% | 24.10% | 24.10% | 23.82% | 24.39% | 23.82% | 23.82% | 23.82% | 23.53% | 25.21% | 25.77% | 25.50% | 24.37% | 24.09% | 24.37% | 26.66% | 28.50% | 24.91% | 7.68% | - | |
| 22 | Lychnorhiza lucerna | Nicaragua | 28.35% | 28.35% | 29.27% | 29.58% | 29.27% | 29.90% | 28.96% | 29.27% | 28.35% | 28.65% | 31.80% | 33.08% | 32.79% | 29.87% | 29.56% | 29.56% | 28.44% | 28.96% | 29.70% | 28.37% | 27.40% | - |
| Species/Population | Location | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Phyllorhiza yurena sp. nov. PV367408-14 | East China Sea | 0 | ||||||||||||||||
| 2 | P. pacifica KY610622, KY610623 | Thailand | 1.22% | 0 | |||||||||||||||
| 3 | P. punctata JN202945 *, JN202946 * | Malaysia | 0.40% | 0 | 0 | P. punctata Group I | |||||||||||||
| 4 | P. punctata OR400205 | Singapore | 0.68% | 0.52% | 0 | - | |||||||||||||
| 5 | P. punctata KT982716 ** | Thailand | 4.68% | 5.01% | - | 4.99% | - | ||||||||||||
| 6 | P. punctata KY610625-7 | Mexico | 3.37% | 3.92% | 2.09% | 3.74% | 0 | 0 | Group II | ||||||||||
| 7 | P. punctata JX393272 | Gulf of Mexico | 3.55% | 3.93% | 2.11% | 3.92% | 0 | 0 | - | ||||||||||
| 8 | P. punctata OR400201, NC_084193 | Singapore | 3.49% | 3.92% | 2.02% | 3.85% | 0 | 0 | 0 | 0 | |||||||||
| 10 | P. punctata MT902932, MT902935 | Mexico | 3.59% | 3.99% | 1.87% | 3.98% | 0 | 0 | 0 | 0 | 0 | ||||||||
| 11 | P. punctata KU901024-5 | Australia | 4.28% | 4.67% | 2.52% | 4.65% | 0.85% | 0.69% | 0.70% | 0.69% | 0.74% | 0 | Group III | ||||||
| 12 | P. punctata KY610624 | Australia | 4.11% | 4.67% | 2.52% | 4.48% | 0.86% | 0.69% | 0.70% | 0.69% | 0.74% | 0 | - | ||||||
| 13 | P. punctata HG931681 | Israel | 4.72% | 6.00% | 2.60% | 5.34% | 2.90% | 1.74% | 2.03% | 2.03% | 2.03% | 1.44% | 1.15% | - | |||||
| 14 | Mastigias papua | Palau | 13.26% | 13.26% | 8.29% | 13.47% | 17.97% | 14.31% | 14.36% | 14.31% | 13.32% | 14.53% | 14.53% | 17.00% | - | ||||
| 15 | Lychnorhiza lucerna | Nicaragua | 17.79% | 17.79% | 11.66% | 18.02% | 22.44% | 18.66% | 18.74% | 18.66% | 18.13% | 18.66% | 18.66% | 24.24% | 20.28% | - | |||
| 16 | Cassiopea andromeda | Mexico | 20.52% | 20.56% | 10.00% | 21.00% | 29.32% | 21.72% | 21.91% | 21.91% | 21.20% | 22.39% | 22.21% | 24.49% | 19.41% | 19.23% | - | ||
| 17 | Cassiopea frondosa | Panama | 20.52% | 20.56% | 10.99% | 20.52% | 28.44% | 21.72% | 21.91% | 21.91% | 21.20% | 22.39% | 22.21% | 24.09% | 18.94% | 20.43% | 2.89% | - | |
| 18 | Versuriga anadyomene | South China Sea | 17.97% | 17.18% | 8.74% | 17.97% | 23.38% | 18.71% | 18.71% | 18.71% | 18.71% | 19.52% | 19.52% | 21.48% | 20.65% | 21.39% | 20.88% | 21.43% | - |
| Species/Population | Location | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Phyllorhiza yurena sp. nov. PV612469-71 | East China Sea | 0 | ||||||||||
| 2 | Phyllorhiza punctata KY610999 | Mexico | 0 | 0 | |||||||||
| 3 | Phyllorhiza pacifica KY610997, KY610998 | Thailand | 0.10% | 0.09% | 0 | ||||||||
| 4 | Phyllorhiza punctata KY611000 | Mexico | 0.10% | 0.09% | 0.43% | - | |||||||
| 5 | Phyllorhiza punctata KY611001 | Mexico | 0.10% | 0.09% | 0.52% | 0.26% | - | ||||||
| 6 | Phyllorhiza punctata HM194825, HG931674 | Australia/Eastern Mediterranean | 1.62% | 1.31% | 1.67% | 1.37% | 1.47% | 0 | |||||
| 7 | Mastigias papua HM194849 | Palau | 12.16% | 10.80% | 11.22% | 11.10% | 10.99% | 11.43% | - | ||||
| 8 | Versuriga anadyomene HM194823 | Palau | 16.47% | 13.85% | 14.39% | 14.06% | 14.17% | 16.06% | 12.74% | - | |||
| 9 | Cassiopea frondosa KY611004 | Panama | 20.88% | 17.43% | 17.62% | 17.47% | 17.60% | 20.13% | 18.41% | 17.05% | - | ||
| 10 | Cassiopea andromeda KY611005 | Mexico | 20.88% | 17.78% | 17.80% | 17.91% | 17.91% | 20.83% | 19.09% | 17.47% | 0.19% | - | |
| 11 | Lychnorhiza lucerna KY610906 | Nicaragua | 17.49% | 15.37% | 15.74% | 15.74% | 15.74% | 17.51% | 15.39% | 14.46% | 15.25% | 15.06% | - |
| Species/Population | Location | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Phyllorhiza yurena sp. nov. PV613528-31 | East China Sea | 0 | |||||||||
| 2 | Phyllorhiza pacifica KY610770 | Thailand | 0 | - | ||||||||
| 3 | Phyllorhiza pacifica KY610774 | Thailand | 0 | 0 | - | |||||||
| 4 | Phyllorhiza punctata KY610771-3 | Mexico | 0.06% | 0.06% | 0.06% | 0 | ||||||
| 5 | Phyllorhiza punctata HM194770, HG931673 | Australia/Eastern Mediterranean | 0.12% | 0.11% | 0.11% | 0.17% | 0 | |||||
| 6 | Mastigias papua HM194796 | Palau | 0.30% | 0.28% | 0.28% | 0.34% | 0.40% | - | ||||
| 7 | Versuriga anadyomene HM194768 | Palau | 0.60% | 0.57% | 0.57% | 0.63% | 0.68% | 0.57% | - | |||
| 8 | Cassiopea andromeda KY610763 | Mexico | 0.96% | 0.91% | 0.92% | 0.91% | 1.02% | 1.02% | 0.85% | - | ||
| 9 | Cassiopea frondosa KY610767 | Panama | 1.08% | 1.02% | 1.03% | 1.03% | 1.14% | 1.14% | 0.97% | 0.23% | - | |
| 10 | Lychnorhiza lucerna KY610785 | Nicaragua | 2.18% | 2.75% | 2.75% | 2.76% | 2.87% | 2.87% | 2.69% | 2.45% | 2.57% | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, X.; Hu, Z.; Zhan, Z.; Chen, Y.; Mu, S.; Xing, B.; Xu, K. Morphological and Phylogenetic Analysis of a New Jellyfish of Phyllorhiza (Scyphozoa, Mastigiidae) from the East China Sea. Biology 2025, 14, 632. https://doi.org/10.3390/biology14060632
Chen X, Hu Z, Zhan Z, Chen Y, Mu S, Xing B, Xu K. Morphological and Phylogenetic Analysis of a New Jellyfish of Phyllorhiza (Scyphozoa, Mastigiidae) from the East China Sea. Biology. 2025; 14(6):632. https://doi.org/10.3390/biology14060632
Chicago/Turabian StyleChen, Xiaoyin, Zhijie Hu, Zifeng Zhan, Yaojun Chen, Sirong Mu, Bingpeng Xing, and Kuidong Xu. 2025. "Morphological and Phylogenetic Analysis of a New Jellyfish of Phyllorhiza (Scyphozoa, Mastigiidae) from the East China Sea" Biology 14, no. 6: 632. https://doi.org/10.3390/biology14060632
APA StyleChen, X., Hu, Z., Zhan, Z., Chen, Y., Mu, S., Xing, B., & Xu, K. (2025). Morphological and Phylogenetic Analysis of a New Jellyfish of Phyllorhiza (Scyphozoa, Mastigiidae) from the East China Sea. Biology, 14(6), 632. https://doi.org/10.3390/biology14060632






