Porcine Ovarian piRNA Dynamics: A Comparative Study During Follicular Atresia
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Follicle Collection and Classification
2.2. Small RNA Library Preparation and Illumina Sequencing
2.3. Identification of piRNAs
2.4. Differential Expression of piRNAs
2.5. Biological Functions Analysis of Potential Target Genes
2.6. qPCR Analysis
3. Results
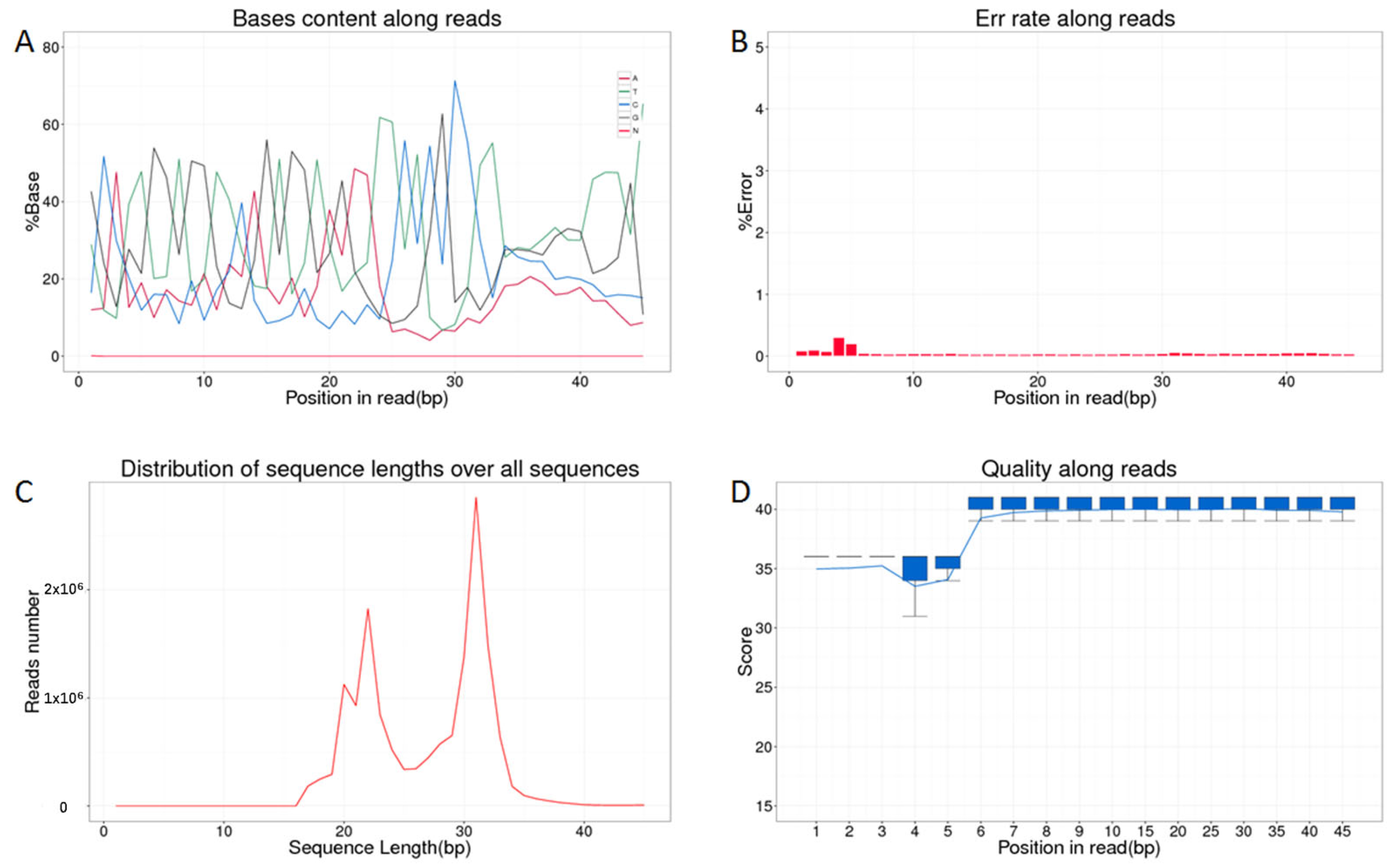
3.1. Sequencing Data Overview and Quality Control Analysis
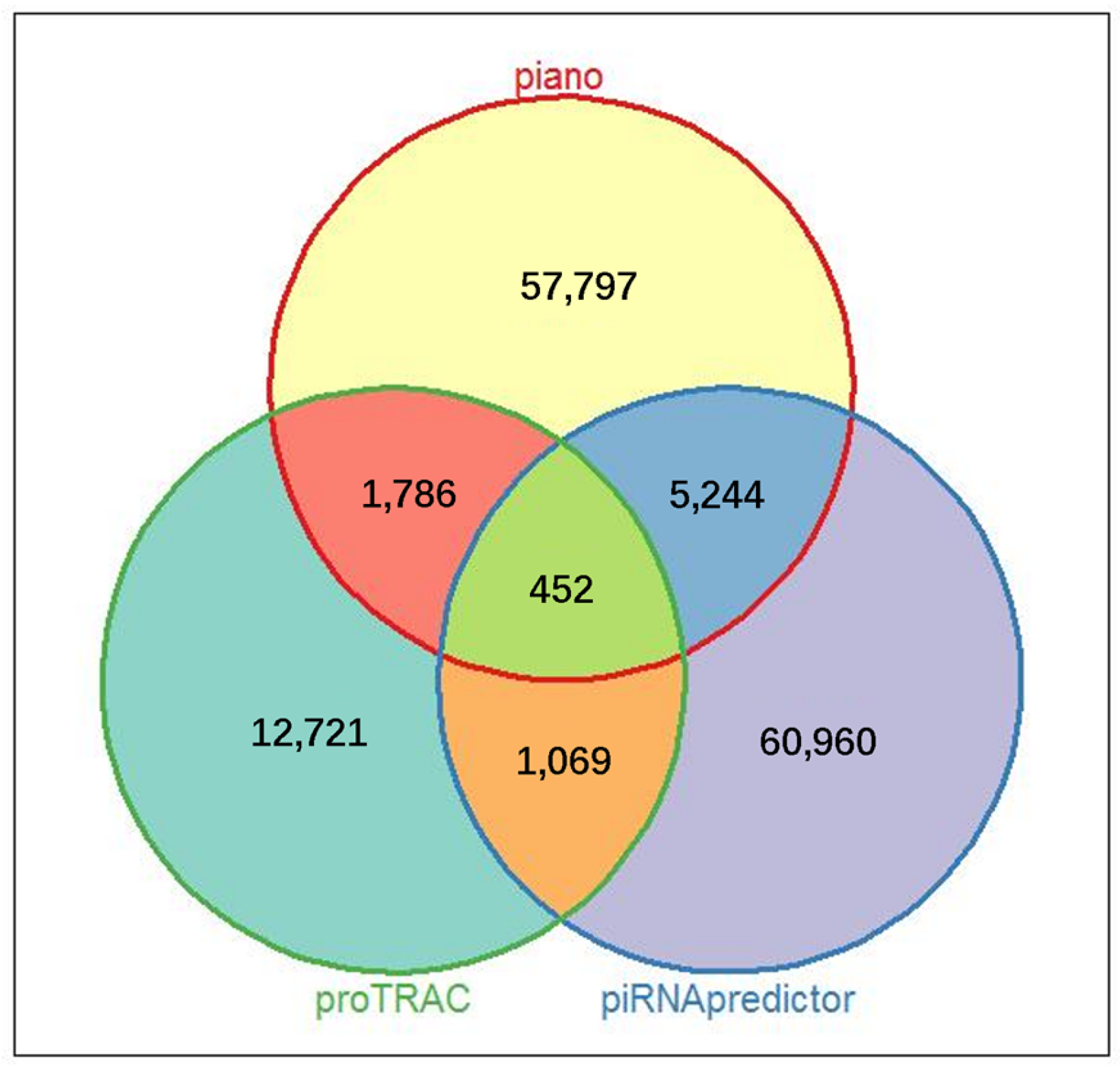
3.2. Screening of Porcine Ovarian piRNAs
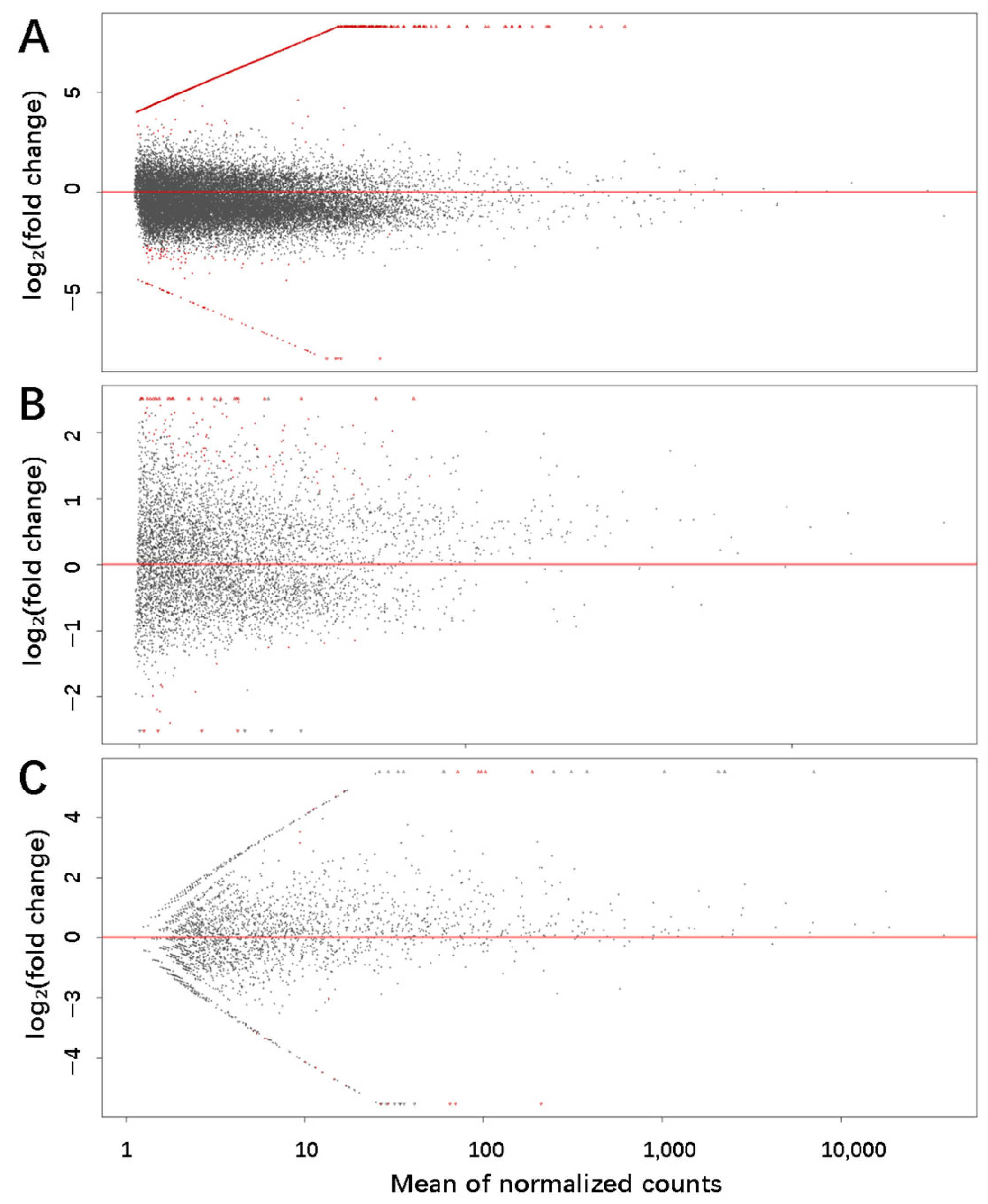
3.3. Differential Expression Profile of piRNAs in Porcine HFs and AFs
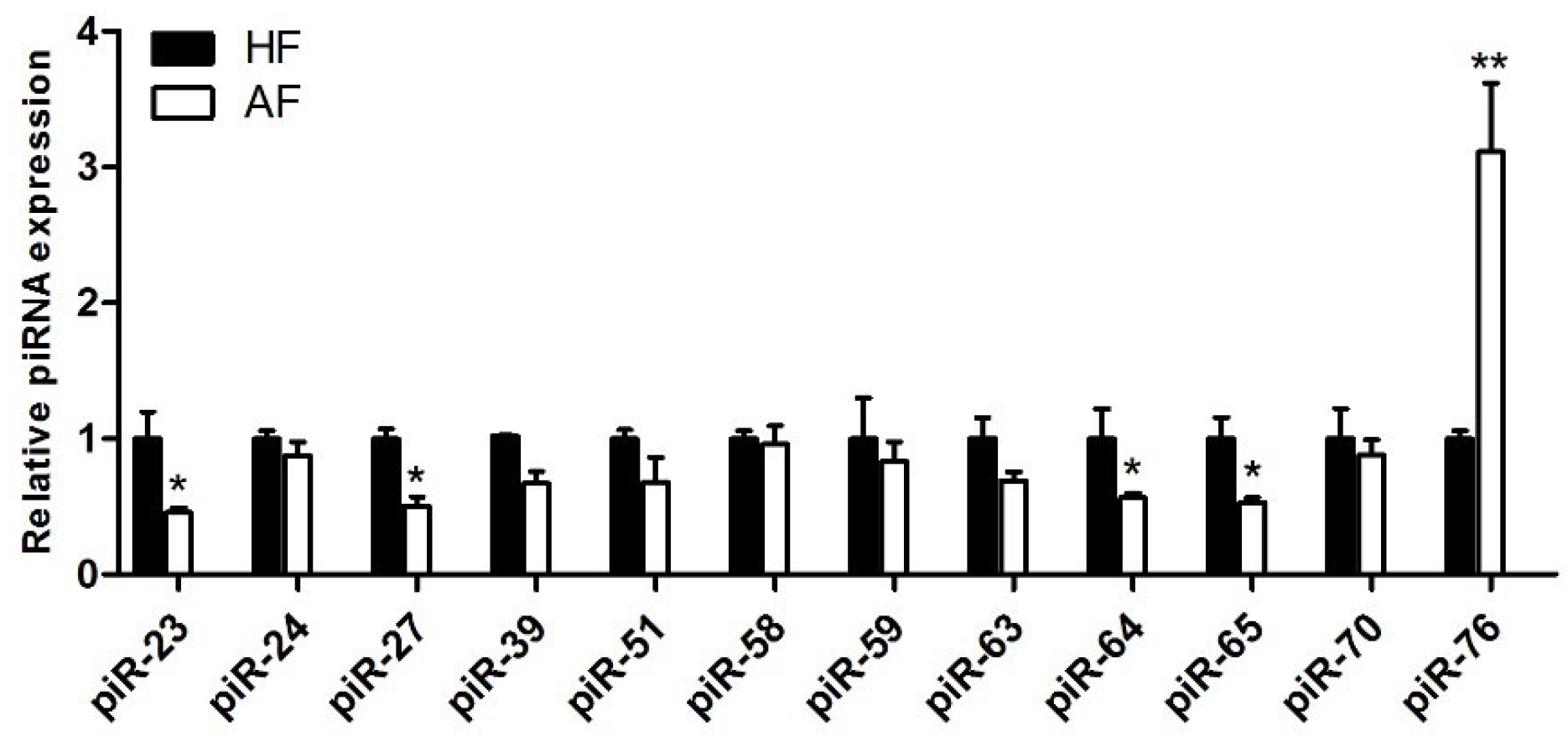
3.4. qPCR Confirmation of Differentially Expressed piRNAs
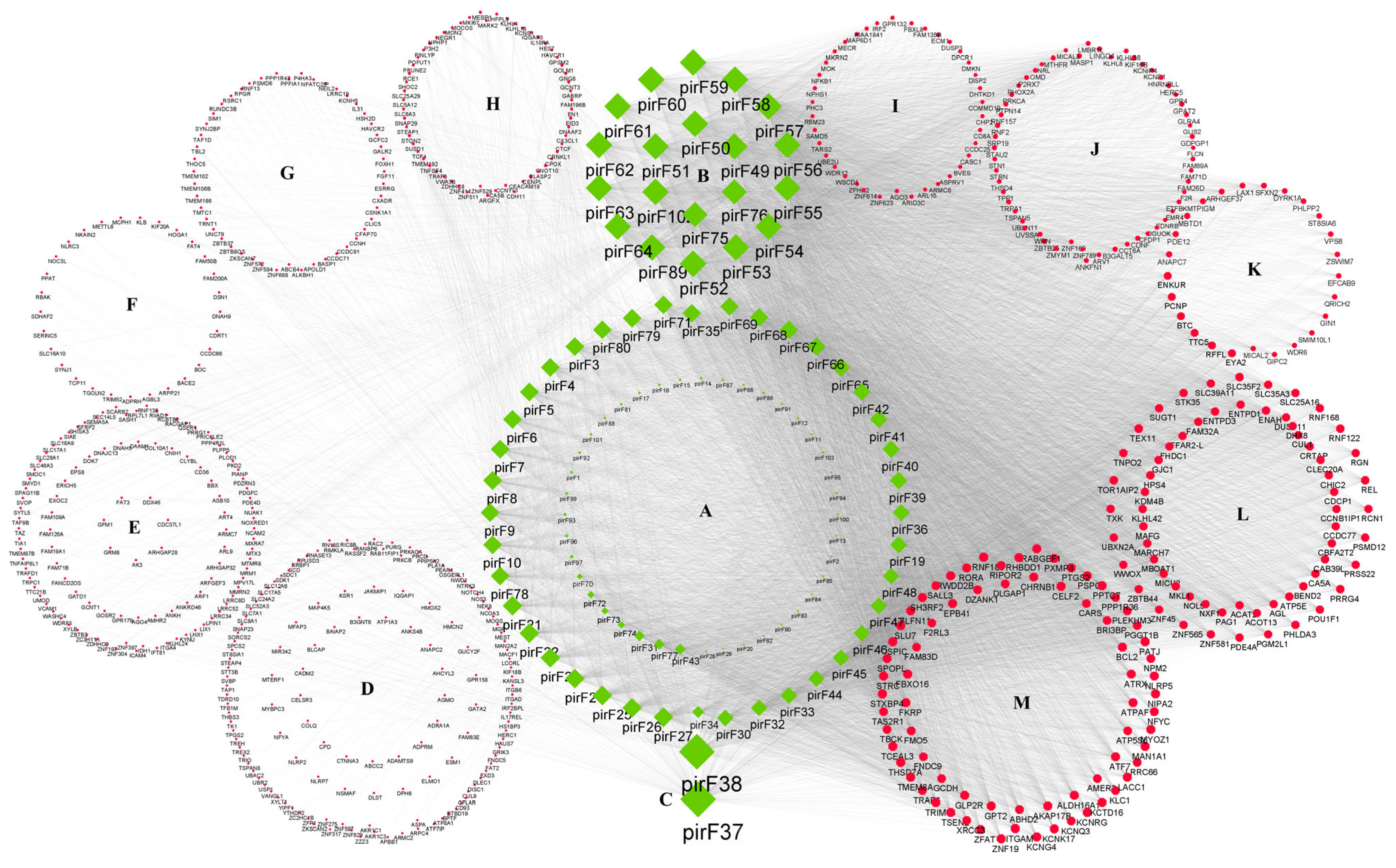
3.5. Prediction of piRNA Target Genes
3.6. Functional Analysis of Target Genes
4. Discussion
4.1. The Regulation of Cell Apoptosis
4.2. Inflammation and Oxidative Stress
4.3. Substance Transport and Signal Transduction
4.4. The Maintenance of Cellular Structural Integrity
4.5. Thoughts on the Methodology
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Evans, A.C.O.; Duffy, P.; Hynes, N.; Boland, M.P. Waves of Follicle Development During the Estrous Cycle in Sheep. Theriogenology 2000, 53, 699–715. [Google Scholar] [CrossRef] [PubMed]
- Marchal, R.; Vigneron, C.; Perreau, C.; Balipapp, A.; Mermillod, P. Effect of Follicular Size on Meiotic and Developmental Competence of Porcine Oocytes. Theriogenology 2002, 57, 1523–1532. [Google Scholar] [CrossRef] [PubMed]
- Mcgee, E.A.; Hsueh, A.J.W. Initial and Cyclic Recruitment of Ovarian Follicles. Endocr. Rev. 2000, 21, 200–214. [Google Scholar]
- Hatzirodos, N.; Irving-Rodgers, H.F.; Hummitzsch, K.; Rodgers, R.J. Transcriptome Profiling of the Theca Interna from Bovine Ovarian Follicles During Atresia. PLoS ONE 2014, 9, e99706. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, Y.; Yao, W.; Li, Q.; Liu, H.; Pan, Z. Initiation of Follicular Atresia: Gene Networks During Early Atresia in Pig Ovaries. Reproduction 2018, 156, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, C.; Jia, C.; Zhang, Y.; Qing, X.; Zhang, Y.; Liu, J.; Xu, S.; Pan, Z. The Role of Circular RNAs in the Physiology and Pathology of the Mammalian Ovary. Int. J. Mol. Sci. 2022, 23, 15204. [Google Scholar] [CrossRef]
- Liu, J.; Du, X.; Zhou, J.; Pan, Z.; Liu, H.; Li, Q. MicroRNA-26b Functions as a Proapoptotic Factor in Porcine Follicular Granulosa cells by Targeting Sma-and Mad-Related Protein 4. Biol. Reprod. 2014, 91, 146–157. [Google Scholar] [CrossRef]
- Guo, T.Y.; Huang, L.; Yao, W.; Du, X.; Li, Q.Q.; Ma, M.L.; Li, Q.F.; Liu, H.L.; Zhang, J.B.; Pan, Z.X. The Potential Biological Functions of Circular RNAs During the Initiation of Atresia in Pig Follicles. Domest. Anim. Endocrinol. 2020, 72, 106401. [Google Scholar] [CrossRef]
- Zhang, J.; Xu, Y.; Liu, H.; Pan, Z. MicroRNAs in Ovarian Follicular Atresia and Granulosa Cell Apoptosis. Reprod. Biol. Endocrinol. 2019, 17, 9. [Google Scholar] [CrossRef]
- Guo, T.; Zhang, J.; Yao, W.; Du, X.; Li, Q.; Huang, L.; Ma, M.; Li, Q.; Liu, H.; Pan, Z. CircINHA Resists Granulosa Cell Apoptosis by Upregulating CTGF as a ceRNA of miR-10a-5p in Pig Ovarian Follicles. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2019, 1862, 194420. [Google Scholar] [CrossRef]
- Aravin, A.; Gaidatzis, D.; Pfeffer, S.; Lagos-Quintana, M.; Landgraf, P.; Iovino, N.; Morris, P.; Brownstein, M.J.; Kuramochi-Miyagawa, S.; Nakano, T.; et al. A Novel Class of Small RNAs Bind to MILI Protein in Mouse Testes. Nature 2006, 442, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Homolka, D.; Pandey, R.R.; Goriaux, C.; Brasset, E.; Vaury, C.; Sachidanandam, R.; Fauvarque, M.-O.; Pillai, R.S. PIWI Slicing and RNA Elements in Precursors Instruct Directional Primary piRNA Biogenesis. Cell Rep. 2015, 12, 418–428. [Google Scholar] [CrossRef] [PubMed]
- Aravin, A.A.; Sachidanandam, R.; Bourc’his, D.; Schaefer, C.; Pezic, D.; Toth, K.F.; Bestor, T.; Hannon, G.J. A piRNA Pathway Primed by Individual Transposons Is Linked to De Novo DNA Methylation in Mice. Mol. Cell 2008, 31, 785–799. [Google Scholar] [CrossRef]
- Halic, M.; Moazed, D. Transposon Silencing by piRNAs. Cell 2009, 138, 1058–1060. [Google Scholar] [CrossRef]
- Dai, P.; Wang, X.; Gou, L.-T.; Li, Z.-T.; Wen, Z.; Chen, Z.-G.; Hua, M.-M.; Zhong, A.; Wang, L.; Su, H.; et al. A Translation-Activating Function of MIWI/piRNA during Mouse Spermiogenesis. Cell 2019, 179, 1566–1581.e16. [Google Scholar] [CrossRef]
- Liu, G.; Lei, B.; Li, Y.; Tong, K.; Ding, Y.; Luo, L.; Xia, X.; Jiang, S.; Deng, C.; Xiong, Y.; et al. Discovery of Potential piRNAs from Next Generation Sequences of the Sexually Mature Porcine Testes. PLoS ONE 2012, 7, e34770. [Google Scholar] [CrossRef]
- Weng, B.; Ran, M.; Chen, B.; Wu, M.; Peng, F.; Dong, L.; He, C.; Zhang, S.; Li, Z. Systematic Identification and Characterization of miRNAs and piRNAs from Porcine Testes. Genes Genom. 2017, 39, 1047–1057. [Google Scholar] [CrossRef]
- Kowalczykiewicz, D.; Świercz, A.; Handschuh, L.; Leśniak, K.; Figlerowicz, M.; Wrzesinski, J. Characterization of Sus scrofa Small Non-Coding RNAs Present in Both Female and Male Gonads. PLoS ONE 2014, 9, e113249. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate Alignment of Transcriptomes in the Presence of Insertions, Deletions and Gene Fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef]
- Wang, K.; Liang, C.; Liu, J.; Xiao, H.; Huang, S.; Xu, J.; Li, F. Prediction of piRNAs Using Transposon Interaction and a Support Vector Machine. BMC Bioinform. 2014, 15, 419. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, X.; Kang, L. A k-mer Scheme to Predict piRNAs and Characterize Locust piRNAs. Bioinformatics 2011, 27, 771–776. [Google Scholar] [CrossRef] [PubMed]
- Rosenkranz, D.; Zischler, H. proTRAC—A Software for Probabilistic piRNA Cluster Detection, Visualization and Analysis. BMC Bioinform. 2012, 13, 5. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Zhang, P.; Kang, J.Y.; Gou, L.T.; Wang, J.; Xue, Y.; Skogerboe, G.; Dai, P.; Huang, D.W.; Chen, R.; Fu, X.D.; et al. MIWI and piRNA-Mediated Cleavage of Messenger RNAs in Mouse Testes. Cell Res. 2015, 25, 193–207. [Google Scholar] [CrossRef]
- Di, R.; He, J.; Song, S.; Tian, D.; Liu, Q.; Liang, X.; Ma, Q.; Sun, M.; Wang, J.; Zhao, W.; et al. Characterization and Comparative Profiling of Ovarian microRNAs During Ovine Anestrus and the Breeding Season. BMC Genom. 2014, 15, 899. [Google Scholar] [CrossRef]
- Yang, C.-X.; Du, Z.-Q.; Wright, E.C.; Selman, B.; Rothschild, M.F.; Ross, J.W. Profile of Porcine Piwi-Interacting RNA During Oocyte Maturation and Early Embryo Development by Deep Sequencing. Biol. Reprod. 2011, 85 (Suppl. S1), 334. [Google Scholar] [CrossRef]
- Li, Z.; Li, Z.; Zhang, Y.; Zhou, L.; Xu, Q.; Li, L.; Zeng, L.; Xue, J.; Niu, H.; Zhong, J.; et al. Mammalian PIWI–piRNA–Target Complexes Reveal Features for Broad and Efficient Target Silencing. Nat. Struct. Mol. Biol. 2024, 31, 1222–1231. [Google Scholar] [CrossRef]
- Yao, W.; Pan, Z.; Du, X.; Zhang, J.; Liu, H.; Li, Q. NORHA, a Novel Follicular Atresia-Related lncRNA, Promotes Porcine Granulosa Cell Apoptosis via the miR-183-96-182 Cluster and FoxO1 Axis. J. Anim. Sci. Biotechnol. 2022, 1, 441–457. [Google Scholar] [CrossRef]
- Ma, M.; Zhang, J.; Gao, X.; Yao, W.; Li, Q.; Pan, Z. miR-361-5p Mediates SMAD4 to Promote Porcine Granulosa Cell Apoptosis Through VEGFA. Biomolecules 2020, 10, 1281. [Google Scholar] [CrossRef]
- Wu, X.Q.; Wang, Y.Q.; Xu, S.M.; Liu, J.F.; Bi, X.Y.; Wang, Z.Q.; Zhang, J.P. The WNT/β-Catenin Signaling Pathway May Be Involved in Granulosa Cell Apoptosis from Patients with PCOS in North China. J. Gynecol. Obstet. Hum. Reprod. 2017, 46, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Cheng, E.H.Y.A.; Wei, M.C.; Weiler, S.; Flavell, R.A.; Mak, T.W.; Lindsten, T.; Korsmeyer, S.J. BCL-2, BCL-XL Sequester BH3 Domain-Only Molecules Preventing BAX- and BAK-Mediated Mitochondrial Apoptosis. Mol. Cell 2001, 8, 705–711. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Zhang, S.; Chao, K.; Feng, R.; Wang, H.; Li, M.; Chen, B.; He, Y.; Zeng, Z.; Chen, M. E3 Ubiquitin ligase RNF183 Is a Novel Regulator in Inflammatory Bowel Disease. J. Crohn’s Colitis 2016, 10, 713–725. [Google Scholar] [CrossRef]
- Liu, S.; Chen, J.; Liu, M.; Zhang, C.; Chao, X.; Yang, H.; Wang, T.; Bi, H.; Ding, Y.; Wang, Z.; et al. miR-107 Suppresses Porcine Granulosa Cell Proliferation and Estradiol Synthesis While Promoting Apoptosis via Targeting PTGS2. Theriogenology 2025, 238, 117367. [Google Scholar] [CrossRef]
- Vasiliou, V.K.; Pappa, A.; Black, W.; Jester, J.; Day, B.; Min, E.; Lassen, N. ALDH3A1 Prevents Apoptosis of Corneal Cells Induced by DNA Damaging Agents and Oxidative Stress. Investig. Ophthalmol. Vis. Sci. 2005, 46, 4777. [Google Scholar]
- Zhang, M.; Zhong, Z.; Hu, Q.; Yan, X.; Wang, Y.; Ye, Q. Relationship Between the Expression of ALDH2 and Apoptosis Level in DBD Liver. Med. J. Wuhan Univ. 2016, 37, 595–597+619. [Google Scholar]
- Boon, N.J.; Livelra, R.A.; Körner, P.-R.; Kochavi, A.; Mertens, S.; Malka, Y.; Voogd, R.; van der Horst, S.E.M.; Huismans, M.A.; Smabers, L.P.; et al. DNA Damage Induces p53-Independent Apoptosis Through Ribosome Stalling. Science 2024, 384, 785–792. [Google Scholar] [CrossRef]
- Sands, J.; Tolaney, S.M.; Ueno, N.T.; Spira, A.I.; Yamamoto, N.; Janjigian, Y.Y.; Naito, Y.; Damodaran, S.; Meric-Bernstam, F.; Modi, S.; et al. A Phase 1b, Multicenter, Open-Label Study of Valemetostat in Combination with DXd Antibody Drug Conjugates (ADCs), Trastuzumab Deruxtecan (T-DXd) or Datopotamab Deruxtecan (Dato-DXd), in Patients with Solid Tumors. J. Clin. Oncol. 2024, 42, TPS4180. [Google Scholar] [CrossRef]
- Karpagavalli, M.; Sivagurunathan, S.; Panda, T.S.; Srikakulam, N.; Arora, R.; Dohadwala, L.; Tiwary, B.K.; Sadras, S.R.; Arunachalam, J.P.; Pandi, G.; et al. piRNAs in the Human Retina and Retinal Pigment Epithelium Reveal a Potential Role in Intracellular Trafficking and Oxidative Stress. Mol. Omics 2024, 20, 248–264. [Google Scholar] [CrossRef]
- Stenmark, H.; Olkkonen, V.M. The rab GTPase family. Genome Biol. 2001, 2, reviews3007.1. [Google Scholar] [CrossRef][Green Version]
- Yusta, B.; Boushey, R.P.; Drucker, D.J. The Glucagon-like Peptide-2 Receptor Mediates Direct Inhibition of Cellular Apoptosis via a cAMP-dependent Protein Kinase-independent Pathway. J. Biol. Chem. 2000, 275, 35345–35352. [Google Scholar] [CrossRef] [PubMed]
- Diviani, D.; Dodge-Kafka, K.L.; Li, J.; Kapiloff, M.S. A-Kinase Anchoring Proteins: Scaffolding Proteins in the Heart. Am. J. Physiol. Heart Circ. Physiol. 2011, 301, 1742–1753. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Li, Y.; Li, Y.; Ji, X.; Li, H.; Wu, D.; Wei, W.; Wang, X. Silencing EPB41 Gene Expression Leads to Cell Cycle Arrest, Migration Inhibition, and Upregulation of Cell Surface Antigen in DC2.4 Cells. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2020, 26, e920594. [Google Scholar] [CrossRef]
- Wu, X.-Y.; Xing, X.-F.; An, M. Polydopamine Nanoparticles Loaded with EPB41L4A-AS2 Promote Ovarian Cancer Cell Apoptosis Through Activation of Mitogen-Activated Protein Kinase Signaling Pathway. Sci. Adv. Mater. 2022, 14, 393–399. [Google Scholar] [CrossRef]
- Turksen, K.; Troy, T.-C. Barriers Built on Claudins. J. Cell Sci. 2004, 117, 2435–2477. [Google Scholar] [CrossRef]
- Resnick, M.B.; Konkin, T.; Routhier, J.; Sabo, E.; Pricolo, V.E. Claudin-1 Is a Strong Prognostic Indicator in Stage II Colonic Cancer: A Tissue Microarray Study. Mod. Pathol. 2005, 18, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Aschenbrenner, J.; Marx, A. Direct and Site-Specific Quantification of RNA 2′-O-Methylation by PCR with an Engineered DNA Polymerase. Nucleic Acids Res. 2016, 44, 3495–3502. [Google Scholar] [CrossRef]
- Wang, N.; Qu, S.; Sun, W.; Zeng, Z.; Liang, H.; Zhang, C.Y.; Chen, X.; Zen, K. Direct Quantification of 3′ Terminal 2′-O-Methylation of Small RNAs by RT-qPCR. RNA 2018, 24, 1520–1529. [Google Scholar] [CrossRef]


| Samples | Total | Clean Reads | Mapping Reads Count | Mapping Rate |
|---|---|---|---|---|
| HF 1 | 16,159,470 | 15,164,669 | 14,862,688 | 98.01% |
| HF 2 | 15,923,735 | 14,443,413 | 14,119,933 | 97.76% |
| HF 3 | 18,060,078 | 16,382,285 | 16,105,126 | 98.31% |
| AF 1 | 16,707,009 | 15,279,119 | 14,948,455 | 97.84% |
| AF 2 | 19,319,035 | 17,165,699 | 16,874,407 | 98.30% |
| AF 3 | 18,171,763 | 16,803,580 | 16,473,089 | 98.03% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Huang, L.; Qin, X.; Li, W.; Cheng, X.; Pan, Z. Porcine Ovarian piRNA Dynamics: A Comparative Study During Follicular Atresia. Biology 2025, 14, 609. https://doi.org/10.3390/biology14060609
Zhang J, Huang L, Qin X, Li W, Cheng X, Pan Z. Porcine Ovarian piRNA Dynamics: A Comparative Study During Follicular Atresia. Biology. 2025; 14(6):609. https://doi.org/10.3390/biology14060609
Chicago/Turabian StyleZhang, Jinbi, Long Huang, Xinxin Qin, Wenjie Li, Xiaolong Cheng, and Zengxiang Pan. 2025. "Porcine Ovarian piRNA Dynamics: A Comparative Study During Follicular Atresia" Biology 14, no. 6: 609. https://doi.org/10.3390/biology14060609
APA StyleZhang, J., Huang, L., Qin, X., Li, W., Cheng, X., & Pan, Z. (2025). Porcine Ovarian piRNA Dynamics: A Comparative Study During Follicular Atresia. Biology, 14(6), 609. https://doi.org/10.3390/biology14060609





