Identifying Molecular Properties of Ataxin-2 Inhibitors for Spinocerebellar Ataxia Type 2 Utilizing High-Throughput Screening and Machine Learning
Simple Summary
Abstract
1. Introduction
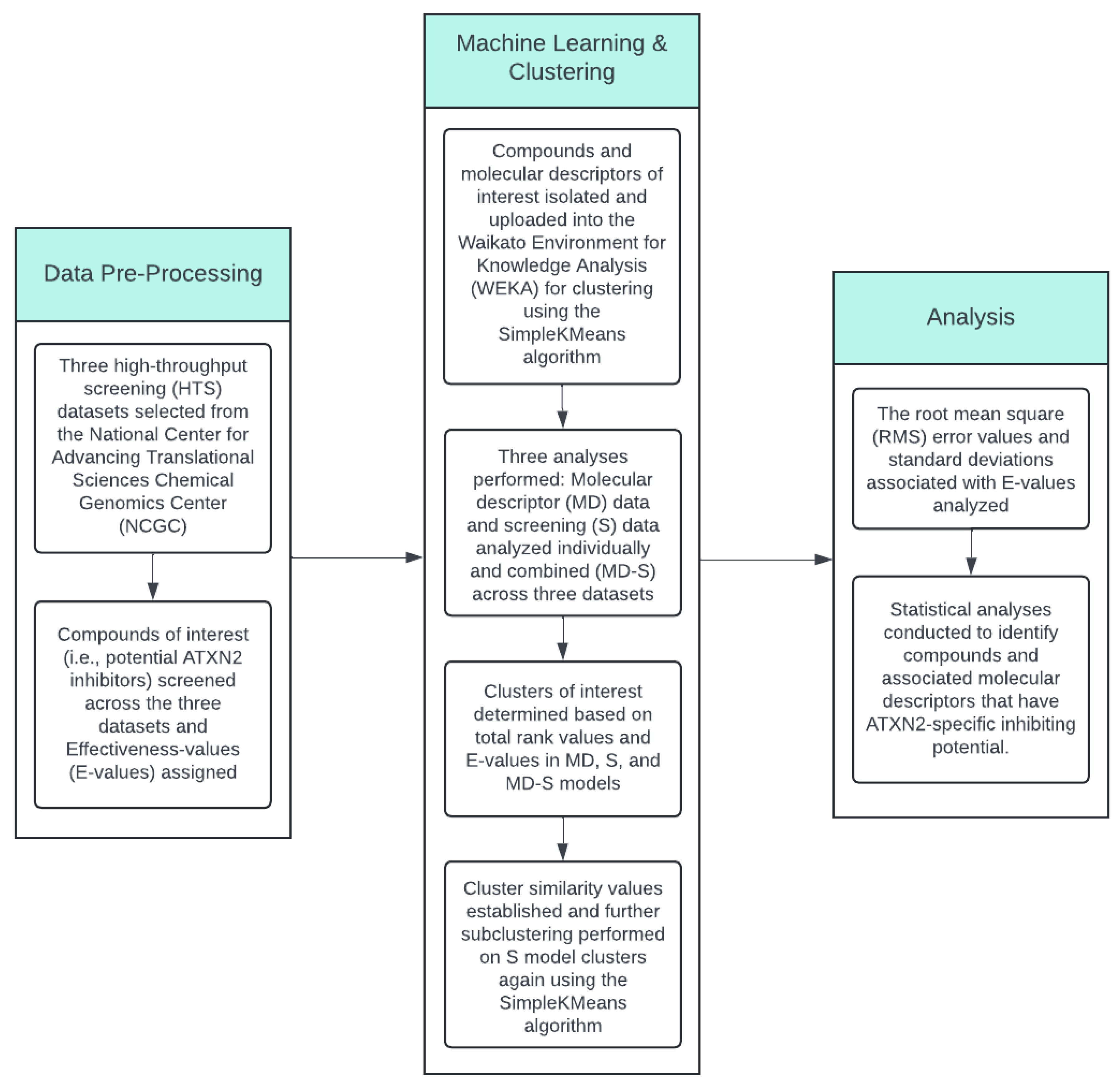
2. Materials and Methods
2.1. Pre-Processing and Selection of High-Throughput Screening (HTS) Data for Analysis
2.2. Machine Learning Application to the HTS Data and Initial Clustering Analysis
2.3. Compounds and Associated Molecular Descriptors Within Clusters of Interest
2.4. Subcluster and HTS Data Analysis
2.5. Statistical Analysis
3. Results
3.1. HTS Data for Analysis
3.2. Data from Machine Learning Application to the HTS Data and Initial Clustering Analysis
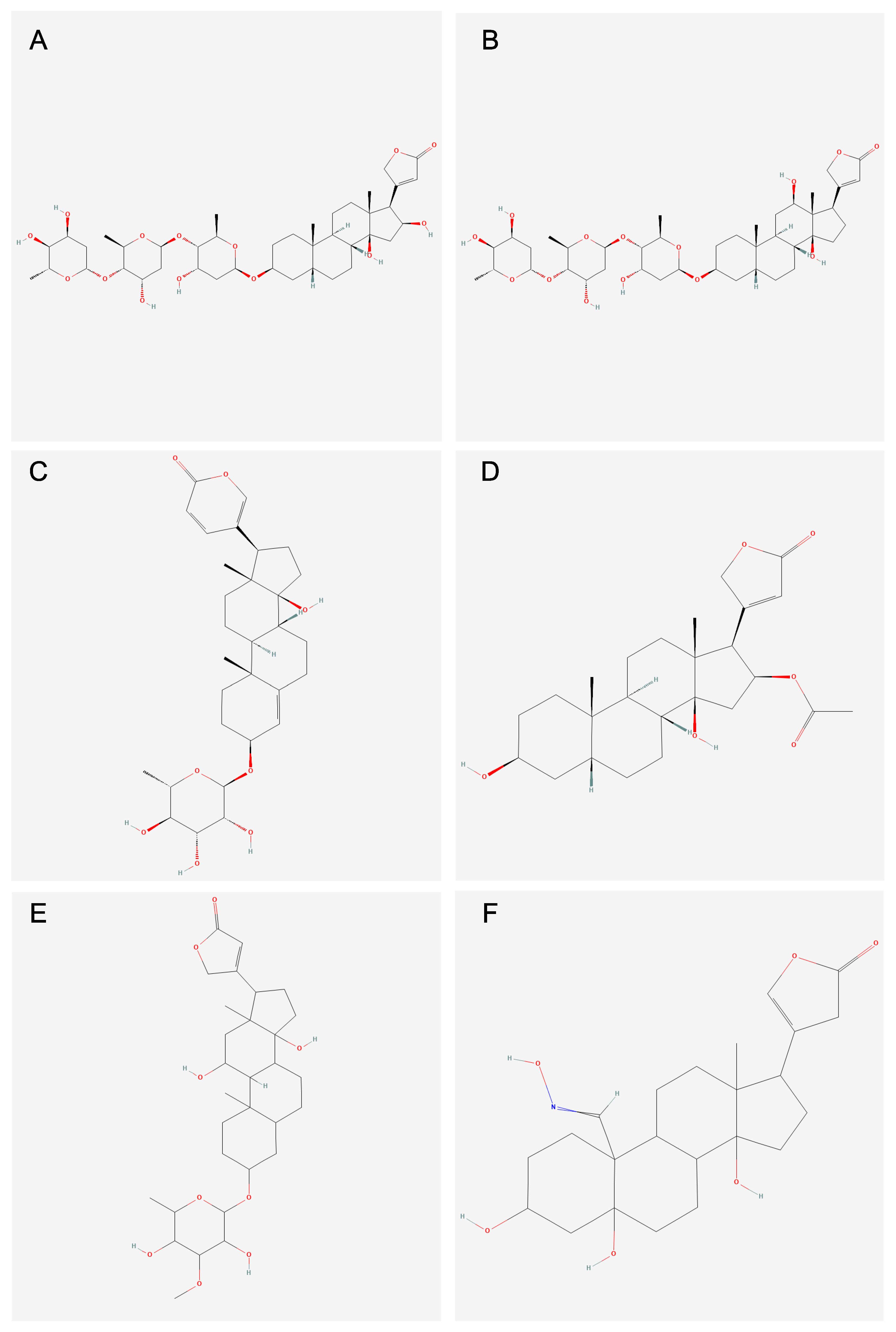
3.3. Analysis of Compounds and Associated Molecular Descriptors Within Clusters of Interest
3.4. Subcluster Analysis
4. Discussion
5. Limitations
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gispert, S.; Twells, R.; Orozco, G.; Brice, A.; Weber, J.; Heredero, L.; Scheufler, K.; Riley, B.; Allotey, R.; Nothers, C.; et al. Chromosomal assignment of the second locus for autosomal dominant cerebellar ataxia (SCA2) to chromosome 12q23–24.1. Nat. Genet. 1993, 4, 295–299. [Google Scholar] [CrossRef] [PubMed]
- Scoles, D.R.; Pulst, S.M. Spinocerebellar Ataxia Type 2. In Polyglutamine Disorders; Nóbrega, C., Pereira de Almeida, L., Eds.; Springer: Cham, Switzerland, 2018; pp. 175–195. [Google Scholar]
- Imbert, G.; Saudou, F.; Yvert, G.; Devys, D.; Trottier, Y.; Garnier, J.-M.; Weber, C.; Mandel, J.-L.; Cancel, G.; Abbas, N.; et al. Cloning of the gene for spinocerebellar ataxia 2 reveals a locus with high sensitivity to expanded CAG/glutamine repeats. Nat. Genet. 1996, 14, 285–291. [Google Scholar] [CrossRef] [PubMed]
- Pulst, S.-M.; Nechiporuk, A.; Nechiporuk, T.; Gispert, S.; Chen, X.-N.; Lopes-Cendes, I.; Pearlman, S.; Starkman, S.; Orozco-Diaz, G.; Lunkes, A.; et al. Moderate expansion of a normally biallelic trinucleotide repeat in spinocerebellar ataxia type 2. Nat. Genet. 1996, 14, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Sanpei, K.; Takano, H.; Igarashi, S.; Sato, T.; Oyake, M.; Sasaki, H.; Wakisaka, A.; Tashiro, K.; Ishida, Y.; Ikeuchi, T.; et al. Identification of the spinocerebellar ataxia type 2 gene using a direct identification of repeat expansion and cloning technique, DIRECT. Nat. Genet 1996, 14, 277–284. [Google Scholar] [CrossRef]
- Paulson, H.L.; Bonini, N.M.; Roth, K.A. Polyglutamine disease and neuronal cell death. Proc. Natl. Acad. Sci. USA 2000, 97, 12957–12958. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ghanekar, S.D.; Kuo, S.-H.; Staffetti, J.S.; Zesiewicz, T.A. Current and emerging treatment modalities for spinocerebellar ataxias. Expert Rev. Neurother. 2022, 22, 101–114. [Google Scholar] [CrossRef]
- Egorova, P.A.; Bezprozvanny, I.B. Molecular Mechanisms and Therapeutics for Spinocerebellar Ataxia Type 2. Neurotherapeutics 2019, 16, 1050–1073. [Google Scholar] [CrossRef]
- Elden, A.C.; Kim, H.-J.; Hart, M.P.; Chen-Plotkin, A.S.; Johnson, B.S.; Fang, X.; Armakola, M.; Geser, F.; Greene, R.; Lu, M.M.; et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature 2010, 466, 1069–1075. [Google Scholar] [CrossRef]
- Pulst, S.M.; Scoles, D.R.; Paul, S. Effects of STAU1/staufen1 on autophagy in neurodegenerative diseases. Autophagy 2023, 19, 2607–2608. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Becker, L.A.; Huang, B.; Bieri, G.; Ma, R.; Knowles, D.A.; Jafar-Nejad, P.; Messing, J.; Kim, H.J.; Soriano, A.; Auburger, G.; et al. Therapeutic reduction of ataxin-2 extends lifespan and reduces pathology in TDP-43 mice. Nature 2017, 544, 367–371. [Google Scholar] [CrossRef]
- Scoles, D.R.; Meera, P.; Schneider, M.D.; Paul, S.; Dansithong, W.; Figueroa, K.P.; Hung, G.; Rigo, F.; Bennett, C.F.; Otis, T.S.; et al. Antisense oligonucleotide therapy for spinocerebellar ataxia type 2. Nature 2017, 544, 362–366. [Google Scholar] [CrossRef] [PubMed]
- Burkhardt, M.F.; Martinez, F.J.; Wright, S.; Ramos, C.; Volfson, D.; Mason, M.; Garnes, J.; Dang, V.; Lievers, J.; Shoukat-Mumtaz, U.; et al. A cellular model for sporadic ALS using patient-derived induced pluripotent stem cells. Mol. Cell Neurosci. 2013, 56, 355–364. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Fang, M.Y.; Markmiller, S.; Vu, A.Q.; Javaherian, A.; Dowdle, W.E.; Jolivet, P.; Bushway, P.J.; Castello, N.A.; Baral, A.; Chan, M.Y.; et al. Small-Molecule Modulation of TDP-43 Recruitment to Stress Granules Prevents Persistent TDP-43 Accumulation in ALS/FTD. Neuron 2019, 103, 802–819.e11. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Inglese, J.; Auld, D.S.; Jadhav, A.; Johnson, R.L.; Simeonov, A.; Yasgar, A.; Zheng, W.; Austin, C.P. Quantitative high-throughput screening: A titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc. Natl. Acad. Sci. USA 2006, 103, 11473–11478. [Google Scholar] [CrossRef]
- Hughes, J.P.; Rees, S.; Kalindjian, S.B.; Philpott, K.L. Principles of early drug discovery. Br. J. Pharmacol. 2011, 162, 1239–1249. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rifaioglu, A.S.; Atas, H.; Martin, M.J.; Cetin-Atalay, R.; Atalay, V.; Doğan, T. Recent applications of deep learning and machine intelligence on in silico drug discovery: Methods, tools and databases. Brief. Bioinform. 2019, 20, 1878–1912. [Google Scholar] [CrossRef]
- Tropsha, A.; Golbraikh, A. Predictive QSAR modeling workflow, model applicability domains, and virtual screening. Curr. Pharm. Des. 2007, 13, 3494–3504. [Google Scholar] [CrossRef] [PubMed]
- Scoles, D.R.; Gandelman, M.; Paul, S.; Dexheimer, T.; Dansithong, W.; Figueroa, K.P.; Pflieger, L.T.; Redlin, S.; Kales, S.C.; Sun, H.; et al. A quantitative high-throughput screen identifies compounds that lower expression of the SCA2-and ALS-associated gene ATXN2. J. Biol. Chem. 2022, 298, 102228. [Google Scholar] [CrossRef]
- ChemAxon. Marvin, version 5.12.4; ChemAxon: Budapest, Hungary, 2013. [Google Scholar]
- Hall, M.; Frank, E.; Holmes, G.; Pfahringer, B.; Reutemann, P.; Witten, I.H. The WEKA Data Mining Software: An Update. SIGKDD Explor. 2009, 11, 10–18. [Google Scholar] [CrossRef]
- Rivera-Borroto, O.M.; Marrero-Ponce, Y.; García-de la Vega, J.M.; Grau-Ábalo, R.d.C. Comparison of Combinatorial Clustering Methods on Pharmacological Data Sets Represented by Machine Learning-Selected Real Molecular Descriptors. J. Chem. Inf. Model. 2011, 51, 3036–3049. [Google Scholar] [CrossRef]
- Karthikeyan, M.; Vyas, R. Machine learning methods in chemoinformatics for drug discovery. In Practical Chemoinformatics; Springer: New Delhi, India, 2014; pp. 133–194. [Google Scholar]
- Syed, A.H.; Khan, T. A supervised classifier-based chemoinformatics model to predict inhibitors essential for sexual reproduction and transmission of the P. falciparum parasite into mosquitoes. Int. J. Adv. Appl. Sci. 2019, 6, 62–72. [Google Scholar] [CrossRef]
- Arthur, D.; Vassilvitskii, S. k-Means++: The Advantages of Careful Seeding. In Proceedings of the Eighteenth Annual ACM-SIAM Symposium on Discrete Algorithms, SODA ’07, New Orleans, LA, USA, 7–9 January 2007; Society for Industrial and Applied Mathematics: Philadelphia, PA, USA, 2007. [Google Scholar]
- Mardia, K.V.; Kent, J.T.; Bibby, J.M. Multivariate Analysis (Probability and Mathematical Statistics), 1st ed.; Academic Press: London, UK, 1980. [Google Scholar]
- Mangat, C.S.; Bharat, A.; Gehrke, S.S.; Brown, E.D. Rank ordering plate data facilitates data visualization and normalization in high-throughput screening. J. Biomol. Screen 2014, 19, 1314–1320. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cendelin, J.; Cvetanovic, M.; Gandelman, M.; Hirai, H.; Orr, H.T.; Pulst, S.M.; Strupp, M.; Tichanek, F.; Tuma, J.; Manto, M. Consensus paper: Strengths and weaknesses of animal models of spinocerebellar ataxias and their clinical implications. Cerebellum 2022, 21, 452–481. [Google Scholar] [CrossRef] [PubMed]
- Dansithong, W.; Paul, S.; Figueroa, K.P.; Rinehart, M.D.; Wiest, S.; Pflieger, L.T.; Scoles, D.R.; Pulst, S.M. Ataxin-2 regulates RGS8 translation in a new BAC-SCA2 transgenic mouse model. PLoS Genet. 2015, 11, e1005182. [Google Scholar] [CrossRef]
- Hansen, S.T.; Meera, P.; Otis, T.S.; Pulst, S.M. Changes in Purkinje cell firing and gene expression precede behavioral pathology in a mouse model of SCA2. Hum. Mol. Genet. 2013, 22, 271–283. [Google Scholar] [CrossRef]
- Huynh, D.P.; Figueroa, K.; Hoang, N.; Pulst, S.-M. Nuclear localization or inclusion body formation of ataxin-2 are not necessary for SCA2 pathogenesis in mouse or human. Nat. Genet. 2000, 26, 44–50. [Google Scholar] [CrossRef]
- Patil, R.; Deshmukh, S.; Rajeswari, K. Analysis of SimpleKMeans with Multiple Dimensions using WEKA. Int. J. Comput. Appl. 2015, 110, 14–17. [Google Scholar] [CrossRef]
- Bauer, M.R.; Di Fruscia, P.; Lucas, S.C.C.; Michaelides, I.N.; Nelson, J.E.; Storer, R.I.; Whitehurst, B.C. Put a ring on it: Application of small aliphatic rings in medicinal chemistry. RSC Med. Chem. 2021, 12, 448–471. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Meyer, E.A.; Castellano, R.K.; Diederich, F. Interactions with aromatic rings in chemical and biological recognition. Angew Chem. Int. Ed. Engl. 2003, 42, 1210–1250. [Google Scholar] [CrossRef] [PubMed]
- Salonen, L.M.; Ellermann, M.; Diederich, F. Aromatic rings in chemical and biological recognition: Energetics and structures. Angew Chem. Int. Ed. Engl. 2011, 50, 4808–4842. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, T.J.; Macdonald, S.J.; Young, R.J.; Pickett, S.D. The impact of aromatic ring count on compound developability: Further insights by examining carbo- and hetero-aromatic and -aliphatic ring types. Drug Discov. Today 2011, 16, 164–171. [Google Scholar] [CrossRef] [PubMed]
- Botelho, A.F.M.; Pierezan, F.; Soto-Blanco, B.; Melo, M.M. A review of cardiac glycosides: Structure, toxicokinetics, clinical signs, diagnosis and antineoplastic potential. Toxicon 2019, 158, 63–68. [Google Scholar] [CrossRef]
- Cornelius, F.; Kanai, R.; Toyoshima, C. A structural view on the functional importance of the sugar moiety and steroid hydroxyls of cardiotonic steroids in binding to Na, K-ATPase. J. Biol. Chem. 2013, 288, 6602–6616. [Google Scholar] [CrossRef]
- Hou, Y.; Shang, C.; Meng, T.; Lou, W. Anticancer potential of cardiac glycosides and steroid-azole hybrids. Steroids 2021, 171, 108852. [Google Scholar] [CrossRef]
- Kumavath, R.; Paul, S.; Pavithran, H.; Paul, M.K.; Ghosh, P.; Barh, D.; Azevedo, V. Emergence of Cardiac Glycosides as Potential Drugs: Current and Future Scope for Cancer Therapeutics. Biomolecules 2021, 11, 1275. [Google Scholar] [CrossRef]
- Morsy, N. Cardiac glycosides in medicinal plants. In Aromatic and Medicinal Plants: Back to Nature; Intechopen: London, UK, 2017; pp. 29–45. [Google Scholar]
- Kupchan, S.M.; Mokotoff, M.; Sandhu, R.S.; Hokin, L.E. The chemistry and biological activity of derivatives of strophanthidin. J. Med. Chem. 1967, 10, 1025–1033. [Google Scholar] [CrossRef]
- Brooks, W.H.; Guida, W.C.; Daniel, K.G. The significance of chirality in drug design and development. Curr. Top Med. Chem. 2011, 11, 760–770. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Meanwell, N.A. Improving drug candidates by design: A focus on physicochemical properties as a means of improving compound disposition and safety. Chem. Res. Toxicol. 2011, 24, 1420–1456. [Google Scholar] [CrossRef] [PubMed]
- Monteleone, S.; Fuchs, J.E.; Liedl, K.R. Molecular Connectivity Predefines Polypharmacology: Aliphatic Rings, Chirality, and sp. Front. Pharmacol. 2017, 8, 552. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Makhova, N.N.; Petukhova, V.Y.; Kuznetsov, V.V. Synthesis of monocyclic diaziridines and their fused derivatives. Arkivoc 2008, 1, 128–152. [Google Scholar] [CrossRef]
- Zaki, M.E.A.; Al-Hussain, S.A.; Bukhari, S.N.A.; Masand, V.H.; Rathore, M.M.; Thakur, S.D.; Patil, V.M. Exploring the Prominent and Concealed Inhibitory Features for Cytoplasmic Isoforms of Hsp90 Using QSAR Analysis. Pharmaceuticals 2022, 15, 303. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kim, S.; Thiessen, P.A.; Bolton, E.E.; Chen, J.; Fu, G.; Gindulyte, A.; Han, L.; He, J.; He, S.; Shoemaker, B.A.; et al. PubChem Substance and Compound databases. Nucleic Acids Res. 2016, 44, D1202–D1213. [Google Scholar] [CrossRef] [PubMed]
- Yap, K.H.; Azmin, S.; Che Hamzah, J.; Ahmad, N.; van de Warrenburg, B.; Mohamed Ibrahim, N. Pharmacological and non-pharmacological management of spinocerebellar ataxia: A systematic review. J. Neurol. 2022, 269, 2315–2337. [Google Scholar] [CrossRef] [PubMed]
- Wätzig, H.; Oltmann-Norden, I.; Steinicke, F.; Nachbar, M.; El-Hady, D.A.; Albishri, H.M.; Baumann, K.; Exner, T.; Böckler, F.M.; El Deeb, S. Data quality in drug discovery: The role of analytical performance in ligand binding assays. J. Comput. Aided Mol. Des. 2015, 29, 847–865. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Fu, A.; Zhang, L. Progress in molecular docking. Quant. Biol. 2019, 7, 83–89. [Google Scholar] [CrossRef]
- Gissinger, J.R.; Jensen, B.D.; Wise, K.E. Modeling chemical reactions in classical molecular dynamics simulations. Polymer 2017, 128, 211–217. [Google Scholar] [CrossRef]
- Singh, H.; Kaur, K. New method for finding initial cluster centroids in K-means algorithm. Int. J. Comput. Appl. 2013, 74, 27–30. [Google Scholar] [CrossRef]
- Matilla-Dueñas, A.; Ashizawa, T.; Brice, A.; Magri, S.; McFarland, K.N.; Pandolfo, M.; Pulst, S.M.; Riess, O.; Rubinsztein, D.C.; Schmidt, J.; et al. Consensus paper: Pathological mechanisms underlying neurodegeneration in spinocerebellar ataxias. Cerebellum 2014, 13, 269–302. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
| 1. If (ATXN ≥ 0) | e = 0 |
| 2. If (ATXN < 0) | |
| and if (CMV ≥ 0) | e = 0 |
| 3. If (ATXN < 0) | |
| and if (CMV < 0) | e = |ATXN| − |CMV| |
| 4. If (e = 0) | E = 0 |
| 5. If (e > 0) | |
| and if (Luc < 0) | E = e − |Luc| |
| 6. If (e > 0) | |
| and if (Luc ≥ 0) | E = e + |Luc| |
| Cluster | ATXN2 Expression Assay | Rank 1 | CMV Control Assay | Rank 2 | Biochemical Control Assay | Rank 3 | Number of Compounds | E | Total Rank |
|---|---|---|---|---|---|---|---|---|---|
| 0 | −70.95 ± 28.08 | 23 | −55.33 ± 32.05 | 6 | −20.78 ± 25.76 | 26 | 40 | −5.16 | 55 |
| 1 | −73.20 ± 17.10 | 19 | −63.87 ± 18.86 | 21 | −6.71 ± 12.11 | 12 | 55 | 2.62 | 52 |
| 2 | −74.91 ± 20.58 | 15 | −61.79 ± 20.82 | 13 | −10.23 ± 16.78 | 16 | 79 | 2.89 | 44 |
| 3 | −68.91 ± 21.68 | 26 | −59.49 ± 22.02 | 10 | −6.46 ± 15.74 | 9 | 66 | 2.96 | 45 |
| 4 | −80.60 ± 12.49 | 6 | −62.06 ± 15.69 | 14 | −11.75 ± 11.65 | 20 | 41 | 6.79 | 40 |
| 5 | −88.56 ± 8.32 | 1 | −43.52 ± 29.75 | 1 | −2.30 ± 10.00 | 2 | 16 | 42.74 | 4 |
| 6 | −80.17 ± 21.66 | 8 | −67.70 ± 25.45 | 25 | −14.04 ± 16.10 | 23 | 45 | −1.57 | 56 |
| 7 | −76.12 ± 17.96 | 12 | −55.06 ± 27.56 | 5 | −5.22 ± 16.44 | 7 | 54 | 15.85 | 24 |
| 8 | −83.22 ± 10.94 | 3 | −65.21 ± 15.89 | 22 | −3.18 ± 14.16 | 4 | 24 | 14.83 | 29 |
| 9 | −73.65 ± 23.88 | 17 | −51.92 ± 24.17 | 2 | −13.56 ± 20.00 | 22 | 70 | 8.17 | 41 |
| 10 | −87.49 ± 5.94 | 2 | −66.02 ± 22.92 | 24 | −0.13 ± 3.23 | 1 | 8 | 21.34 | 27 |
| 11 | −80.73 ± 15.56 | 5 | −63.36 ± 18.92 | 20 | −15.47 ± 16.58 | 24 | 92 | 1.89 | 49 |
| 12 | −75.96 ± 24.82 | 13 | −62.54 ± 28.54 | 16 | −18.31 ± 21.93 | 25 | 52 | −4.88 | 54 |
| 13 | −71.68 ± 18.85 | 21 | −61.17 ± 19.09 | 12 | −6.59 ± 13.02 | 11 | 67 | 3.92 | 44 |
| 14 | −73.37 ± 20.13 | 18 | −58.32 ± 18.60 | 9 | −11.51 ± 17.49 | 19 | 58 | 3.53 | 46 |
| 15 | −71.15 ± 24.88 | 22 | −56.03 ± 22.68 | 7 | −12.81 ± 18.74 | 21 | 76 | 2.31 | 50 |
| 16 | −78.14 ± 19.35 | 10 | −65.68 ± 23.21 | 23 | −9.67 ± 15.88 | 15 | 67 | 2.79 | 48 |
| 17 | −79.95 ± 21.80 | 9 | −62.77 ± 29.69 | 17 | −3.53 ± 9.14 | 5 | 17 | 13.66 | 31 |
| 18 | −74.63 ± 18.67 | 16 | −57.77 ± 19.27 | 8 | −11.35 ± 18.14 | 17 | 98 | 5.51 | 41 |
| 19 | −70.79 ± 18.05 | 24 | −62.89 ± 17.83 | 19 | −6.51 ± 13.14 | 10 | 53 | 1.39 | 53 |
| 20 | −69.21 ± 22.92 | 25 | −54.83 ± 24.57 | 4 | −3.56 ± 8.39 | 6 | 23 | 10.82 | 35 |
| 21 | −75.62 ± 23.16 | 14 | −62.52 ± 24.31 | 15 | −8.92 ± 10.92 | 14 | 27 | 4.18 | 43 |
| 22 | −72.00 ± 20.00 | 20 | −54.04 ± 23.87 | 3 | −3.03 ± 6.77 | 3 | 70 | 14.93 | 26 |
| 23 | −80.19 ± 13.71 | 7 | −62.82 ± 13.26 | 18 | −11.37 ± 13.88 | 18 | 38 | 6.00 | 43 |
| 24 | −76.41 ± 24.14 | 11 | −61.05 ± 23.66 | 11 | −8.04 ± 12.60 | 13 | 49 | 7.32 | 35 |
| 25 | −81.66 ± 14.10 | 4 | −71.27 ± 19.48 | 26 | −5.79 ± 13.92 | 8 | 36 | 4.60 | 38 |
| Cluster | ATXN2 Expression Assay | Rank 1 | CMV Control Assay | Rank 2 | Biochemical Control Assay | Rank 3 | Number of Compounds | E | Total Rank |
|---|---|---|---|---|---|---|---|---|---|
| 0 | −95.30 ± 7.22 | 1 | −91.73 ± 9.19 | 26 | −26.53 ± 31.41 | 25 | 50 | −22.96 | 52 |
| 1 | −70.27 ± 14.38 | 18 | −53.95 ± 17.21 | 9 | −4.74 ± 8.99 | 8 | 48 | 11.58 | 35 |
| 2 | −81.17 ± 11.20 | 12 | −62.26 ± 15.96 | 14 | −8.42 ± 14.50 | 15 | 77 | 10.49 | 41 |
| 3 | −63.42 ± 17.29 | 23 | −53.86 ± 12.23 | 8 | −7.62 ± 10.81 | 14 | 74 | 1.94 | 45 |
| 4 | −64.23 ± 18.26 | 22 | −28.10 ± 14.27 | 3 | −2.92 ± 9.71 | 3 | 56 | 33.21 | 28 |
| 5 | −87.94 ± 9.63 | 3 | −43.63 ± 29.00 | 6 | −2.65 ± 9.68 | 2 | 18 | 41.66 | 11 |
| 6 | −68.12 ± 20.87 | 21 | −40.64 ± 17.02 | 5 | −13.03 ± 16.69 | 20 | 35 | 14.44 | 46 |
| 7 | −85.69 ± 12.36 | 6 | −71.50 ± 19.28 | 20 | −5.31 ± 17.96 | 10 | 43 | 8.88 | 36 |
| 8 | −82.96 ± 9.64 | 10 | −66.85 ± 11.52 | 18 | −5.27 ± 12.71 | 9 | 55 | 10.84 | 37 |
| 9 | −78.08 ± 12.77 | 15 | −56.08 ± 16.81 | 10 | −16.18 ± 19.49 | 24 | 68 | 5.81 | 49 |
| 10 | −83.05 ± 8.27 | 9 | −60.48 ± 16.58 | 12 | −4.12 ± 9.12 | 5 | 18 | 18.45 | 26 |
| 11 | −86.72 ± 8.04 | 4 | −71.86 ± 15.09 | 22 | −14.00 ± 15.95 | 21 | 115 | 0.86 | 47 |
| 12 | −15.33 ± 13.66 | 26 | −12.87 ± 10.45 | 1 | −3.08 ± 8.84 | 4 | 22 | −0.62 | 31 |
| 13 | −72.96 ± 15.81 | 17 | −63.07 ± 15.48 | 16 | −4.17 ± 7.63 | 6 | 69 | 5.72 | 39 |
| 14 | −69.10 ± 17.06 | 20 | −48.02 ± 16.85 | 7 | −42.99 ± 28.42 | 26 | 17 | −21.91 | 53 |
| 15 | −29.47 ± 14.34 | 25 | −25.71 ± 16.14 | 2 | −5.97 ± 8.25 | 11 | 53 | −2.21 | 38 |
| 16 | −84.98 ± 10.92 | 7 | −71.52 ± 15.56 | 21 | −16.00 ± 21.24 | 23 | 59 | −2.55 | 51 |
| 17 | −94.83 ± 6.92 | 2 | −89.92 ± 10.17 | 25 | −1.87 ± 4.93 | 1 | 39 | 3.04 | 28 |
| 18 | −78.27 ± 13.21 | 14 | −62.09 ± 16.59 | 13 | −11.47 ± 15.57 | 17 | 66 | 4.70 | 44 |
| 19 | −81.81 ± 11.80 | 11 | −72.52 ± 13.38 | 23 | −7.10 ± 10.45 | 13 | 61 | 2.19 | 47 |
| 20 | −53.81 ± 19.74 | 24 | −29.08 ± 13.79 | 4 | −11.54 ± 12.92 | 18 | 37 | 13.18 | 46 |
| 21 | −86.69 ± 13.26 | 5 | −75.62 ± 15.68 | 24 | −10.03 ± 15.17 | 16 | 27 | 1.04 | 45 |
| 22 | −83.43 ± 12.34 | 8 | −68.40 ± 13.84 | 19 | −14.18 ± 17.41 | 22 | 65 | 0.85 | 49 |
| 23 | −78.62 ± 10.66 | 13 | −62.66 ± 10.96 | 15 | −12.11 ± 13.55 | 19 | 53 | 3.85 | 47 |
| 24 | −70.25 ± 15.49 | 19 | −59.12 ± 16.38 | 11 | −4.27 ± 10.47 | 7 | 65 | 6.86 | 37 |
| 25 | −77.31 ± 16.90 | 16 | −65.69 ± 19.06 | 17 | −6.45 ± 14.83 | 12 | 31 | 5.16 | 45 |
| Cluster | ATXN2 Expression Assay | Rank 1 | CMV Control | Rank 2 | Biochemical Control | Rank 3 | Number of Compounds | E | Total Rank |
|---|---|---|---|---|---|---|---|---|---|
| 0 | −94.39 ± 6.96 | 4 | −93.02 ± 7.79 | 25 | −88.17 ± 10.78 | 26 | 15 | −86.80 | 55 |
| 1 | −30.70 ± 7.61 | 25 | −39.44 ± 7.62 | 7 | −5.76 ± 9.41 | 17 | 43 | −14.51 | 49 |
| 2 | −70.55 ± 5.02 | 17 | −53.78 ± 4.71 | 11 | −4.34 ± 5.61 | 13 | 69 | 12.43 | 41 |
| 3 | −47.15 ± 6.30 | 23 | −55.00 ± 4.67 | 13 | −4.69 ± 5.91 | 14 | 44 | −12.53 | 50 |
| 4 | −88.57 ± 6.81 | 5 | −12.84 ± 5.72 | 2 | −0.68 ± 7.39 | 1 | 24 | 75.06 | 8 |
| 5 | −85.39 ± 3.80 | 9 | −73.68 ± 4.22 | 21 | −19.17 ± 5.18 | 19 | 74 | −7.45 | 49 |
| 6 | −86.65 ± 6.21 | 7 | −34.77 ± 4.20 | 6 | −1.88 ± 4.03 | 4 | 34 | 50.00 | 17 |
| 7 | −86.73 ± 3.40 | 6 | −71.17 ± 2.43 | 20 | −2.74 ± 4.06 | 7 | 127 | 12.82 | 33 |
| 8 | −83.19 ± 5.32 | 12 | −65.60 ± 6.04 | 17 | −38.68 ± 7.78 | 24 | 32 | −21.09 | 53 |
| 9 | −69.34 ± 5.10 | 18 | −26.73 ± 7.09 | 5 | −1.77 ± 5.11 | 3 | 38 | 40.84 | 26 |
| 10 | −79.04 ± 7.55 | 14 | −41.79 ± 6.61 | 9 | −30.33 ± 9.41 | 20 | 33 | 6.93 | 43 |
| 11 | −95.41 ± 5.41 | 2 | −90.50 ± 7.49 | 24 | −36.86 ± 9.23 | 23 | 34 | −31.96 | 49 |
| 12 | −13.14 ± 7.97 | 26 | −12.08 ± 8.07 | 1 | −5.66 ± 8.79 | 16 | 38 | −4.60 | 43 |
| 13 | −57.09 ± 4.95 | 21 | −66.84 ± 4.14 | 18 | −3.91 ± 6.36 | 11 | 52 | −13.67 | 50 |
| 14 | −70.98 ± 9.53 | 16 | −53.81 ± 10.13 | 12 | −68.46 ± 12.65 | 25 | 19 | −51.29 | 53 |
| 15 | −41.83 ± 7.31 | 24 | −16.49 ± 8.09 | 4 | −2.39 ± 7.13 | 6 | 36 | 22.96 | 34 |
| 16 | −71.70 ± 4.26 | 15 | −70.37 ± 4.81 | 19 | −1.50 ± 5.02 | 2 | 87 | −0.17 | 36 |
| 17 | −95.03 ± 3.37 | 3 | −83.68 ± 4.52 | 23 | −3.72 ± 5.76 | 10 | 46 | 7.63 | 36 |
| 18 | −82.70 ± 4.41 | 13 | −59.62 ± 4.39 | 15 | −18.64 ± 5.42 | 18 | 53 | 4.43 | 46 |
| 19 | −85.75 ± 4.75 | 8 | −49.89 ± 3.94 | 10 | −3.92 ± 4.88 | 12 | 67 | 31.95 | 30 |
| 20 | −67.87 ± 12.39 | 19 | −15.96 ± 7.71 | 3 | −32.55 ± 15.15 | 22 | 15 | 19.35 | 44 |
| 21 | −98.83 ± 2.18 | 1 | −96.97 ± 2.68 | 26 | −3.21 ± 6.01 | 9 | 96 | −1.35 | 36 |
| 22 | −62.51 ± 8.13 | 20 | −57.90 ± 6.65 | 14 | −31.95 ± 8.35 | 21 | 23 | −27.34 | 55 |
| 23 | −55.05 ± 5.93 | 22 | −40.79 ± 5.93 | 8 | −5.52 ± 8.90 | 15 | 54 | 8.75 | 45 |
| 24 | −85.38 ± 3.58 | 10 | −79.59 ± 3.18 | 22 | −2.95 ± 5.10 | 8 | 68 | 2.84 | 40 |
| 25 | −85.14 ± 4.71 | 11 | −61.87 ± −61.87 | 16 | −2.16 ± 6.54 | 5 | 100 | 21.10 | 32 |
| Molecular Descriptor | Subcluster 4_0 Mean ± SD | Subcluster 24_1 Mean ± SD | Statistical Test Result | |
|---|---|---|---|---|
| 1 | Aliphatic ring count | 6.17 ± 1.47 | 3.57 ± 1.90 | Mann–Whitney U = 5.50, p = 0.023 |
| 2 | Aromatic atom count | 1.00 ± 2.45 | 10.43 ± 7.83 | Welch’s corrected t(7) = 3.02, p = 0.018 |
| 3 | Aromatic bond count | 1.00 ± 2.45 | 10.71 ± 7.93 | Welch’s corrected t(7) = 3.07, p = 0.017 |
| 4 | Aromatic ring count | 0.17 ± 0.41 | 1.86 ± 1.35 | Welch’s corrected t(7) = 3.16, p = 0.015 |
| 5 | Asymmetric atom count | 14.17 ± 5.71 | 6.00 ± 3.74 | Student’s t(11) = 3.100, p = 0.010 |
| 6 | Carboaromatic ring count | 0.00 ± 0.00 | 1.43 ± 1.27 | Welch’s corrected t(6) = 2.97, p = 0.025 |
| 7 | Carbo ring count | 4.00 ± 0.00 | 2.57 ± 1.40 | Welch’s corrected t(6) = 2.71, p = 0.035 |
| 8 | Chiral center count | 14.17 ± 5.71 | 6.57 ± 3.51 | Student’s t(7) = 2.94, p = 0.013 |
| 9 | Aliphatic ring ratio | 6.17 ± 1.47 | 3.57 ± 1.90 | Welch’s corrected t(7) = 2.93, p = 0.022 |
| 10 | Aromatic atom ratio | 0.01 ± 0.03 | 0.11 ± 0.08 | Welch’s corrected t(8) = 2.95, p = 0.019 |
| 11 | Aromatic bond ratio | 0.01 ± 0.03 | 0.11 ± 0.08 | Welch’s corrected t(8) = 3.00, p = 0.018 |
| 12 | Aromatic ring ratio | 0.03 ± 0.07 | 0.32 ± 0.26 | Welch’s corrected t(7) = 2.93, p = 0.022 |
| 13 | Asymmetric atom ratio | 0.12 ± 0.02 | 0.07 ± 0.06 | Student’s t(11) = 3.419, p = 0.006 |
| 14 | Carboaliphatic ring ratio | 0.66 ± 0.13 | 0.22 ± 0.28 | Mann–Whitney U = 3.00, p = 0.008 |
| 15 | Carboaromatic ring ratio | 0.00 ± 0.00 | 0.27 ± 0.27 | Welch’s corrected t(6) = 2.64, p = 0.038 |
| 16 | Chiral center ratio | 0.16 ± 0.02 | 0.08 ± 0.05 | Student’s t(11) = 3.35, p = 0.007 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sahay, S.; Wen, J.; Scoles, D.R.; Simeonov, A.; Dexheimer, T.S.; Jadhav, A.; Kales, S.C.; Sun, H.; Pulst, S.M.; Facelli, J.C.; et al. Identifying Molecular Properties of Ataxin-2 Inhibitors for Spinocerebellar Ataxia Type 2 Utilizing High-Throughput Screening and Machine Learning. Biology 2025, 14, 522. https://doi.org/10.3390/biology14050522
Sahay S, Wen J, Scoles DR, Simeonov A, Dexheimer TS, Jadhav A, Kales SC, Sun H, Pulst SM, Facelli JC, et al. Identifying Molecular Properties of Ataxin-2 Inhibitors for Spinocerebellar Ataxia Type 2 Utilizing High-Throughput Screening and Machine Learning. Biology. 2025; 14(5):522. https://doi.org/10.3390/biology14050522
Chicago/Turabian StyleSahay, Smita, Jingran Wen, Daniel R. Scoles, Anton Simeonov, Thomas S. Dexheimer, Ajit Jadhav, Stephen C. Kales, Hongmao Sun, Stefan M. Pulst, Julio C. Facelli, and et al. 2025. "Identifying Molecular Properties of Ataxin-2 Inhibitors for Spinocerebellar Ataxia Type 2 Utilizing High-Throughput Screening and Machine Learning" Biology 14, no. 5: 522. https://doi.org/10.3390/biology14050522
APA StyleSahay, S., Wen, J., Scoles, D. R., Simeonov, A., Dexheimer, T. S., Jadhav, A., Kales, S. C., Sun, H., Pulst, S. M., Facelli, J. C., & Jones, D. E. (2025). Identifying Molecular Properties of Ataxin-2 Inhibitors for Spinocerebellar Ataxia Type 2 Utilizing High-Throughput Screening and Machine Learning. Biology, 14(5), 522. https://doi.org/10.3390/biology14050522






