Targeting Methionine Metabolism Reveals AMPK-SAMTOR Signaling as a Therapeutic Vulnerability in Prostate Cancer
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. TCGA Data Analysis
2.2. Cell Lines and Reagents
2.3. MTT Cell Viability and Cell Survival Assay
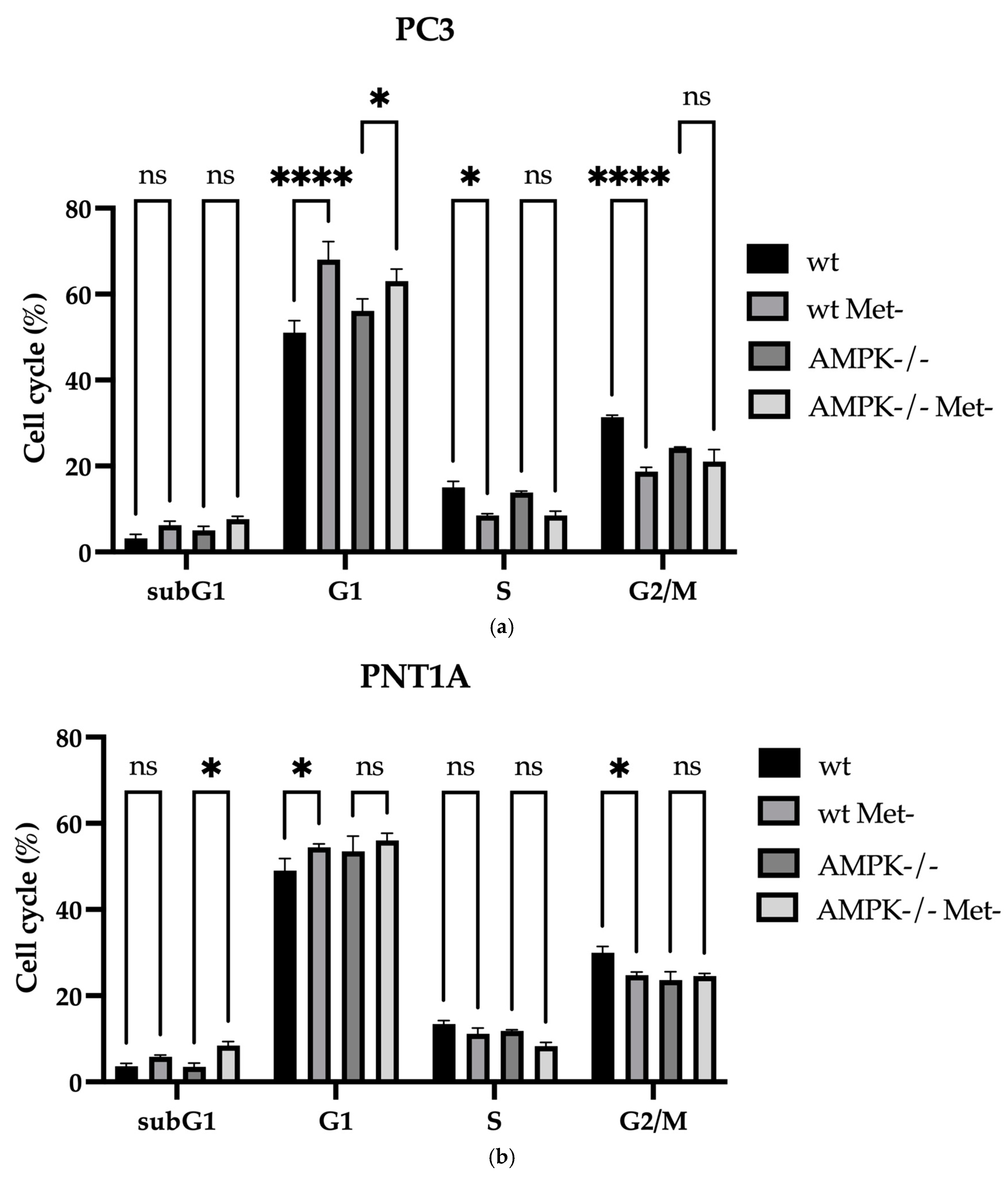
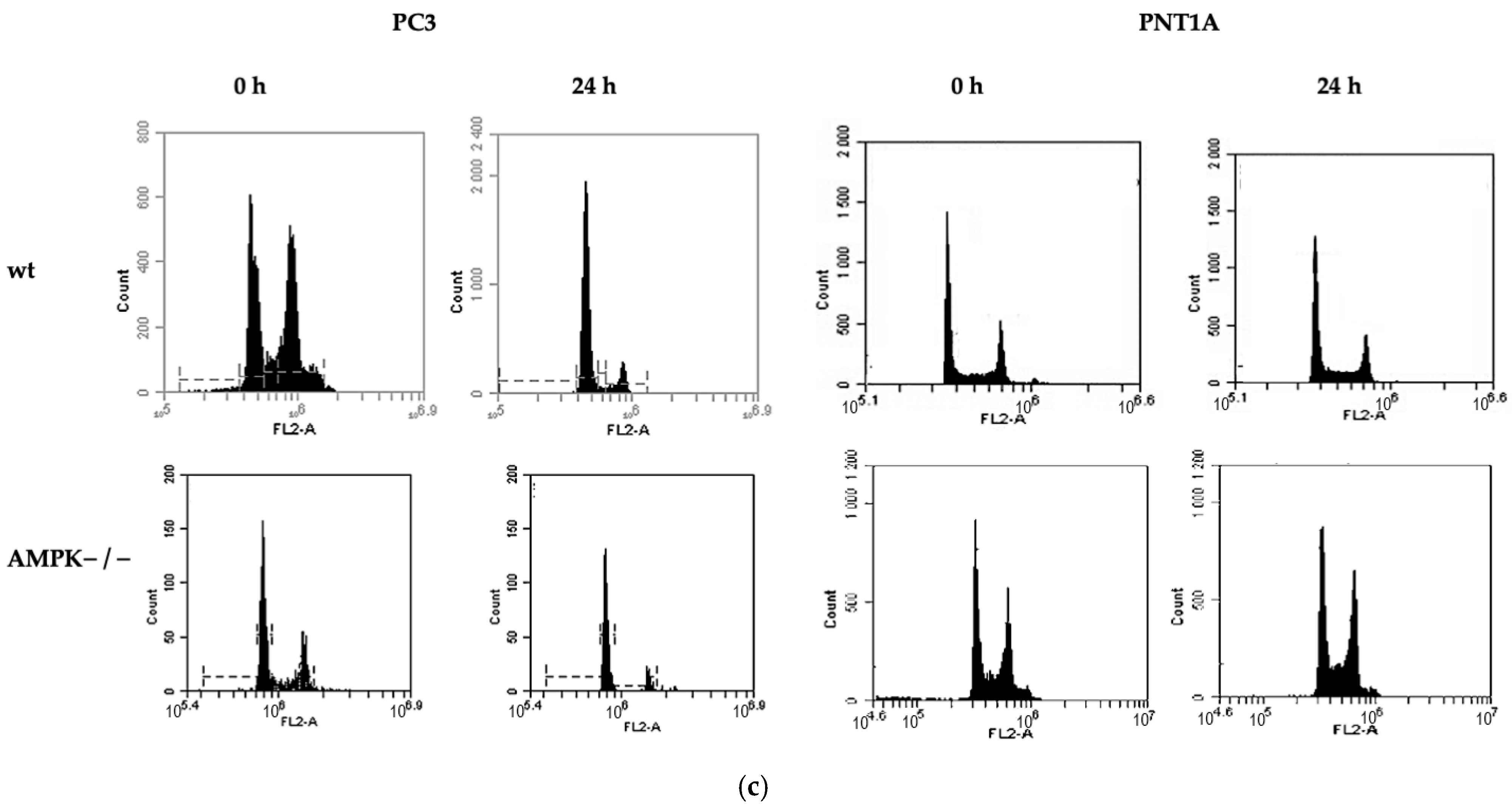
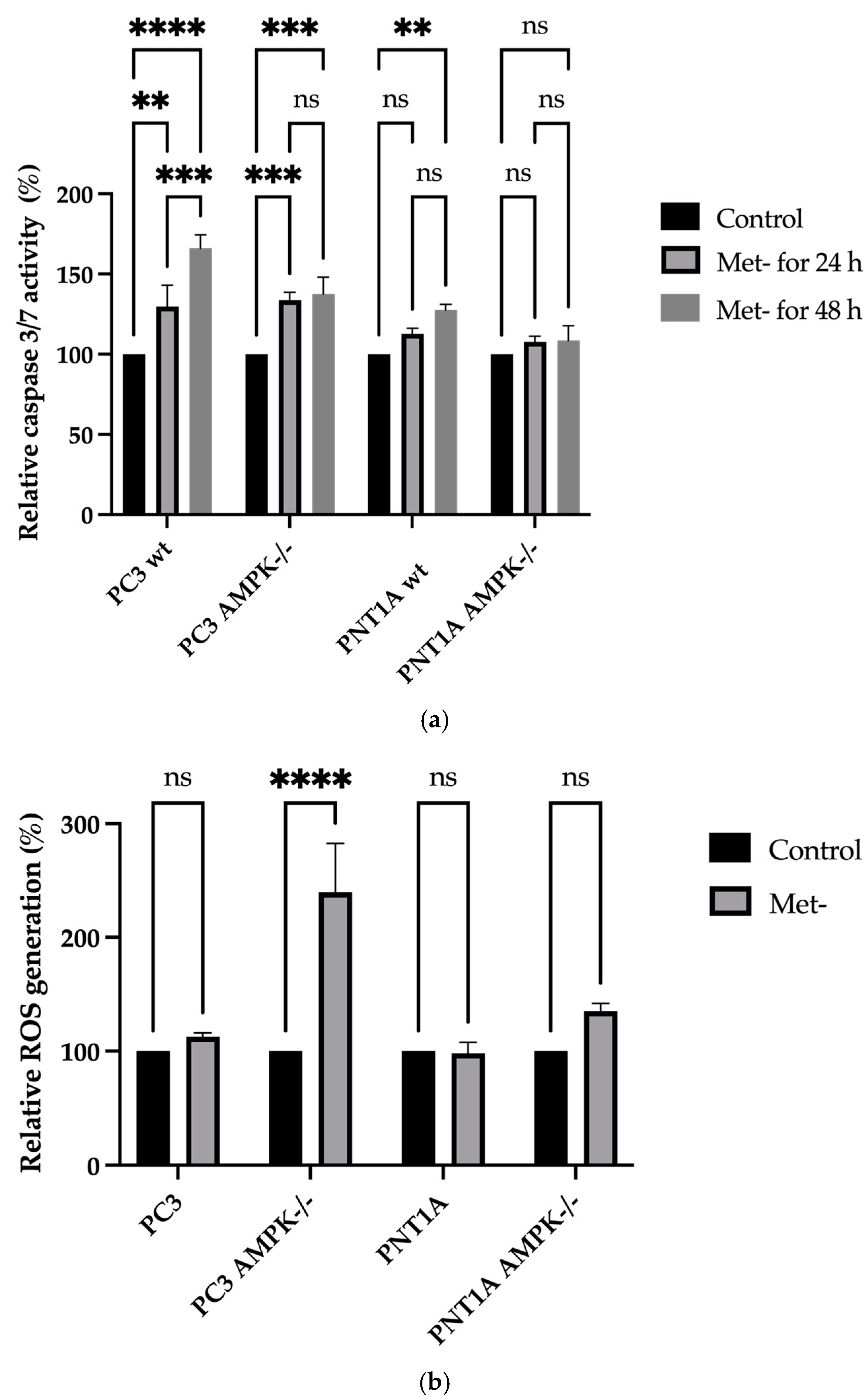
2.4. Cell Cycle and Cell Death Analysis
2.5. Colony Formation Assay
2.6. Western Blot Analysis and Immunoprecipitation (IP)
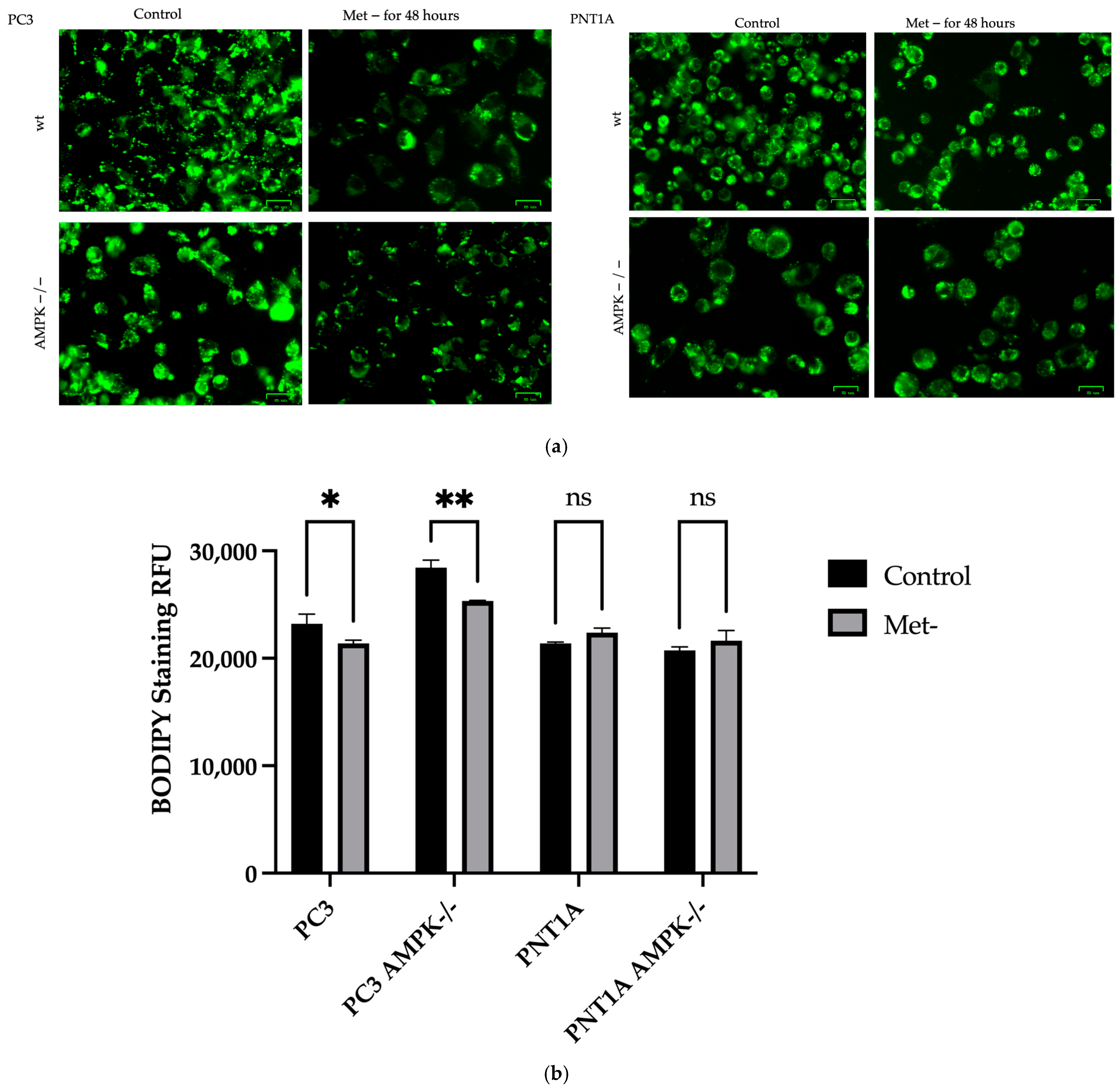
2.7. Oil-Red O Staining and BODIPY® 493/503 Staining-Fluorescence Measurement
2.8. Total Reactive Oxygen Species Detection
2.9. Data Analysis
3. Results
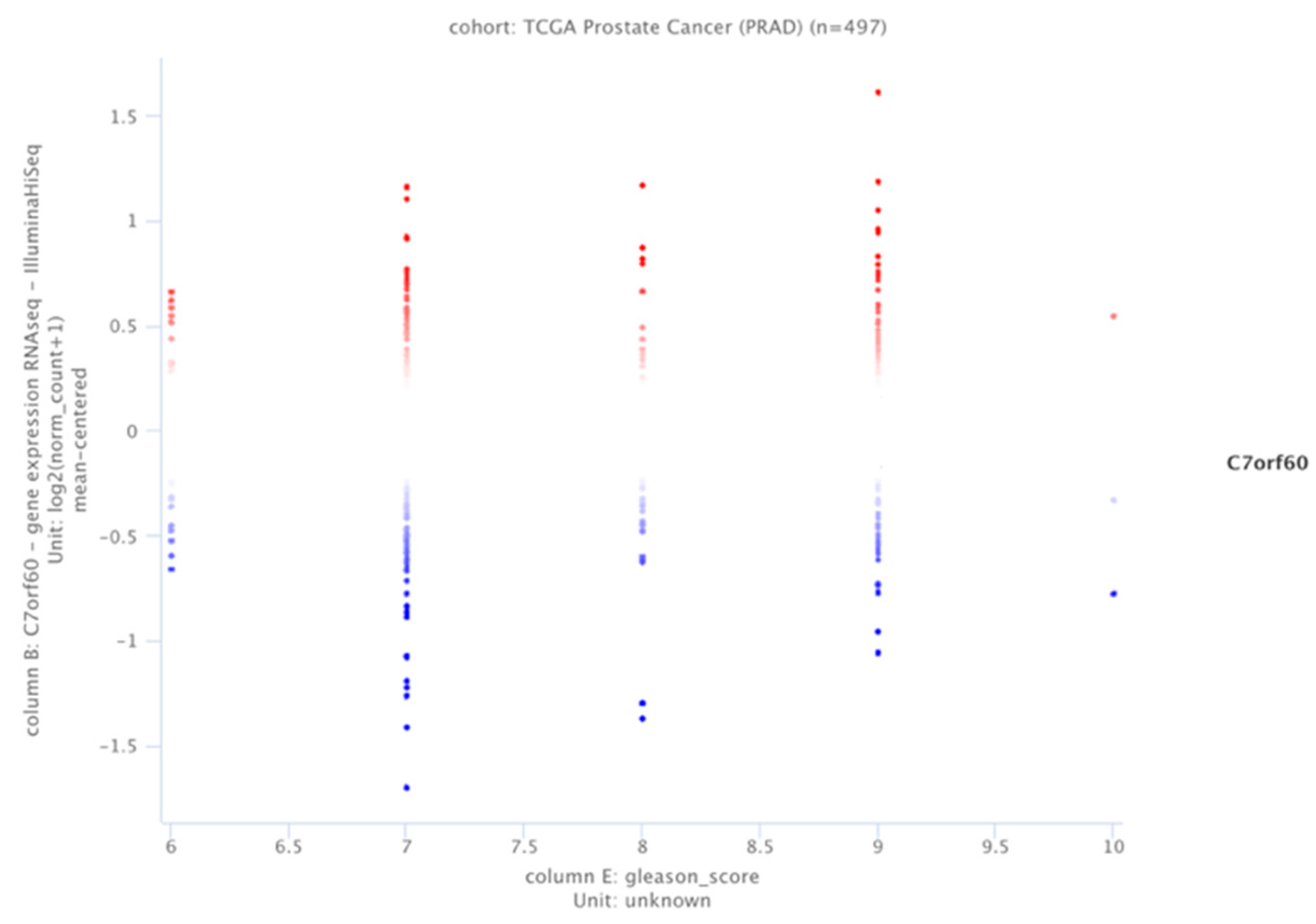
3.1. SAMTOR (C7orf60) Expression Profile Is Related to the Gleason Index Distribution
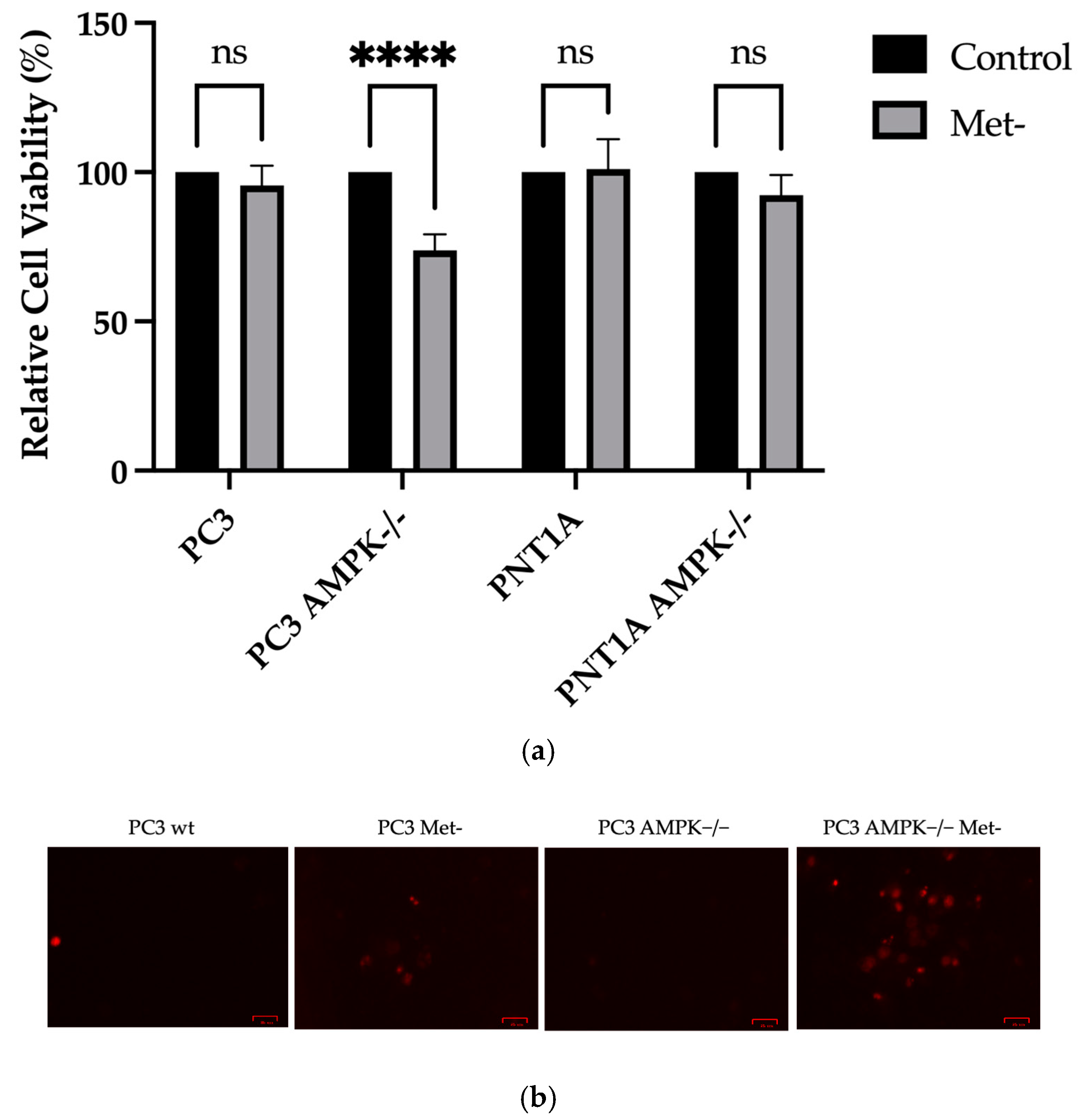
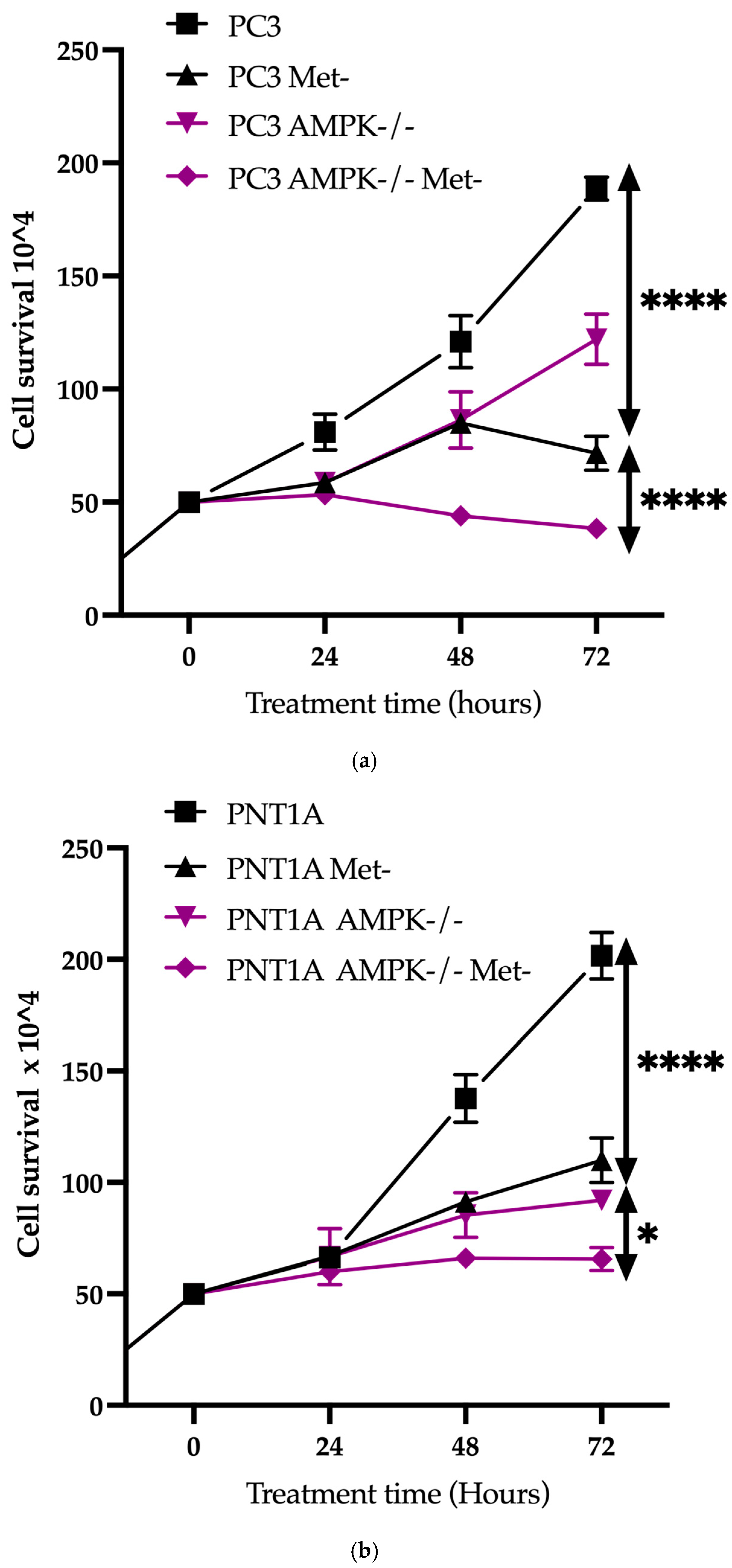
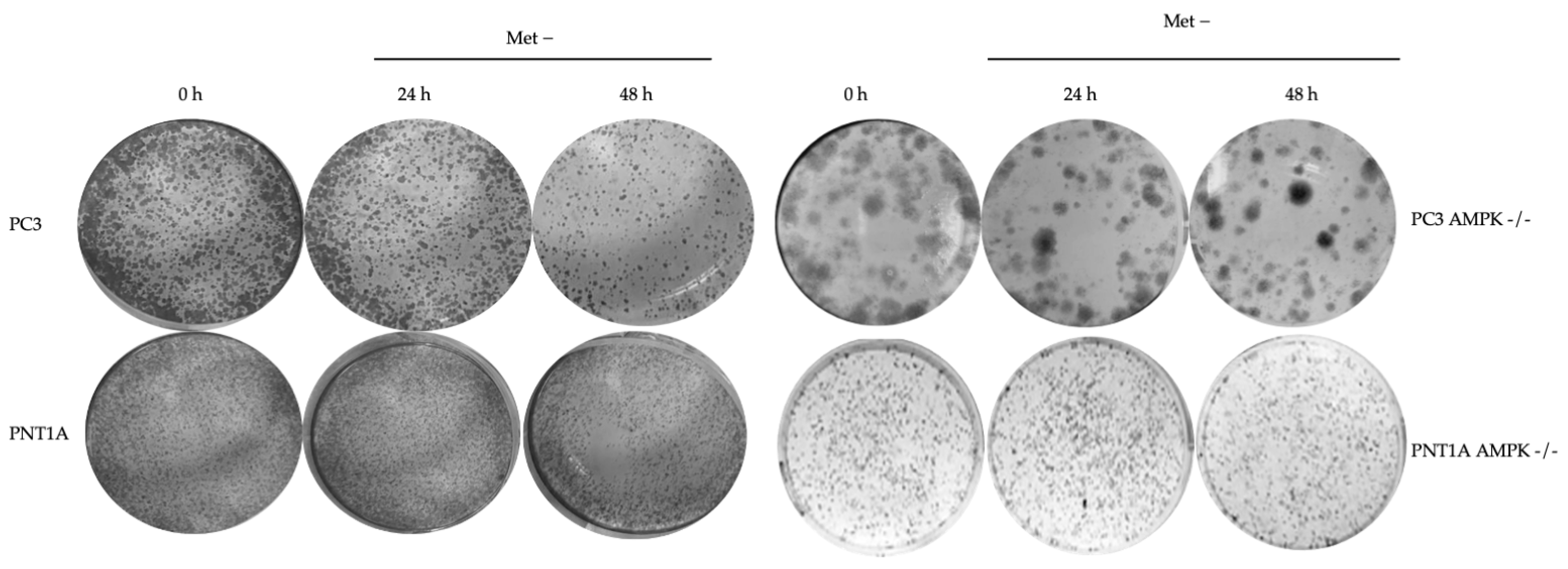
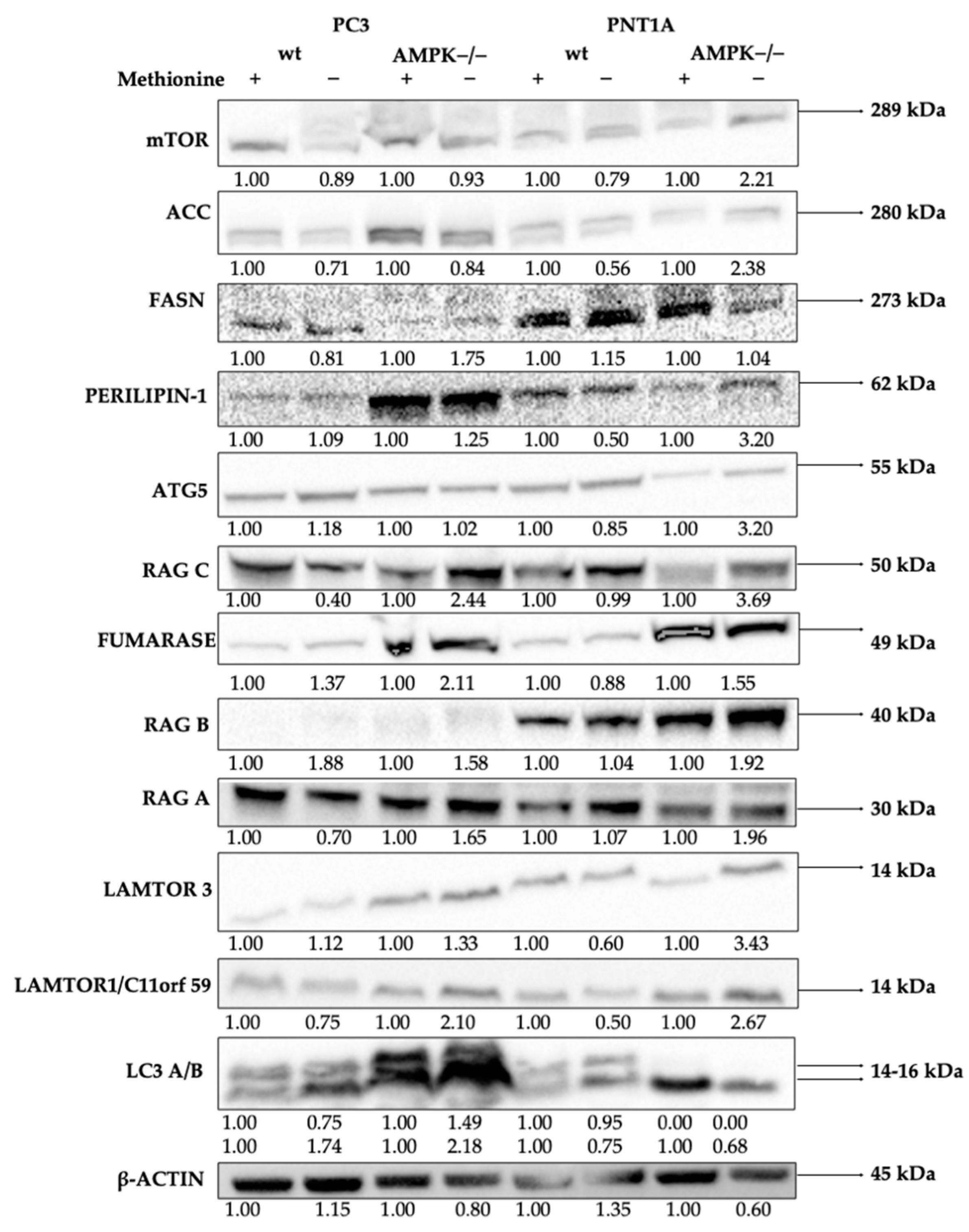
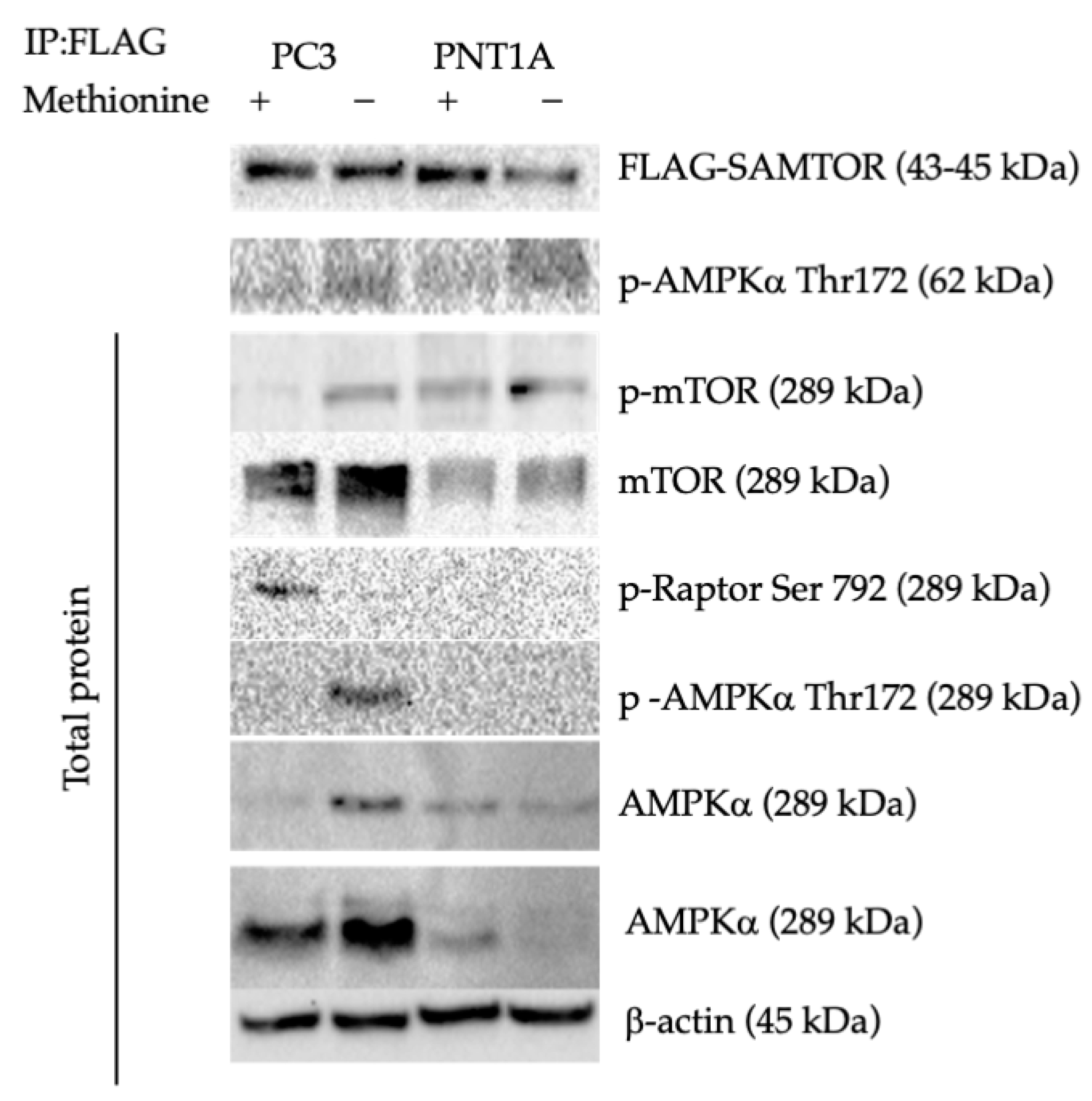
3.2. Methionine Starvation Led to Cell Viability Loss in Only AMPK−/− PC3 PCa Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hanahan, D.; Weinberg, R.A. The Hallmarks of Cancer Review Douglas. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Cavuoto, P.; Fenech, M.F. A review of methionine dependency and the role of methionine restriction in cancer growth control and life-span extension. Cancer Treat. Rev. 2012, 38, 726–736. [Google Scholar] [CrossRef]
- Overgaard, A.J.; Weir, J.M.; De Souza, D.P.; Tull, D.; Haase, C.; Meikle, P.J.; Pociot, F. Lipidomic and metabolomic characterization of a genetically modified mouse model of the early stages of human type 1 diabetes pathogenesis. Metabolomics 2016, 12, 13. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, P. Methionine dependence of cancer. Biomolecules 2020, 10, 568. [Google Scholar] [CrossRef]
- Rutherfurd, S.M.; Moughan, P.J. Determination of sulfur amino acids in foods as related to bioavailability. Proc. J. AOAC Int. 2008, 91, 907–913. [Google Scholar] [CrossRef]
- Hoffman, R.M. The wayward methyl group and the cascade to cancer. Cell Cycle 2017, 16, 825–829. [Google Scholar] [CrossRef]
- Séité, S.; Mourier, A.; Camougrand, N.; Salin, B.; Figueiredo-Silva, A.C.; Fontagné-Dicharry, S.; Panserat, S.; Seiliez, I. Dietary methionine deficiency affects oxidative status, mitochondrial integrity and mitophagy in the liver of rainbow trout (Oncorhynchus mykiss). Sci. Rep. 2018, 8, 10151. [Google Scholar] [CrossRef] [PubMed]
- Aubel, C.; Dehez, S.; Chabanon, H.; Seva, C.; Ferrara, M.; Brachet, P. Activation of c-Jun N-terminal kinase 1 (JNK-1) after amino acid deficiency in HeLa cells. Cell. Signal. 2001, 13, 417–423. [Google Scholar] [CrossRef]
- Cooke, D.; Mattocks, D.; Nichenametla, S.N.; Anunciado-Koza, R.P.; Koza, R.A.; Ables, G.P. Weight Loss and Concomitant Adipose Autophagy under methionine-Restricted Obese Mice is Not Dependent on Adiponectin or FGF21. Obesity 2020, 28, 1075–1085. [Google Scholar] [CrossRef]
- Zakikhani, M.; Dowling, R.J.O.; Sonenberg, N.; Pollak, M.N. The effects of adiponectin and metformin on prostate and colon neoplasia involve the activation of AMP-activated protein kinase. Cancer Prev. Res. 2008, 1, 369–375. [Google Scholar] [CrossRef]
- Ruan, T.; Li, L.; Lyu, Y.; Luo, Q.; Wu, B. Effect of methionine deficiency on oxidative stress and apoptosis in the small intestine of broilers. Acta Vet. Hung. 2018, 66, 52–65. [Google Scholar] [CrossRef] [PubMed]
- Kitada, M.; Xu, J.; Ogura, Y.; Monno, I.; Koya, D. Mechanism of Activation of Mechanistic Target of Rapamycin Complex 1 by Methionine. Front. Cell Dev. Biol. 2020, 8, 715. [Google Scholar] [CrossRef]
- Ruckenstuhl, C.; Netzberger, C.; Entfellner, I.; Carmona-Gutierrez, D.; Kickenweiz, T.; Stekovic, S.; Gleixner, C.; Schmid, C.; Klug, L.; Sorgo, A.G.; et al. Lifespan Extension by Methionine Restriction Requires Autophagy-Dependent Vacuolar Acidification. PLoS Genet. 2014, 10, e1004347. [Google Scholar] [CrossRef]
- Borrego, S.L.; Fahrmann, J.; Datta, R.; Stringari, C.; Grapov, D.; Zeller, M.; Chen, Y.; Wang, P.; Baldi, P.; Gratton, E.; et al. Metabolic changes associated with methionine stress sensitivity in MDA-MB-468 breast cancer cells. Cancer Metab. 2016, 4, 9. [Google Scholar] [CrossRef]
- Aledo, J.C.; Li, Y.; de Magalhães, J.P.; Ruíz-Camacho, M.; Pérez-Claros, J.A. Mitochondrially encoded methionine is inversely related to longevity in mammals. Aging Cell 2011, 10, 198–207. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Orozco, J.M.; Saxton, R.A.; Condon, K.J.; Liu, G.Y.; Krawczyk, P.A.; Scaria, S.M.; Wade Harper, J.; Gygi, S.P.; Sabatini, D.M. SAMTOR is an S-adenosylmethionine sensor for the mTORC1 pathway. Science 2017, 358, 813–818. [Google Scholar] [CrossRef]
- Parmigiani, A.; Nourbakhsh, A.; Ding, B.; Wang, W.; Kim, Y.C.; Akopiants, K.; Guan, K.L.; Karin, M.; Budanov, A.V. Sestrins Inhibit mTORC1 Kinase Activation through the GATOR Complex. Cell Rep. 2014, 9, 1281–1291. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.; Ye, Q.; Zeng, X.; Yang, G.; Ye, C.; Chen, M.; Yu, H.; Wang, Y.; Wang, G.; Huang, S.; et al. CBS and MAT2A improve methionine-mediated DNA synthesis through SAMTOR/mTORC1/S6K1/CAD pathway during embryo implantation. Cell Prolif. 2021, 54, e12950. [Google Scholar] [CrossRef]
- Wolfson, R.L.; Chantranupong, L.; Wyant, G.A.; Gu, X.; Orozco, J.M.; Shen, K.; Condon, K.J.; Petri, S.; Kedir, J.; Scaria, S.M.; et al. KICSTOR recruits GATOR1 to the lysosome, which is necessary for nutrients to regulate mTORC1. Nature 2017, 543, 438–442. [Google Scholar] [CrossRef]
- Hurtley, S.M. SAMTOR joins the family. Science 2017, 358, 758–760. [Google Scholar] [CrossRef]
- Parkhitko, A.A.; Favorova, O.O.; Khabibullin, D.I.; Anisimov, V.N.; Henske, E.P. Kinase mTOR: Regulation and role in maintenance of cellular homeostasis, tumor development, and aging. Biochemistry 2014, 79, 88–101. [Google Scholar] [CrossRef]
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef]
- Villa, E.; Sahu, U.; O’Hara, B.P.; Ali, E.S.; Helmin, K.A.; Asara, J.M.; Gao, P.; Singer, B.D.; Ben-Sahra, I. mTORC1 stimulates cell growth through SAM synthesis and m6A mRNA-dependent control of protein synthesis. Mol. Cell 2021, 81, 2076–2093.e9. [Google Scholar] [CrossRef]
- Corbin, J.M.; Ruiz-Echevarría, M.J. One-carbon metabolism in prostate cancer: The role of androgen signaling. Int. J. Mol. Sci. 2016, 17, 1208. [Google Scholar] [CrossRef]
- Pardo, J.C.; Ruiz de Porras, V.; Gil, J.; Font, A.; Puig-Domingo, M.; Jordà, M. Lipid Metabolism and Epigenetics Crosstalk in Prostate Cancer. Nutrients 2022, 14, 851. [Google Scholar] [CrossRef]
- Reina-Campos, M.; Linares, J.F.; Duran, A.; Cordes, T.; L’Hermitte, A.; Badur, M.G.; Bhangoo, M.S.; Thorson, P.K.; Richards, A.; Rooslid, T.; et al. Increased Serine and One-Carbon Pathway Metabolism by PKCλ/ι Deficiency Promotes Neuroendocrine Prostate Cancer. Cancer Cell 2019, 35, 385–400.e9. [Google Scholar] [CrossRef]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repečka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef]
- Wolfson, R.L.; Chantranupong, L.; Saxton, R.A.; Shen, K.; Scaria, S.M.; Cantor, J.R.; Sabatini, D.M. Sestrin2 is a leucine sensor for the mTORC1 pathway. Science 2016, 351, 43–48. [Google Scholar] [CrossRef]
- Peterson, T.R.; Sengupta, S.S.; Harris, T.E.; Carmack, A.E.; Kang, S.A.; Balderas, E.; Guertin, D.A.; Madden, K.L.; Carpenter, A.E.; Finck, B.N.; et al. MTOR complex 1 regulates lipin 1 localization to control the srebp pathway. Cell 2011, 146, 408–420. [Google Scholar] [CrossRef]
- O’Reilly, K.E.; Rojo, F.; She, Q.B.; Solit, D.; Mills, G.B.; Smith, D.; Lane, H.; Hofmann, F.; Hicklin, D.J.; Ludwig, D.L.; et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006, 66, 1500–1508. [Google Scholar] [CrossRef]
- Martin, D.; Nguyen, Q.; Molinolo, A.; Gutkind, J.S. Accumulation of dephosphorylated 4EBP after mTOR inhibition with rapamycin is sufficient to disrupt paracrine transformation by the KSHV vGPCR oncogene. Oncogene 2014, 33, 2405–2412. [Google Scholar] [CrossRef]
- Sugimura, T.; Birnbaum, S.M.; Winitz, M.; Greenstein, J.P. Quantitative nutritional studies with water-soluble, chemically defined diets. VIII. The forced feeding of diets each lacking in one essential amino acid. Arch. Biochem. Biophys. 1959, 81, 448–455. [Google Scholar] [CrossRef]
- Pacana, T.; Cazanave, S.; Verdianelli, A.; Patel, V.; Min, H.K.; Mirshahi, F.; Quinlivan, E.; Sanyal, A.J. Dysregulated hepatic methionine metabolism drives homocysteine elevation in diet-induced non-alcoholic fatty liver disease. PLoS ONE 2015, 10, e0136822. [Google Scholar] [CrossRef]
- Elshorbagy, A.K.; Valdivia-Garcia, M.; Mattocks, D.A.L.; Plummer, J.D.; Smith, A.D.; Drevon, C.A.; Refsum, H.; Perrone, C.E. Cysteine supplementation reverses methionine restriction effects on rat adiposity: Significance of stearoyl-coenzyme a desaturase. J. Lipid Res. 2011, 52, 104–112. [Google Scholar] [CrossRef]
- Guo, H.; Lishko, V.K.; Herrera, H.; Groce, A.; Kubota, T.; Hoffman, R.M. Therapeutic Tumor-specific Cell Cycle Block Induced by Methionine Starvation in Vivo. Cancer Res. 1993, 53, 5676–5679. [Google Scholar]
- Wang, X.; Song, Y.; Shi, Y.; Yang, D.; Li, J.; Yin, B. SNHG3 could promote prostate cancer progression through reducing methionine dependence of PCa cells. Cell. Mol. Biol. Lett. 2022, 27, 13. [Google Scholar] [CrossRef]
- Park, H.S.; Song, J.W.; Park, J.H.; Lim, B.K.; Moon, O.S.; Son, H.Y.; Lee, J.H.; Gao, B.; Won, Y.S.; Kwon, H.J. TXNIP/VDUP1 attenuates steatohepatitis via autophagy and fatty acid oxidation. Autophagy 2020, 17, 2549–2564. [Google Scholar] [CrossRef]
- Zhang, C.S.; Jiang, B.; Li, M.; Zhu, M.; Peng, Y.; Zhang, Y.L.; Wu, Y.Q.; Li, T.Y.; Liang, Y.; Lu, Z.; et al. The lysosomal v-ATPase-regulator complex is a common activator for AMPK and mTORC1, acting as a switch between catabolism and anabolism. Cell Metab. 2014, 20, 526–540. [Google Scholar] [CrossRef]
- Chantranupong, L.; Scaria, S.M.; Saxton, R.A.; Gygi, M.P.; Shen, K.; Wyant, G.A.; Wang, T.; Harper, J.W.; Gygi, S.P.; Sabatini, D.M. The CASTOR Proteins Are Arginine Sensors for the mTORC1 Pathway. Cell 2016, 165, 153–164. [Google Scholar] [CrossRef]
- Meng, D.; Yang, Q.; Wang, H.; Melick, C.H.; Navlani, R.; Frank, A.R.; Jewell, J.L. Glutamine and asparagine activate mTORC1 independently of Rag GTPases. J. Biol. Chem. 2020, 295, 2890–2899. [Google Scholar] [CrossRef]
- Liteplo, R.G. Altered methionine metabolism in metastatic variants of a human melanoma cell line. Cancer Lett. 1989, 44, 23–31. [Google Scholar] [CrossRef]
- Liteplo, R.G. Reversion to a homocysteine-responsive phenotype in a human melanoma cell line is associated with diminished growth potential and increased methionine biosynthesis. Exp. Cell Res. 1990, 186, 340–345. [Google Scholar] [CrossRef]
- Strain, J.J.; Lynch, S.M. Excess dietary methionine decreases indices of copper statusin the rat. Ann. Nutr. Metab. 1990, 34, 93–97. [Google Scholar] [CrossRef]
- Dahlhoff, C.; Worsch, S.; Sailer, M.; Hummel, B.A.; Fiamoncini, J.; Uebel, K.; Obeid, R.; Scherling, C.; Geisel, J.; Bader, B.L.; et al. Methyl-donor supplementation in obese mice prevents the progression of NAFLD, activates AMPK and decreases acyl-carnitine levels. Mol. Metab. 2014, 3, 565–580. [Google Scholar] [CrossRef]
- Sajan, M.P.; Bandyopadhyay, G.; Miura, A.; Standaert, M.L.; Nimal, S.; Longnus, S.L.; Van Obberghen, E.; Hainault, I.; Foufelle, F.; Kahn, R.; et al. AICAR and metformin, but not exercise, increase muscle glucose transport through AMPK-, ERK-, and PDK1-dependent activation of atypical PKC. Am. J. Physiol. Endocrinol. Metab. 2010, 298, E179–E192. [Google Scholar] [CrossRef]
- Gwinn, D.M.; Shackelford, D.B.; Egan, D.F.; Mihaylova, M.M.; Mery, A.; Vasquez, D.S.; Turk, B.E.; Shaw, R.J. AMPK Phosphorylation of Raptor Mediates a Metabolic Checkpoint. Mol. Cell 2008, 30, 214–226. [Google Scholar] [CrossRef]
- Hoffman, R.M.; Yano, S. Tumor-Specific S/G 2-Phase Cell Cycle Arrest of Cancer Cells by Methionine Restriction. Methods Mol. Biol. 2019, 1866, 49–60. [Google Scholar] [CrossRef]
- Helliwell, K.E.; Wheeler, G.L.; Leptos, K.C.; Goldstein, R.E.; Smith, A.G. Insights into the evolution of vitamin B12 auxotrophy from sequenced algal genomes. Mol. Biol. Evol. 2011, 28, 2921–2933. [Google Scholar] [CrossRef]
- Wallis, K.; Bird, J.T.; Gies, A.; Mackintosh, S.G.; Tackett, A.J.; Byrum, S.; Miousse, I.R. Methionine stress induces ferroptosis under methioninedependent cancer cells. bioRxiv 2020. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, L.; Pan, J. The role of L-type amino acid transporter 1 in human tumors. Intractable Rare Dis. Res. 2015, 4, 165–169. [Google Scholar] [CrossRef]
- Lu, S.; Epner, D.E. Molecular mechanisms of cell cycle block by methionine restriction in human prostate cancer cells. Nutr. Cancer 2000, 38, 123–130. [Google Scholar] [CrossRef]
- Song, B.; Zeng, Q.; Liu, Y.; Wu, B. Effect of methionine deficiency on the apoptosis and cell cycle of kidney in broilers. Res. Vet. Sci. 2019, 135, 228–236. [Google Scholar] [CrossRef]
- Bardella, C.; Olivero, M.; Lorenzato, A.; Geuna, M.; Adam, J.; O’Flaherty, L.; Rustin, P.; Tomlinson, I.; Pollard, P.J.; Di Renzo, M.F. Cells Lacking the Fumarase Tumor Suppressor Are Protected from Apoptosis through a Hypoxia-Inducible Factor-Independent, AMPK-Dependent Mechanism. Mol. Cell. Biol. 2012, 32, 3081–3094. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, S.; Zhai, A.; Zhang, B.; Tian, G. AMPK-mediated regulation of lipid metabolism by phosphorylation. Biol. Pharm. Bull. 2018, 41, 985–993. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arisan, S.; Sever, A.; Obakan-Yerlikaya, P.; Arisan, E.D.; Uysal-Onganer, P. Targeting Methionine Metabolism Reveals AMPK-SAMTOR Signaling as a Therapeutic Vulnerability in Prostate Cancer. Biology 2025, 14, 507. https://doi.org/10.3390/biology14050507
Arisan S, Sever A, Obakan-Yerlikaya P, Arisan ED, Uysal-Onganer P. Targeting Methionine Metabolism Reveals AMPK-SAMTOR Signaling as a Therapeutic Vulnerability in Prostate Cancer. Biology. 2025; 14(5):507. https://doi.org/10.3390/biology14050507
Chicago/Turabian StyleArisan, Serdar, Ayyuce Sever, Pinar Obakan-Yerlikaya, Elif Damla Arisan, and Pinar Uysal-Onganer. 2025. "Targeting Methionine Metabolism Reveals AMPK-SAMTOR Signaling as a Therapeutic Vulnerability in Prostate Cancer" Biology 14, no. 5: 507. https://doi.org/10.3390/biology14050507
APA StyleArisan, S., Sever, A., Obakan-Yerlikaya, P., Arisan, E. D., & Uysal-Onganer, P. (2025). Targeting Methionine Metabolism Reveals AMPK-SAMTOR Signaling as a Therapeutic Vulnerability in Prostate Cancer. Biology, 14(5), 507. https://doi.org/10.3390/biology14050507





