Significant Changes in Low-Abundance Protein Content Detected by Proteomic Analysis of Urine from Patients with Renal Stones After Extracorporeal Shock Wave Lithotripsy
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. ESWL Procedure
2.3. Sample Preparation
2.4. Two-Dimensional Analysis
2.5. Protein Digestion and MALDI-TOF/TOF-MS Analysis
2.6. Database MS/MS Searching
2.7. Protein Validation by Western Blot Analysis
2.8. Bioinformatic Analysis of Proteomic Data
2.9. Statistical Analysis
3. Results
3.1. Urine Sample Collection and Removal of High-Abundance Proteins
3.2. Urine Proteomic Analyses
3.3. Validation of the Protein Sequence and Urinary Content, Respectively, by LIFT-MALDI TOF (MS/MS) and Western Blot Analyses of Some Peculiar Proteins Identified by MS and Selected Among the Others in Post-ESWL Urine Samples
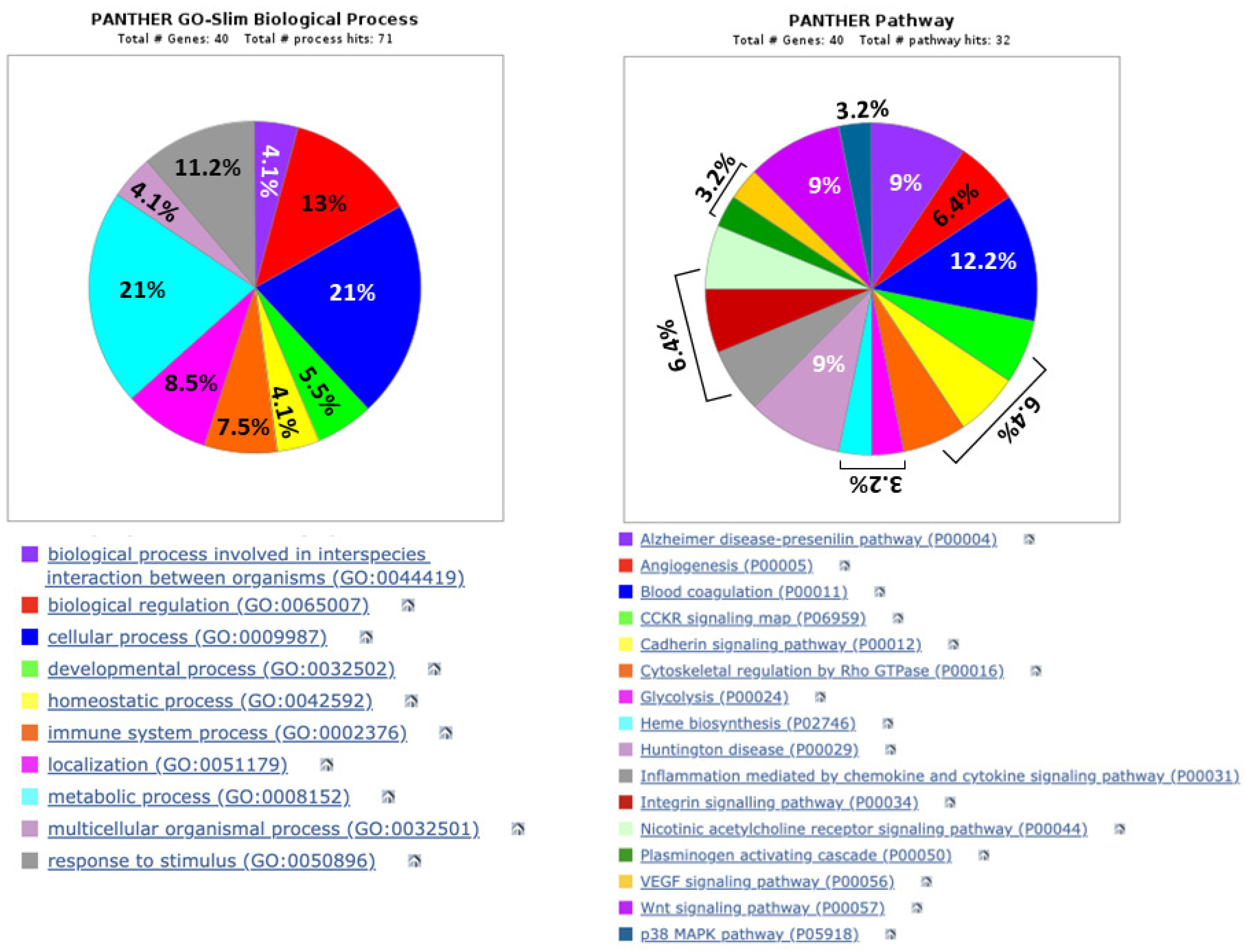
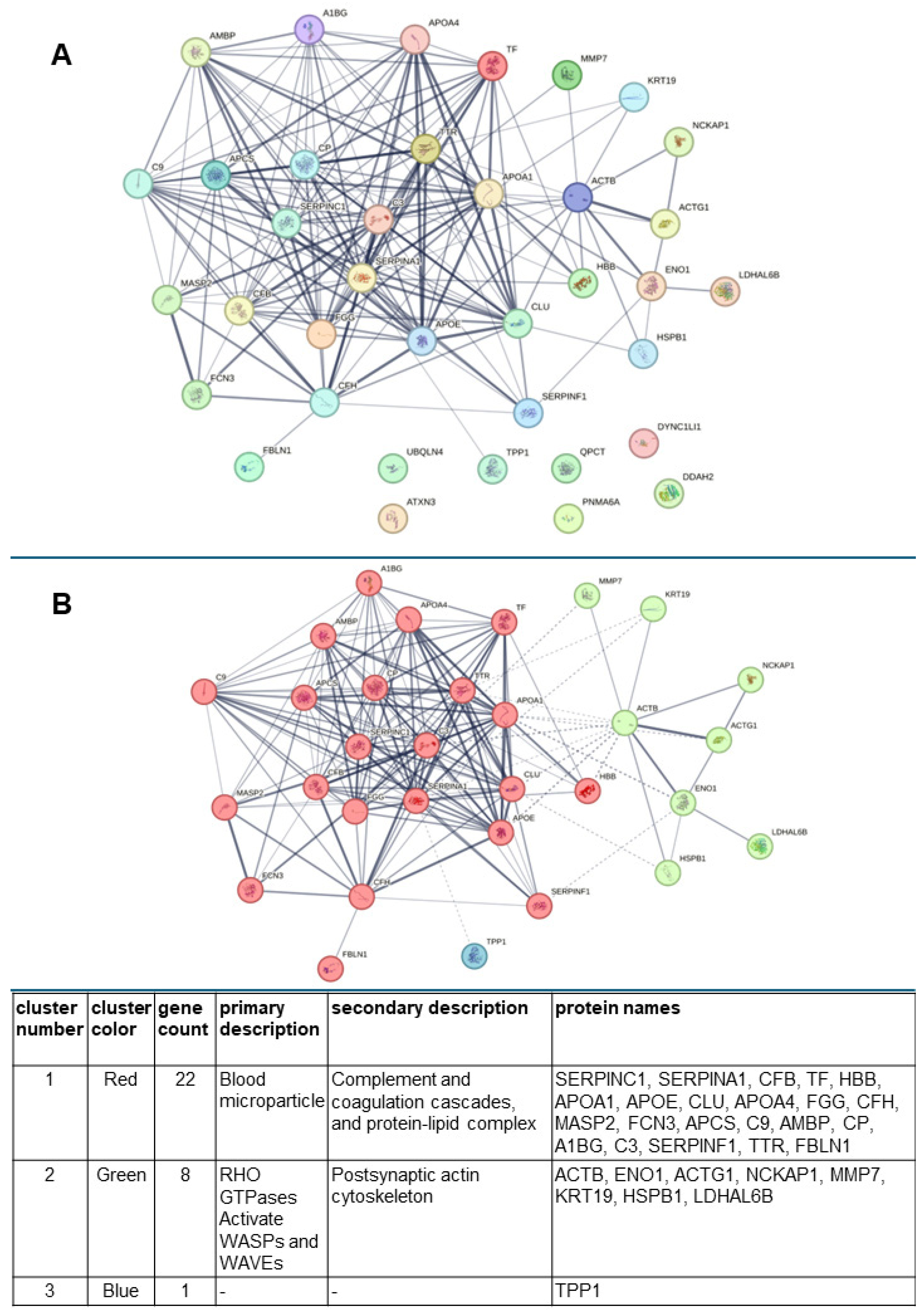
3.4. Computer Analysis of the Biological Functions and Related Pathways, as Well as Possible Networks, Between the Urinary Proteins Whose Levels Were Modified by Exposure of Patients to Lithotripsy
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Clark, D.L.; Connors, B.A.; Evan, A.P.; Handa, R.K.; Gao, S. Effect of shock wave number on renal oxidative stress and inflammation. BJU Int. 2011, 107, 318–322. [Google Scholar] [CrossRef] [PubMed]
- Gecit, İ.; Kavak, S.; Meral, I.; Pirinçci, N.; Güneş, M.; Demir, H.; Cengiz, N.; Ceylan, K. Effects of shock waves on oxidative stress, antioxidant enzyme and element levels in kidney of rats. Biol. Trace Elem. Res. 2011, 144, 1069–1076. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Liu, J.; Li, Z.; Ge, C.; Guo, H.; Song, S.; Li, Z.; Bai, S. The association between renal pelvis urine density and the risk of severe infectious complications in patient with symptom-free hydronephrosis after shock wave lithotripsy: A multi-center prospective study. Urolithiasis 2024, 2, 72. [Google Scholar] [CrossRef]
- Shao, Y.; Connors, B.; Evan, A.P.; Willis, L.R.; Lifshitz, D.; Lingeman, J.E. Morphological changes induced in the pig kidney by extracorporeal shock wave lithotripsy: Nephron injury. Anat. Rec. Part A Discov. Mol. Cell. Evol. Biol. 2003, 275, 979–989. [Google Scholar] [CrossRef]
- Clark, D.L.; Connors, B.A.; Evan, A.P.; Willis, L.R.; Handa, R.K.; Gao, S. Localization of renal oxidative stress and inflammatory response after lithotripsy. BJU Int. 2009, 103, 1562–1568. [Google Scholar] [CrossRef]
- Singh, D.P.; Mondal, S.; Sarkar, D. Assessment of Plasma Cystatin C as a Marker of Acute Renal Injury in Patients Undergoing Extracorporeal Shock Wave Lithotripsy for Renal Stone Disease. Cureus 2024, 16, e67293. [Google Scholar] [CrossRef]
- Dzięgała, M.; Krajewski, W.; Kołodziej, A.; Dembowski, J.; Zdrojowy, R. Evaluation and physiopathology of minor transient shock wave lithotripsy—Induced renal injury based on urinary biomarkers levels. Central Eur. J. Urol. 2018, 71, 214–220. [Google Scholar] [CrossRef]
- Suttapitugsakul, S.; Sassanarakkit, S.; Peerapen, P.; Thongboonkerd, V. Integrated proteomics reveals enrichment of oxidative stress and inflammatory proteins in the urine and stone matrix of calcium oxalate stone formers. Urolithiasis 2025, 53, 25. [Google Scholar] [CrossRef]
- Vinaiphat, A.; Thongboonkerd, V. Prospects for proteomics in kidney stone disease. Expert Rev. Proteom. 2017, 14, 185–187. [Google Scholar] [CrossRef]
- Rodríguez-Suárez, E.; Siwy, J.; Zürbig, P.; Mischak, H. Urine as a source for clinical proteome analysis: From discovery to clinical application. Biochim. Biophys. Acta 2014, 1844, 884–898. [Google Scholar] [CrossRef]
- Vaidya, V.S.; Waikar, S.S.; Ferguson, M.A.; Collings, F.B.; Sunderland, K.; Gioules, C.; Bradwin, G.; Matsouaka, R.; Betensky, R.A.; Curhan, G.C.; et al. Urinary biomarkers for sensitive and specific detection of acute kidney injury in humans. Clin. Transl. Sci. 2008, 1, 200–208. [Google Scholar] [CrossRef]
- Zhu, W.; Liu, M.; Wang, G.C.; Peng, B.; Yan, Y.; Che, J.P.; Ma, Q.W.; Yao, X.D.; Zheng, J.H. Fibrinogen alpha chain precursor and apolipoprotein A-I in urine as biomarkers for noninvasive diagnosis of calcium oxalate nephrolithiasis: A proteomics study. BioMed Res. Int. 2014, 2014, 415651. [Google Scholar] [CrossRef] [PubMed]
- Wright, C.A.; Howles, S.; Trudgian, D.C.; Kessler, B.M.; Reynard, J.M.; Noble, J.G.; Hamdy, F.C.; Turney, B.W. Label-free quantitative proteomics reveals differentially regulated proteins influencing urolithiasis. Mol. Cell. Proteom. 2011, 10, M110.005686. [Google Scholar] [CrossRef]
- Wood, K.; Keys, T.; Mufarrij, P.; Assimos, D.G. Impact of stone removal on renal function: A review. Rev. Urol. 2011, 13, 73–89. [Google Scholar] [PubMed]
- Santucci, L.; Bruschi, M.; Candiano, G.; Lugani, F.; Petretto, A.; Bonanni, A.; Ghiggeri, G.M. Urine Proteome Biomarkers in Kidney Diseases. I. Limits, Perspectives, and First Focus on Normal Urine. Biomark. Insights 2016, 11, 41–48. [Google Scholar] [CrossRef]
- Aitekenov, S.; Gaipov, A.; Bukasov, R. Review: Detection and quantification of proteins in human urine. Talanta 2021, 223 Pt 1, 121718. [Google Scholar] [CrossRef] [PubMed]
- Candiano, G.; Santucci, L.; Bruschi, M.; Petretto, A.; D’Ambrosio, C.; Scaloni, A.; Righetti, P.G.; Ghiggeri, G.M. “Cheek-to-cheek” urinary proteome profiling via combinatorial peptide ligand libraries: A novel, unexpected elution system. J. Proteom. 2012, 75, 796–805. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Sinha, P.; Poland, J.; Schnölzer, M.; Rabilloud, T. A new silver staining apparatus and procedure for matrix-assisted laser desorption/ionization-time of flight analysis of proteins after two-dimensional electrophoresis. Proteomics 2001, 1, 835–840. [Google Scholar] [CrossRef]
- Eleuterio, E.; Trubiani, O.; Sulpizio, M.; Di Giuseppe, F.; Pierdomenico, L.; Marchisio, M.; Giancola, R.; Giammaria, G.; Miscia, S.; Caputi, S.; et al. Proteome of human stem cells from periodontal ligament and dental pulp. PLoS ONE 2013, 8, e71101. [Google Scholar] [CrossRef]
- Di Giuseppe, F.; Ricci-Vitiani, L.; Pallini, R.; Di Pietro, R.; Di Iorio, P.; Ascani, G.; Ciccarelli, R.; Angelucci, S. Changes Induced by P2X7 Receptor Stimulation of Human Glioblastoma Stem Cells in the Proteome of Extracellular Vesicles Isolated from Their Secretome. Cells 2024, 13, 571. [Google Scholar] [CrossRef] [PubMed]
- Suckau, D.; Resemann, A.; Schuerenberg, M.; Hufnagel, P.; Franzen, J.; Holle, A. A novel MALDI LIFT-TOF/TOF mass spectrometer for proteomics. Anal. Bioanal. Chem. 2003, 376, 952–965. [Google Scholar] [CrossRef]
- Castagna, A.; Cecconi, D.; Sennels, L.; Rappsilber, J.; Guerrier, L.; Fortis, F.; Boschetti, E.; Lomas, L.; Righetti, P.G. Exploring the hidden human urinary proteome via ligand library beads. J. Proteome Res. 2005, 4, 1917–1930. [Google Scholar] [CrossRef] [PubMed]
- de Jesus, J.R.; da Silva Fernandes, R.; de Souza Pessôa, G.; Raimundo, I.M., Jr.; Arruda, M.A.Z. Depleting high-abundant and enriching low-abundant proteins in human serum: An evaluation of sample preparation methods using magnetic nanoparticle, chemical depletion and immunoaffinity techniques. Talanta 2017, 170, 199–209. [Google Scholar] [CrossRef] [PubMed]
- Bomanji, J.; Boddy, S.A.; Britton, K.E.; Nimmon, C.C.; Whitfield, H.N. Radionuclide evaluation pre- and post-extracorporeal shock wave lithotripsy for renal calculi. J. Nucl. Med. 1987, 28, 1284–1289. [Google Scholar]
- Lingeman, J.E.; McAteer, J.A.; Gnessin, E.; Evan, A.P. Shock wave lithotripsy: Advances in technology and technique. Nat. Rev. Urol. 2009, 6, 660–670. [Google Scholar] [CrossRef]
- McAteer, J.A.; Evan, A.P. The acute and long-term adverse effects of shock wave lithotripsy. Semin. Nephrol. 2008, 28, 200–213. [Google Scholar] [CrossRef]
- Al-Awadi, K.A.; Kehinde, E.O.; Loutfi, I.; Mojiminiyi, O.A.; Al-Hunayan, A.; Abdul-Halim, H.; Al-Sarraf, A.; Memon, A.; Abraham, M.P. Treatment of renal calculi by lithotripsy: Minimizing short-term shock wave induced renal damage by using antioxidants. Urol. Res. 2008, 36, 51–60. [Google Scholar] [CrossRef]
- Skolarikos, A.; Alivizatos, G.; de la Rosette, J. Extracorporeal shock wave lithotripsy 25 years later: Complications and their prevention. Eur. Urol. 2006, 50, 981–990; discussion 990. [Google Scholar] [CrossRef]
- Gabert, B.J.; Kültz, D. Osmoprotective proteome adjustments in mouse kidney papilla. Biochim. Biophys. Acta. 2011, 1814, 435–448. [Google Scholar] [CrossRef][Green Version]
- Magalhães, P.; Mischak, H.; Zürbig, P. Urinary proteomics using capillary electrophoresis coupled to mass spectrometry for diagnosis and prognosis in kidney diseases. Curr. Opin. Nephrol. Hypertens. 2016, 25, 494–501. [Google Scholar] [CrossRef] [PubMed]
- Siwy, J.; Zürbig, P.; Argiles, A.; Beige, J.; Haubitz, M.; Jankowski, J.; Julian, B.A.; Linde, P.G.; Marx, D.; Mischak, H.; et al. Noninvasive diagnosis of chronic kidney diseases using urinary proteome analysis. Nephrol. Dial. Transplant. 2017, 32, 2079–2089. [Google Scholar] [CrossRef]
- Zou, C.; Wang, C.; Lu, L. Advances in the study of subclinical AKI biomarkers. Front. Physiol. 2022, 13, 960059. [Google Scholar] [CrossRef] [PubMed]
- Sarma, J.V.; Ward, P.A. The complement system. Cell Tissue Res. 2011, 343, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Kościelska-Kasprzak, K.; Bartoszek, D.; Myszka, M.; Zabińska, M.; Klinger, M. The complement cascade and renal disease. Arch. Immunol. Ther. Exp. 2014, 62, 47–57. [Google Scholar] [CrossRef]
- Tringali, E.; Vetrano, D.; Tondolo, F.; Maritati, F.; Fabbrizio, B.; Pasquinelli, G.; Provenzano, M.; La Manna, G.; Baraldi, O. Role of serum complement C3 and C4 on kidney outcomes in IgA nephropathy. Sci. Rep. 2024, 14, 16224. [Google Scholar] [CrossRef]
- Villacorta, J.; Diaz-Crespo, F.; Acevedo, M.; Cavero, T.; Guerrero, C.; Praga, M.; Fernandez-Juarez, G. Circulating C3 levels predict renal and global outcome in patients with renal vasculitis. Clin. Rheumatol. 2016, 35, 2733–2740. [Google Scholar] [CrossRef]
- Gaya da Costa, M.; Poppelaars, F.; Berger, S.P.; Daha, M.R.; Seelen, M.A. The lectin pathway in renal disease: Old concept and new insights. Nephrol. Dial. Transplant. 2018, 33, 2073–2079. [Google Scholar] [CrossRef]
- Petr, V.; Thurman, J.M. The role of complement in kidney disease. Nat. Rev. Nephrol. 2023, 19, 771–787. [Google Scholar] [CrossRef]
- Lu, Z.; Wang, F.; Liang, M. SerpinC1/Antithrombin III in kidney-related diseases. Clin. Sci. 2017, 131, 823–831. [Google Scholar] [CrossRef]
- Zhou, F.; Luo, Q.; Han, L.; Shen, G.; Huang, L.; Ye, H. Proteomics reveals urine apolipoprotein A-I as a potential biomarker of acute kidney injury following percutaneous coronary intervention in elderly patients. Exp. Ther. Med. 2021, 22, 745. [Google Scholar] [CrossRef] [PubMed]
- Stangl, S.; Kollerits, B.; Lamina, C.; Meisinger, C.; Huth, C.; Stöckl, A.; Dähnhardt, D.; Böger, C.A.; Krämer, B.K.; Peters, A.; et al. Association between apolipoprotein A-IV concentrations and chronic kidney disease in two large population-based cohorts: Results from the KORA studies. J. Intern. Med. 2015, 278, 410–423. [Google Scholar] [CrossRef]
- Vivekanandan-Giri, A.; Slocum, J.L.; Buller, C.L.; Basrur, V.; Ju, W.; Pop-Busui, R.; Lubman, D.M.; Kretzler, M.; Pennathur, S. Urine glycoprotein profile reveals novel markers for chronic kidney disease. Int. J. Proteom. 2011, 2011, 214715. [Google Scholar] [CrossRef]
- Nemtsova, Y.; Wiseman, J.A.; El-Banna, M.; Lobel, P.; Sleat, D.E. Inducible transgenic expression of tripeptidyl peptidase 1 in a mouse model of late-infantile neuronal ceroid lipofuscinosis. PLoS ONE 2018, 13, e0192286. [Google Scholar] [CrossRef]
- Morawski, M.; Schilling, S.; Kreuzberger, M.; Waniek, A.; Jäger, C.; Koch, B.; Cynis, H.; Kehlen, A.; Arendt, T.; Hartlage-Rübsamen, M.; et al. Glutaminyl cyclase in human cortex: Correlation with (pGlu)-amyloid-β load and cognitive decline in Alzheimer’s disease. J. Alzheimer's Dis. 2014, 39, 385–400. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, Y.; Zhao, Z.; Peng, W.; Wang, P.; Xu, X.; Zhao, C. Glutaminyl cyclases, the potential targets of cancer and neurodegenerative diseases. Eur. J. Pharmacol. 2022, 931, 175178. [Google Scholar] [CrossRef]
- Sowa, A.S.; Popova, T.G.; Harmuth, T.; Weber, J.J.; Pereira Sena, P.; Schmidt, J.; Hübener-Schmid, J.; Schmidt, T. Neurodegenerative phosphoprotein signaling landscape in models of SCA3. Mol. Brain 2021, 14, 57. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Han, J.; Fan, J.; Song, J.; Wang, S. ATXN3 promotes proliferation, stemness and motility of clear cell renal cell carcinoma cells by regulating S100A8 ubiquitination. Physiol. Int. 2023, 110, 311–325. [Google Scholar] [CrossRef]
- Pang, S.W.; Lahiri, C.; Poh, C.L.; Tan, K.O. PNMA family: Protein interaction network and cell signalling pathways implicated in cancer and apoptosis. Cell. Signal. 2018, 45, 54–62. [Google Scholar] [CrossRef]
- Wu, T.; Fu, Y.; Brekken, D.; Yan, M.; Zhou, X.J.; Vanarsa, K.; Deljavan, N.; Ahn, C.; Putterman, C.; Mohan, C. Urine proteome scans uncover total urinary protease, prostaglandin D synthase, serum amyloid P, and superoxide dismutase as potential markers of lupus nephritis. J. Immunol. 2010, 184, 2183–2193. [Google Scholar] [CrossRef]
- Zager, R.A. Alpha 1 Microglobulin: A Potentially Paradoxical Anti-Oxidant Agent. Adv. Tech. Biol. Med. 2017, 5, 238. [Google Scholar] [CrossRef] [PubMed]
- Penders, J.; Delanghe, J.R. Alpha 1-microglobulin: Clinical laboratory aspects and applications. Clin. Chim. Acta 2004, 346, 107–118. [Google Scholar] [CrossRef]
- Makridakis, M.; Kontostathi, G.; Petra, E.; Stroggilos, R.; Lygirou, V.; Filip, S.; Duranton, F.; Mischak, H.; Argiles, A.; Zoidakis, J.; et al. Multiplexed MRM-based protein quantification of putative prognostic biomarkers for chronic kidney disease progression in plasma. Sci. Rep. 2020, 10, 4815. [Google Scholar] [CrossRef]
- Haubitz, M.; Good, D.M.; Woywodt, A.; Haller, H.; Rupprecht, H.; Theodorescu, D.; Dakna, M.; Coon, J.J.; Mischak, H. Identification and validation of urinary biomarkers for differential diagnosis and evaluation of therapeutic intervention in anti-neutrophil cytoplasmic antibody-associated vasculitis. Mol. Cell. Proteom. 2009, 8, 2296–2307. [Google Scholar] [CrossRef] [PubMed]
- Casanova, A.G.; Vicente-Vicente, L.; Hernández-Sánchez, M.T.; Prieto, M.; Rihuete, M.I.; Ramis, L.M.; Del Barco, E.; Cruz, J.J.; Ortiz, A.; Cruz-González, I.; et al. Urinary transferrin pre-emptively identifies the risk of renal damage posed by subclinical tubular alterations. Biomed. Pharmacother. 2020, 121, 109684. [Google Scholar] [CrossRef]
- Neiman, M.; Hedberg, J.J.; Dönnes, P.R.; Schuppe-Koistinen, I.; Hanschke, S.; Schindler, R.; Uhlén, M.; Schwenk, J.M.; Nilsson, P. Plasma profiling reveals human fibulin-1 as candidate marker for renal impairment. J. Proteome Res. 2011, 10, 4925–4934. [Google Scholar] [CrossRef] [PubMed]
- Bruschi, M.; Granata, S.; Candiano, G.; Fabris, A.; Petretto, A.; Ghiggeri, G.M.; Gambaro, G.; Zaza, G. Proteomic Analysis of Urinary Extracellular Vesicles Reveals a Role for the Complement System in Medullary Sponge Kidney Disease. Int. J. Mol. Sci. 2019, 20, 5517. [Google Scholar] [CrossRef]
- Yi, H.; Ye, R.; Wang, J.; Gao, L.; Zhang, W.; Liu, C. Diagnostic Value of Serum Ficolin-3 and Gal-3 in Sepsis Complicated with Acute Kidney Injury. Int. J. Gen. Med. 2024, 17, 5299–5307. [Google Scholar] [CrossRef]
- Gaipov, A.; Jackson, C.D.; Talwar, M.; Balaraman, V.; Chakravarty, A.; Cseprekal, O.; Mathe, Z.; Remport, A.; Kovesdy, C.P.; Eason, J.D.; et al. Association between serum prealbumin level and outcomes in prevalent kidney transplant recipients. J. Renal Nutr. 2019, 29, 188–195. [Google Scholar] [CrossRef]
- Ngai, H.H.; Sit, W.H.; Jiang, P.P.; Thongboonkerd, V.; Wan, J.M. Markedly increased urinary preprohaptoglobin and haptoglobin in passive Heymann nephritis: A differential proteomics approach. J. Proteome Res. 2007, 6, 3313–3320. [Google Scholar] [CrossRef]
- Dugina, V.B.; Shagieva, G.S.; Shakhov, A.S.; Alieva, I.B. The Cytoplasmic Actins in the Regulation of Endothelial Cell Function. Int. J. Mol. Sci. 2021, 22, 7836. [Google Scholar] [CrossRef] [PubMed]
- Karpushev, A.V.; Levchenko, V.; Ilatovskaya, D.V.; Pavlov, T.S.; Staruschenko, A. Novel role of Rac1/WAVE signaling mechanism in regulation of the epithelial Na+ channel. Hypertension 2011, 57, 996–1002. [Google Scholar] [CrossRef]
- Blaine, J.; Dylewski, J. Regulation of the Actin Cytoskeleton in Podocytes. Cells 2020, 9, 1700. [Google Scholar] [CrossRef]
- Ahmadian, E.; Eftekhari, A.; Atakishizada, S.; Valiyeva, M.; Ardalan, M.; Khalilov, R.; Kavetskyy, T. Podocytopathy: The role of actin cytoskeleton. Biomed. Pharmacother. 2022, 156, 113920. [Google Scholar] [CrossRef] [PubMed]
- Nogueira, C.W.; Soares, F.A.; Nascimento, P.C.; Muller, D.; Rocha, J.B. 2.;3-Dimercaptopropane-1-sulfonic acid and meso-2,3-dimercaptosuccinic acid increase mercury- and cadmium-induced inhibition of delta-aminolevulinate dehydratase. Toxicology 2003, 184, 85–95. [Google Scholar] [CrossRef]
- Matsumoto, T.; Urushido, M.; Ide, H.; Ishihara, M.; Hamada-Ode, K.; Shimamura, Y.; Ogata, K.; Inoue, K.; Taniguchi, Y.; Taguchi, T.; et al. Small heat shock protein beta-1 (HSPB1)is upregulated and regulates autophagy and apoptosis of renal tubular cells in acute kidney injury. PLoS ONE 2015, 10, e0126229. [Google Scholar] [CrossRef]
- Fong-Ngern, K.; Thongboonkerd, V. Alpha-enolase on apical surface of renal tubular epithelial cells serves as a calcium oxalate crystal receptor. Sci. Rep. 2016, 6, 36103. [Google Scholar] [CrossRef] [PubMed]
- Djudjaj, S.; Papasotiriou, M.; Bülow, R.D.; Wagnerova, A.; Lindenmeyer, M.T.; Cohen, C.D.; Strnad, P.; Goumenos, D.S.; Floege, J.; Boor, P. Keratins are novel markers of renal epithelial cell injury. Kidney Int. 2016, 89, 792–808. [Google Scholar] [CrossRef]
- Tan, R.J.; Liu, Y. Matrix metalloproteinase in kidney homeostasis diseases. Am. J. Physiol. Renal Physiol. 2012, 302, F1351–F1361. [Google Scholar] [CrossRef]
- Ho, J.; Rush, D.N.; Krokhin, O.; Antonovici, M.; Gao, A.; Bestland, J.; Wiebe, C.; Hiebert, B.; Rigatto, C.; Gibson, I.W.; et al. Elevated Urinary Matrix Metalloproteinase-7 Detects Underlying Renal Allograft Inflammation and Injury. Transplantation 2016, 100, 648–654. [Google Scholar] [CrossRef]
- He, W.; Tan, R.J.; Li, Y.; Wang, D.; Nie, J.; Hou, F.F.; Liu, Y. Matrix metalloproteinase-7 as a surrogate marker predicts renal Wnt/β-catenin activity in CKD. J. Am. Soc. Nephrol. 2012, 23, 294–304. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Dong, Y.; Tian, X.; Tan, T.K.; Liu, Z.; Zhao, Y.; Zhang, Y.; Harris, D.C.; Zheng, G. Matrix metalloproteinases contribute to kidney fibrosis in chronic kidney diseases. World J. Nephrol. 2013, 2, 84–89. [Google Scholar] [CrossRef] [PubMed]
- van Swelm, R.P.; Laarakkers, C.M.; Pertijs, J.C.; Verweij, V.; Masereeuw, R.; Russel, F.G. Urinary proteomic profiling reveals diclofenac-induced renal injury and hepatic regeneration in mice. Toxicol. Appl. Pharmacol. 2013, 269, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Osis, G.; Traylor, A.M.; Black, L.M.; Spangler, D.; George, J.F.; Zarjou, A.; Verlander, J.W.; Agarwal, A. Expression of lactate dehydrogenase A and B isoforms in the mouse kidney. Am. J. Physiol. Renal Physiol. 2021, 320, F706–F718. [Google Scholar] [CrossRef]
- Tryggvason, S.; Nukui, M.; Oddsson, A.; Tryggvason, K.; Jörnvall, H. Glomerulus proteome analysis with two-dimensional gel electrophoresis and mass spectrometry. Cell. Mol. Life Sci. 2007, 64, 3317–3335. [Google Scholar] [CrossRef]
- Baylis, C. Nitric oxide synthase derangements and hypertension in kidney disease. Curr. Opin. Nephrol. Hypertens. 2012, 21, 1–6. [Google Scholar] [CrossRef]
- Palm, F.; Onozato, M.L.; Luo, Z.; Wilcox, C.S. Dimethylarginine dimethylaminohydrolase (DDAH): Expression, regulation, and function in the cardiovascular and renal systems. Am. J. Physiol. Heart Circ. Physiol. 2007, 293, H3227–H3245. [Google Scholar] [CrossRef]
- Jantrapirom, S.; Piccolo, L.L.; Pruksakorn, D.; Potikanond, S.; Nimlamool, W. Ubiquilin Networking in Cancers. Cancers 2020, 12, 1586. [Google Scholar] [CrossRef]
- Shah, P.P.; Lockwood, W.W.; Saurabh, K.; Kurlawala, Z.; Shannon, S.P.; Waigel, S.; Zacharias, W.; Beverly, L.J. Ubiquilin1 represses migration and epithelial-to-mesenchymal transition of human non-small cell lung cancer cells. Oncogene 2015, 34, 1709–1717. [Google Scholar] [CrossRef]
- Marples, D.; Schroer, T.A.; Ahrens, N.; Taylor, A.; Knepper, M.A.; Nielsen, S. Dynein and dynactin colocalize with AQP2 water channels in intracellular vesicles from kidney collecting duct. Am. J. Physiol. Renal Physiol. 1998, 274, F384–F394. [Google Scholar] [CrossRef]
- Sörensen, I.; Susnik, N.; Inhester, T.; Degen, J.L.; Melk, A.; Haller, H.; Schmitt, R. Fibrinogen, acting as a mitogen for tubulointerstitial fibroblasts, promotes renal fibrosis. Kidney Int. 2011, 80, 1035–1044. [Google Scholar] [CrossRef] [PubMed]
- Sörensen-Zender, I.; Rong, S.; Susnik, N.; Lange, J.; Gueler, F.; Degen, J.L.; Melk, A.; Haller, H.; Schmitt, R. Role of fibrinogen in acute ischemic kidney injury. Am. J. Physiol. Renal Physiol. 2013, 305, F777–F785. [Google Scholar] [CrossRef] [PubMed]
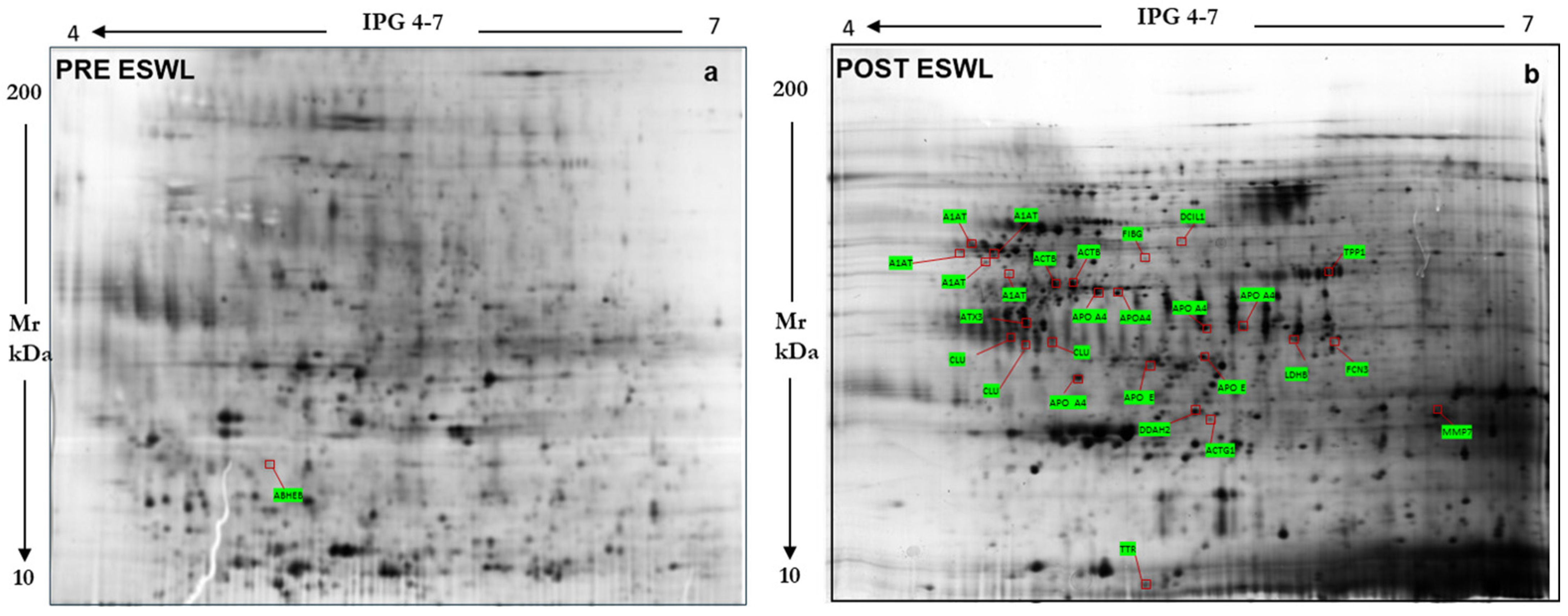
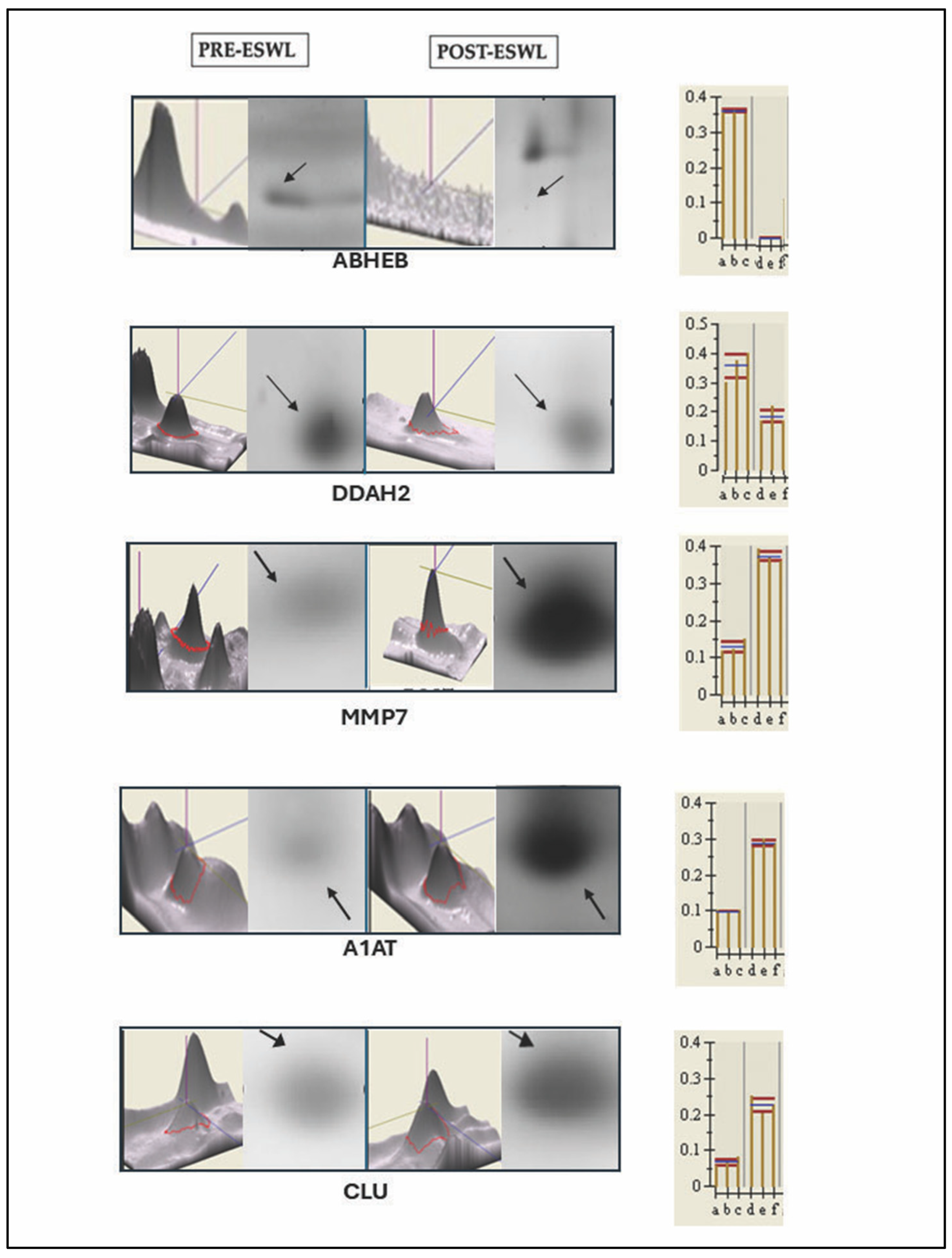




| Protein Name | Abbreviation | Swiss- Prot/NCBInr AC a | SCORE b _SC c | Matching Peptides # | Theoretical Mr_pI | n-Fold Variation d (p < 0.001) |
|---|---|---|---|---|---|---|
| Alpha-1 antitrypsin variant | A1AT | P01009 | 61_19 | 7 | 46,751-5.43 | +3.42 |
| Actin, cytoplasmic 1 | ACTB | gi|501885 | 155_46 | 20 | 42,058_5.29 | +2.50 |
| Apolipoprotein A-IV precursor | APOA4 | gi|178757 | 126_35 | 18 | 45,353_5.33 | +1.32 |
| Ficolin-3 | FCN3 | O00602 | 47_18 | 5 | 33,404_6.20 | +2.13 |
| Apolipoprotein E | APOE | P02649 | 54_30 | 9 | 36,248_5.65 | +1.69 |
| Serpin peptidase inhibitor | A1AT | gi|15080499 | 62_23 | 8 | 46,864_5.36 | +0.91 |
| N(G),N(G)-dimethylarginine dimethylaminohydrolase 2 | DDAH2 | O95865 | 80_22 | 7 | 29,915_5.66 | −2.40 |
| Actin, cytoplasmic 1 G | ACTG1 | gi|40226101 | 67_7 | 21 | 29,878_5.5 | −2.12 |
| Matrix metalloproteinase 7 | MMP7 | gi|348021 | 82_26 | 9 | 29,810_6.56 | +4.18 |
| Delta-aminolevulinic acid dehydratase | Hem2 | P13716 | 40_19 | 6 | 36,728_6.32 | +2.76 |
| Ubiquilin-4 | UBQL4 | Q9NRR5 | 67_24 | 9 | 53,205_5.67 | −1.88 |
| Complement C3 | C3 | gi|78101268 | 88_15 | 8 | 187,148_6.02 | −2.02 |
| Hemoglobin subunit beta | HBB | P68871 | 59_40 | 5 | 16,102_6.75 | +3.01 |
| Alpha-1 antitrypsin | A1AT | gi|50363217 | 112_37 | 14 | 46,790_5.42 | +1.66 |
| Ataxin 3 variant | ATX3 | gi|28951768 | 54_26 | 6 | 38,978_4.81 | +1.27 |
| Clusterin | CLU | gi|338305 | 110_41 | 12 | 37,002_5.74 | +2.69 |
| Apolipoprotein E | APOE | gi|178853 | 61_29 | 9 | 36,243_5.81 | +1.78 |
| Cytoplasmic dynein | DC1L1 | Q9Y6G9 | 55_13 | 5 | 56,834_6.01 | −1.93 |
| Fibrinogen gamma | FIBG | gi|70906437 | 75_41 | 8 | 50,103_5.70 | −1.18 |
| Apolipoprotein A-IV | APOA4 | P06727 | 66_12 | 23 | 45,371_5.28 | +1.39 |
| Lactate Dehydrogenase A like 6B | LDHAL6B | Q9BYZ2 | 131_36 | 14 | 41,943_5.86 | +3.01 |
| Tripeptidyl Peptidase-I | TPP1 | gi|215261288 | 51_11 | 5 | 62,797_6.26 | +1.19 |
| Transthyretin | TTR | gi|212374952 | 66_56 | 5 | 13,799_5.35 | +1.65 |
| Apolipoprotein A4 | APOA4 | gi|93163358 | 123_37 | 17 | 45,371_5.28 | +2.77 |
| Protein Name | Abbreviation | Swiss- Prot/NCBInr AC a | SCORE b _SC c | Matching Peptides # | Theoretical Mr_pI |
|---|---|---|---|---|---|
| Actin, cytoplasmic 1 | ACTB | P60709 | 109_37 | 11 | 42,052/5.28 |
| Alpha-1-antitrypsin | A1AT | P01009 | 190_5555 | 23 | 46,878/5.37 |
| S Variant of Human Alpha1-Antitrypsin | A1AT | gi|231240 | 200_57 | 22 | 39,099/5.27 |
| Alpha1-Antitrypsin | A1AT | gi|253723069 | 115_67 | 27 | 44,273/5.43 |
| Cleaved Antitrypsin Polymer | A1AT | gi|7546268 | 200_55 | 19 | 37,622/5.43 |
| Antitrypsin alpha-1 mutant | A1AT | gi|224224 | 229_45 | 22 | 46,873/5.35 |
| Serpin-Proteinase Complex | A1AT | gi|83754916 | 132_36 | 14 | 40,041/5.20 |
| Cleaved Antitrypsin P10 Pro | A1AT | gi|301598706 | 201_53 | 19 | 38,558/5.11 |
| Cleaved Antitrypsin Polymer | A1AT | gi|7245932 | 98_54 | 17 | 36,612/5.44 |
| A1AT Alpha-1-antitrypsin | A1AT | gi|28637 | 54_29 | 8 | 22,871/6.11 |
| Cleaved Antitrypsin P10 Pro, P9-P6 Asp | A1AT | gi|301598708 | 76_27 | 9 | 38,474/5.16 |
| Inhibitor BrCN fragment lI, alpha1 protein | A1AT | gi|223039 | 104_63 | 7 | 11,785/5.76 |
| Alpha-1B-glycoprotein | A1BG | P04217 | 90_23 | 10 | 54,790/5.56 |
| Alpha- 1- microglobulin | AMBP | P02760 | 59_23 | 7 | 38,999/5.95 |
| Antithrombin-III | ANT3 | P01008 | 226_46 | 25 | 53,025/6.32 |
| Apolipoprotein A-I | APOA1 | P02647 | 280_73 | 30 | 23,00/5.22 |
| Chain A Human Apolipoprotein A-I | APOA1 | gi|2914175 | 219_88 | 25 | 23,389/5.55 |
| Truncated Human Apolipoprotein A-I | APOA1 | gi|347447518 | 271_84 | 25 | 21,611/5.02 |
| Apolipoprotein A-IV | APOA4 | P06727 | 254_56 | 28 | 45,371/5.28 |
| Apolipoprotein A-IV | APOA4 | gi|563320 | 291_33 | 62 | 28,141/5.39 |
| Apolipoprotein E | APOE | P02649 | 53_28 | 10 | 36,246/5.46 |
| Apolipoprotein E precursor | APOE | gi|4557325 | 195_62 | 24 | 36,246/5.56 |
| Alpha-enolase OS | ENOA | P03733 | 53_16 | 7 | 47,481/7.01 |
| Crystal Structure of Human Enolase 1 | ENOA | gi|203282367 | 89_23 | 10 | 47,350/6.99 |
| Ceruloplasmin | CERU | P00450 | 245_28 | 28 | 122,983/5.44 |
| Complement protein 9 | C9 | gi|179726 | 175_24 | 18 | 64,399/5.49 |
| Complement component C9 | CO9 | P02748 | 98_13 | 10 | 64,615/5.43 |
| Complement factor B | CB | gi|57209925 | 55_14 | 10 | 86,648/6.44 |
| Complement factor B preproprotein | GB | gi|67782358 | 55_11 | 9 | 86,847/6.67 |
| Human Complement Component C3 | C3 | gi|78101268 | 153_22 | 22 | 11,4238/5.55 |
| Complement factor H | CFAH | P08603 | 267_25 | 35 | 14,3680/6.21 |
| FicoIin-3 | FCN3 | O75636 | 82_36 | 11 | 33,395/6.20 |
| Fibulin-1 | FBLN1 | P23142 | 56_16 | 10 | 81,268/5.07 |
| Glutaminyl-peptide cyclotransferase | QPGT | Q16769 | 191_44 | 18 | 40,965/6.12 |
| Glutaminyl Cyclase Mutant E201 | QPCT | gi|185177689 | 195_54 | 19 | 37,590/5.78 |
| Glutaminyl-peptide cyclotransferase | QPCT | gi|75766183 | 150_48 | 18 | 37,606/5.69 |
| Glycosytated Human Glutaminyl Cyclase | QPCT | gi|345101018 | 127_40 | 15 | 37,916/5.96 |
| Glutaminyl-peptide cyclotransferase | QPCT | Q16769 | 171_45 | 15 | 40,965/5.69 |
| Glutaminyl CycIase Mutant | QPCT | gi|185177695 | 175_48 | 17 | 37,619/5.78 |
| Glutaminyl CycIase with Glutamine | QPCT | gi|75766189 | 162_53 | 19 | 37,605/5.78 |
| Heat shock protein beta-1 | HSPB1 | P04792 | 150_11 | 50 | 22,826/5.98 |
| Keratin C 19 | K1C19 | P08727 | 61_19 | 8 | 44,079/5.04 |
| Human MbI-Associated Protein 19 | Map19 | gi|50513645 | 103_44 | 8 | 19,531/5.44 |
| Paraneoplastic antigen-like protein 6C | PNM6C | P0CW26 | 46_10 | 5 | 44,304/5,24 |
| Pigment epithelial-differentiating factor | PEDF | gi|189778 | 131_18 | 9 | 46,471/5.84 |
| Crystal Structure of PEDF | PEDF | gi|15988024 | 164_37 | 15 | 44,306/5.77 |
| Serum amyloid P-component | SAMP | P02743 | 135_31 | 10 | 25,485/6.10 |
| Serpin-Proteinase Complex | A1AT | gi|83764916 | 131_36 | 14 | 40,041/5.20 |
| Serotransferrin | TF | P02787 | 390_50 | 40 | 79,294/6.81 |
| Transthyretin | TTR | P02766 | 89_53 | 8 | 15,991/5.52 |
| ABBR. Name | Mw/ pI Theor. | PMF Score a | Peptide Matched/ Peptide Searched | SC b % | Lift (MS2) Ion Parent Masses (m/z) | Score c Tof-Tof | Peptide Sequence |
|---|---|---|---|---|---|---|---|
| MMP7 | 29810/ 6.56 | 82 | 9/18 | 50 | 1784.9653 1492.8482 1139.6207 | 125 | IVSYTRDLPHITVDR DLPHITVDRLVSK WTSKVVTYR |
| DDAH2 | 29915/ 5.66 | 89 | 7/19 | 36 | 2497.2438 459.7863 | 200 | DFAVSTVPVSGPSHLRGLCGMGGI TVVAGSSDAAQKAVR |
| CLU | 37002/ 5.74 | 110 | 12/28 | 42 | 2551.2517 1714.8468 | 167 | FFTREPQDTYHYLPFSLPHR FMETVAEKALQEYR |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carestia, E.; Di Giuseppe, F.; Kazemi, M.; Ramahi, M.; Priyadarshi, U.; Giuliani, P.; De Francesco, P.; Schips, L.; Di Ilio, C.; Ciccarelli, R.; et al. Significant Changes in Low-Abundance Protein Content Detected by Proteomic Analysis of Urine from Patients with Renal Stones After Extracorporeal Shock Wave Lithotripsy. Biology 2025, 14, 482. https://doi.org/10.3390/biology14050482
Carestia E, Di Giuseppe F, Kazemi M, Ramahi M, Priyadarshi U, Giuliani P, De Francesco P, Schips L, Di Ilio C, Ciccarelli R, et al. Significant Changes in Low-Abundance Protein Content Detected by Proteomic Analysis of Urine from Patients with Renal Stones After Extracorporeal Shock Wave Lithotripsy. Biology. 2025; 14(5):482. https://doi.org/10.3390/biology14050482
Chicago/Turabian StyleCarestia, Elena, Fabrizio Di Giuseppe, Mohammad Kazemi, Massoumeh Ramahi, Uditanshu Priyadarshi, Patricia Giuliani, Piergustavo De Francesco, Luigi Schips, Carmine Di Ilio, Renata Ciccarelli, and et al. 2025. "Significant Changes in Low-Abundance Protein Content Detected by Proteomic Analysis of Urine from Patients with Renal Stones After Extracorporeal Shock Wave Lithotripsy" Biology 14, no. 5: 482. https://doi.org/10.3390/biology14050482
APA StyleCarestia, E., Di Giuseppe, F., Kazemi, M., Ramahi, M., Priyadarshi, U., Giuliani, P., De Francesco, P., Schips, L., Di Ilio, C., Ciccarelli, R., Di Iorio, P., & Angelucci, S. (2025). Significant Changes in Low-Abundance Protein Content Detected by Proteomic Analysis of Urine from Patients with Renal Stones After Extracorporeal Shock Wave Lithotripsy. Biology, 14(5), 482. https://doi.org/10.3390/biology14050482








