Mapping the Interactome of KRAS and Its G12C/D/V Mutants by Integrating TurboID Proximity Labeling with Quantitative Proteomics
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
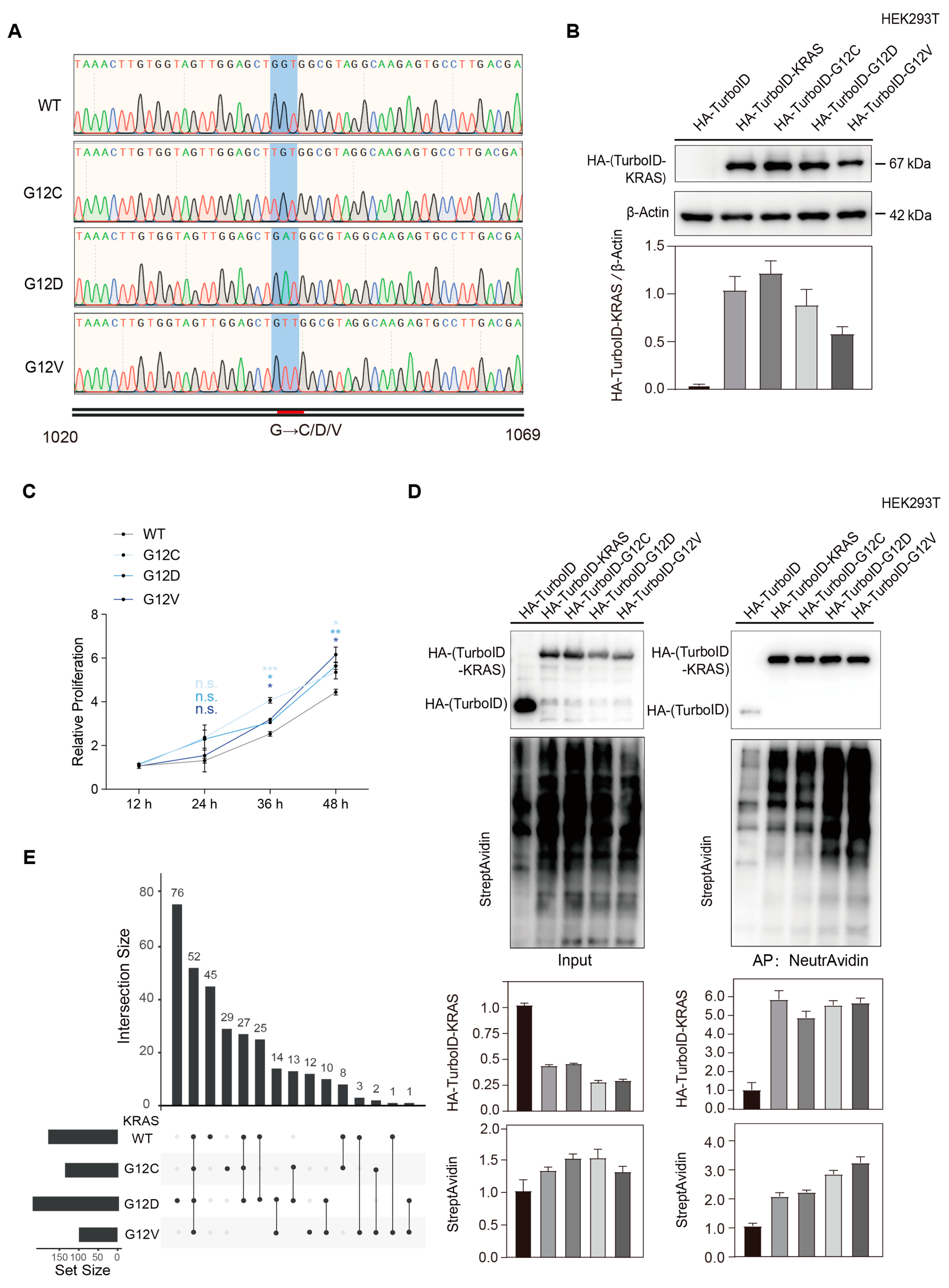
2.1. Construction of KRAS Wild-Type and G12 C/D/V Mutant Stable Cell Lines
2.1.1. Plasmid Construction and Sequence Verification
2.1.2. Plasmid Transformation and Amplification
2.1.3. Lentivirus Infection and Biological Activity Verification
2.2. TurboID Proximity Labeling for Enrichment of KRAS Interacting Proteins
2.3. Quantitative Proteomic Characterization of KRAS Interacting Proteins
2.3.1. Sample Preparation
2.3.2. LC-MS/MS Analysis
2.3.3. Data Processing and Bioinformatics Analysis
2.4. Statistical Analysis
3. Results
3.1. An Integrated Strategy of Mapping KRAS-Interacting Proteins
3.2. KRAS G12C/D/V Mutations Induce Intracellular Metabolic Reprogramming
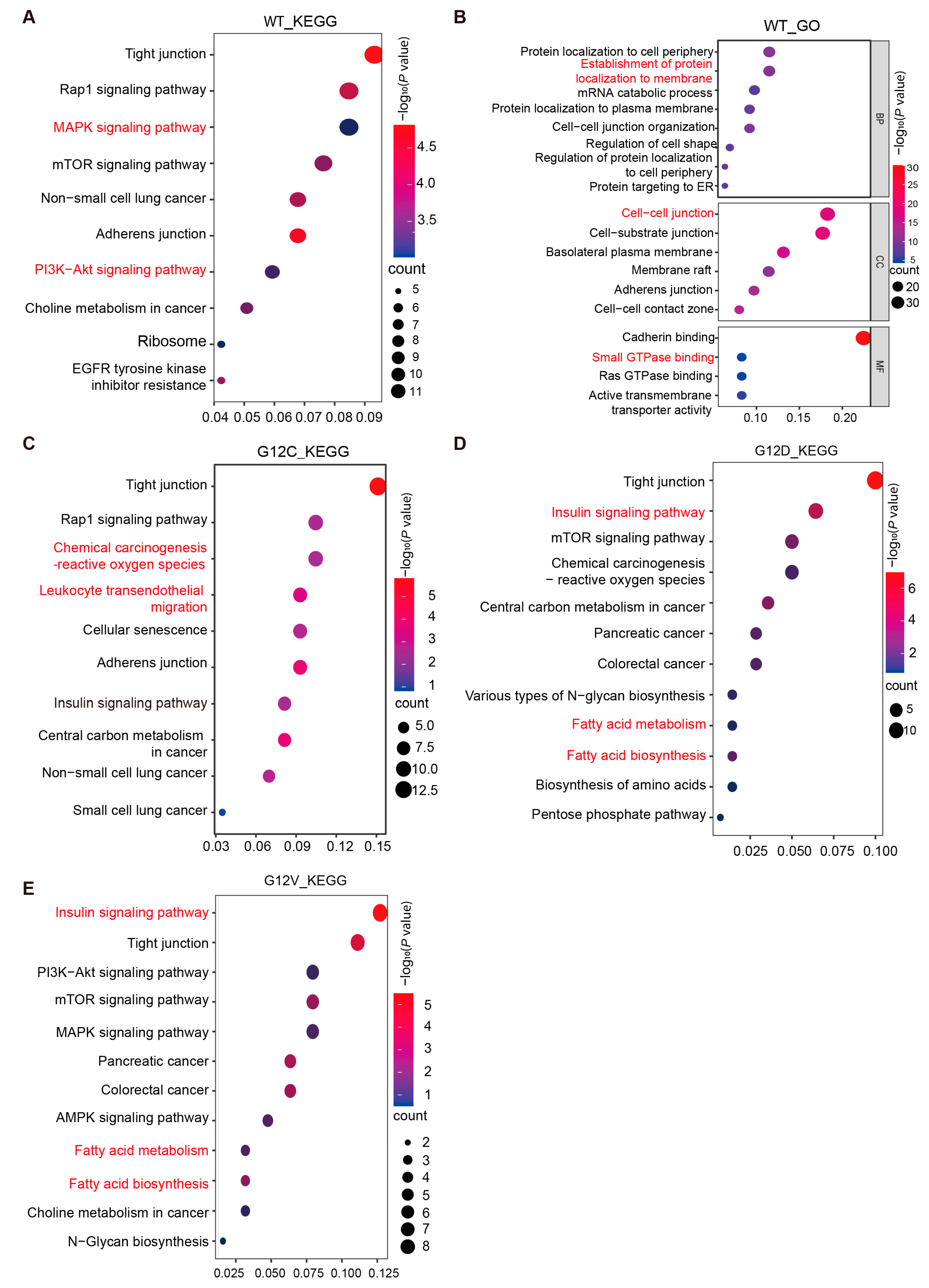
3.2.1. Pathway and Functional Enrichment Analysis of KRAS WT-Interacting Proteins
3.2.2. Dynamic Changes in KRAS Interacting Proteins Caused by KRAS G12C/D/V Mutations
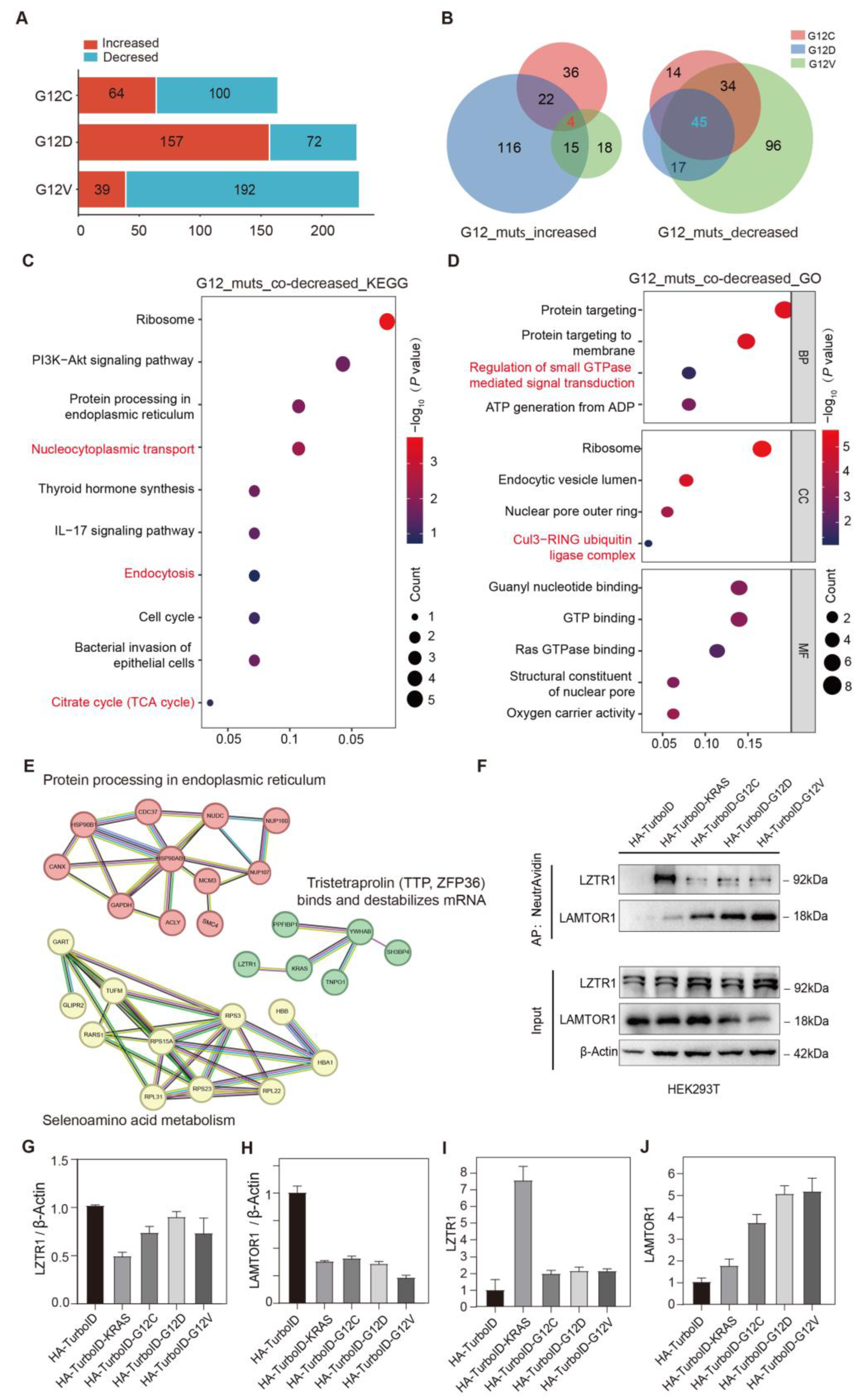
3.3. Simultaneously Changed Interacting Proteins in All KRAS G12C/D/V Mutants
3.3.1. Simultaneously Decreased Interacting Proteins in All KRAS G12C/D/V Mutants
3.3.2. Simultaneously Increased Interacting Proteins in All KRAS G12C/D/V Mutants
3.3.3. Validation of Simultaneously Changed Interacting Proteins in KRAS G12C/D/V Mutants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| KRAS | Kirsten rat sarcoma viral oncogene homolog; |
| PL | Proximity labeling; |
| PAAD | Pancreatic adenocarcinoma; |
| PanIN | Pancreatic intraepithelial neoplasia; |
| CRC | Colorectal cancer; |
| CHOL | Cholangiocarcinoma; |
| LUAD | Lung adenocarcinoma; |
| GTP | Guanosine triphosphate; |
| GDP | Guanosine diphosphate; |
| PI3K | Phosphatidylinositol 3-kinase; |
| AKT | Protein kinase B; |
| MAPK | Mitogen-activated protein kinase; |
| LZTR1 | Zipper-like transcription regulator 1; |
| LAMTOR1 | Late endosomal/lysosomal adaptor, MAPK, and mTOR activator 1; |
| GLUD1 | Glutamate dehydrogenase 1; |
| GOT1 | Aspartate aminotransferase 1; |
| FASN | Fatty acid synthase; |
| ACCA | Acetyl-CoA carboxylase alpha; |
| CUL3 | Cullin 3; |
| mTORC1 | Mammalian target of rapamycin 1; |
| PPI | Protein–protein interaction; |
| GO | Gene Ontology; |
| BP | Biological process; |
| CC | Cellular component; |
| MF | Molecular function; |
| KEGG | Kyoto Encyclopedia of Genes and Genomes; |
| ROS | Reactive oxygen species. |
Appendix A

References
- Punekar, S.R.; Velcheti, V.; Neel, B.G.; Wong, K.-K. The current state of the art and future trends in RAS-targeted cancer therapies. Nat. Rev. Clin. Oncol. 2022, 19, 637–655. [Google Scholar] [CrossRef] [PubMed]
- Simanshu, D.K.; Nissley, D.V.; McCormick, F. RAS Proteins and Their Regulators in Human Disease. Cell 2017, 170, 17–33. [Google Scholar] [CrossRef]
- Arrington, A.K.; Heinrich, E.L.; Lee, W.; Duldulao, M.; Patel, S.; Sanchez, J.; Garcia-Aguilar, J.; Kim, J. Prognostic and predictive roles of KRAS mutation in colorectal cancer. Int. J. Mol. Sci. 2012, 13, 12153–12168. [Google Scholar] [CrossRef]
- The AACR Project GENIE Consortium; The AACR Project GENIE Consortium; André, F.; Arnedos, M.; Baras, A.S.; Baselga, J.; Bedard, P.L.; Berger, M.F.; Bierkens, M.; Calvo, F.; et al. AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov. 2017, 7, 818–831. [Google Scholar]
- Moore, A.R.; Rosenberg, S.C.; McCormick, F.; Malek, S. RAS-targeted therapies: Is the undruggable drugged? Nat. Rev. Drug Discov. 2020, 19, 533–552. [Google Scholar] [CrossRef]
- Prior, I.A.; Hood, F.E.; Hartley, J.L. The Frequency of Ras Mutations in Cancer. Cancer Res. 2020, 80, 2969–2974. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014, 511, 543–550. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell 2017, 32, 185–203.e13. [Google Scholar] [CrossRef]
- Prior, I.A.; Lewis, P.D.; Mattos, C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012, 72, 2457–2467. [Google Scholar] [CrossRef]
- Lu, S.; Jang, H.; Nussinov, R.; Zhang, J. The Structural Basis of Oncogenic Mutations G12, G13 and Q61 in Small GTPase K-Ras4B. Sci. Rep. 2016, 6, 21949. [Google Scholar] [CrossRef]
- McIntyre, C.A.; Grimont, A.; Park, J.; Meng, Y.; Sisso, W.J.; Seier, K.; Jang, G.H.; Walch, H.; Aveson, V.G.; Falvo, D.J. Distinct clinical outcomes and biological features of specific KRAS mutants in human pancreatic cancer. Cancer Cell 2024, 42, 1614–1629.e5. [Google Scholar] [CrossRef]
- Bournet, B.; Muscari, F.; Buscail, C.; Assenat, E.; Barthet, M.; Hammel, P.; Selves, J.; Guimbaud, R.; Cordelier, P.; Buscail, L. KRAS G12D Mutation Subtype Is A Prognostic Factor for Advanced Pancreatic Adenocarcinoma. Clin. Transl. Gastroenterol. 2016, 7, e157. [Google Scholar] [CrossRef] [PubMed]
- Giehl, K. Oncogenic Ras in tumour progression and metastasis. Biol. Chem. 2005, 386, 193–205. [Google Scholar] [CrossRef]
- Isermann, T.; Sers, C.; Der, C.J.; Papke, B. KRAS inhibitors: Resistance drivers and combinatorial strategies. Trends Cancer 2025, 11, 91–116. [Google Scholar] [CrossRef] [PubMed]
- Hunter, J.C.; Manandhar, A.; Carrasco, M.A.; Gurbani, D.; Gondi, S.; Westover, K.D. Biochemical and Structural Analysis of Common Cancer-Associated KRAS Mutations. Mol. Cancer Res. 2015, 13, 1325–1335. [Google Scholar] [CrossRef] [PubMed]
- Cherfils, J.; Zeghouf, M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol. Rev. 2013, 93, 269–309. [Google Scholar] [CrossRef]
- Buday, L.; Downward, J. Epidermal growth factor regulates p21ras through the formation of a complex of receptor, Grb2 adapter protein, and Sos nucleotide exchange factor. Cell 1993, 73, 611–620. [Google Scholar] [CrossRef]
- Jeong, A.; Suazo, K.F.; Wood, W.G.; Distefano, M.D.; Li, L. Isoprenoids and protein prenylation: Implications in the pathogenesis and therapeutic intervention of Alzheimer’s disease. Crit. Rev. Biochem. Mol. Biol. 2018, 53, 279–310. [Google Scholar] [CrossRef]
- Cargnello, M.; Roux, P.P. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol. Mol. Biol. Rev. 2011, 75, 50–83. [Google Scholar] [CrossRef]
- Rossi Sebastiano, M.; Konstantinidou, G. Targeting Long Chain Acyl-CoA Synthetases for Cancer Therapy. Int. J. Mol. Sci. 2019, 20, 3624. [Google Scholar] [CrossRef] [PubMed]
- Lito, P. KRAS Oncoprotein Signaling in Cancer. N. Engl. J. Med. 2025, 392, 296–298. [Google Scholar] [CrossRef]
- Huang, L.; Guo, Z.; Wang, F.; Fu, L. KRAS mutation: From undruggable to druggable in cancer. Signal Transduct. Target. Ther. 2021, 6, 386. [Google Scholar] [CrossRef]
- Singhal, A.; Li, B.T.; O’Reilly, E.M. Targeting KRAS in cancer. Nat. Med. 2024, 30, 969–983. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-P.; Zhou, W.; Wang, J.; Huang, X.; Zuo, Y.; Wang, T.-S.; Gao, X.; Xu, Y.-Y.; Zou, S.-W.; Liu, Y.-B.; et al. Arginine Methylation of MDH1 by CARM1 Inhibits Glutamine Metabolism and Suppresses Pancreatic Cancer. Mol. Cell 2016, 64, 673–687. [Google Scholar] [CrossRef] [PubMed]
- Bigenzahn, J.W.; Collu, G.M.; Kartnig, F.; Pieraks, M.; Vladimer, G.I.; Heinz, L.X.; Sedlyarov, V.; Schischlik, F.; Fauster, A.; Rebsamen, M.; et al. LZTR1 is a regulator of RAS ubiquitination and signaling. Science 2018, 362, 1171–1177. [Google Scholar] [CrossRef]
- Damianou, A.; Liang, Z.; Lassen, F.; Vendrell, I.; Vere, G.; Hester, S.; Charles, P.D.; Pinto-Fernandez, A.; Santos, A.; Fischer, R.; et al. Oncogenic mutations of KRAS modulate its turnover by the CUL3/LZTR1 E3 ligase complex. Life Sci. Alliance 2024, 7, e202302245. [Google Scholar] [CrossRef]
- Zhao, L.; Gao, N.; Peng, X.; Chen, L.; Meng, T.; Jiang, C.; Jin, J.; Zhang, J.; Duan, Q.; Tian, H.; et al. TRAF4-Mediated LAMTOR1 Ubiquitination Promotes mTORC1 Activation and Inhibits the Inflammation-Induced Colorectal Cancer Progression. Adv. Sci. 2024, 11, e2301164. [Google Scholar] [CrossRef]
- Hertel, A.; Alves, L.M.; Dutz, H.; Tascher, G.; Bonn, F.; Kaulich, M.; Dikic, I.; Eimer, S.; Steinberg, F.; Bremm, A. USP32-regulated LAMTOR1 ubiquitination impacts mTORC1 activation and autophagy induction. Cell Rep. 2022, 41, 111653. [Google Scholar] [CrossRef]
- Wu, B.; Wang, Q.; Li, B.; Jiang, M. LAMTOR1 degrades MHC-II via the endocytic in hepatocellular carcinoma. Carcinogenesis 2022, 43, 1059–1070. [Google Scholar] [CrossRef]
- Hobbs, G.A.; Der, C.J.; Rossman, K.L. RAS isoforms and mutations in cancer at a glance. J. Cell Sci. 2016, 129, 1287–1292. [Google Scholar] [CrossRef]
- Haigis, K.M. KRAS Alleles: The Devil Is in the Detail. Trends Cancer 2017, 3, 686–697. [Google Scholar] [CrossRef]
- Poulin, E.J.; Bera, A.K.; Lu, J.; Lin, Y.-J.; Strasser, S.D.; Paulo, J.A.; Huang, T.Q.; Morales, C.; Yan, W.; Cook, J.; et al. Tissue-Specific Oncogenic Activity of KRAS(A146T). Cancer Discov. 2019, 9, 738–755. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.I.; Roux, K.J. Filling the Void: Proximity-Based Labeling of Proteins in Living Cells. Trends Cell Biol. 2016, 26, 804–817. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.I.; KC, B.; Zhu, W.; Roux, K.J. Probing nuclear pore complex architecture with proximity-dependent biotinylation. Proc. Natl. Acad. Sci. USA 2014, 111, E2453–E2461. [Google Scholar] [CrossRef] [PubMed]
- Gross, A.J.; Sizer, I.W. The oxidation of tyramine, tyrosine, and related compounds by peroxidase. J. Biol. Chem. 1959, 234, 1611–1614. [Google Scholar] [CrossRef]
- Kiel, C.; Matallanas, D.; Kolch, W. The Ins and Outs of RAS Effector Complexes. Biomolecules 2021, 11, 236. [Google Scholar] [CrossRef]
- Kargl, J.; Busch, S.E.; Yang, G.H.Y.; Kim, K.-H.; Hanke, M.L.; Metz, H.E.; Hubbard, J.J.; Lee, S.M.; Madtes, D.K.; McIntosh, M.W.; et al. Neutrophils dominate the immune cell composition in non-small cell lung cancer. Nat. Commun. 2017, 8, 14381. [Google Scholar] [CrossRef]
- Sun, S.; Pei, X.; Zhang, K.; Zhang, C.; Zhang, J.; Liu, K.; Cui, W.; Pan, F.; Zhang, F. Significance of interstitial tumor-associated macrophages in the progression of lung adenocarcinoma. Oncol. Lett. 2016, 12, 4467–4476. [Google Scholar] [CrossRef][Green Version]
- Zhang, A.M.Y.; Xia, Y.H.; Lin, J.S.H.; Chu, K.H.; Wang, W.C.K.; Ruiter, T.J.J.; Yang, J.C.C.; Chen, N.; Chhuor, J.; Patil, S.; et al. Hyperinsulinemia acts via acinar insulin receptors to initiate pancreatic cancer by increasing digestive enzyme production and inflammation. Cell Metab. 2023, 35, 2119–2135.e5. [Google Scholar] [CrossRef]
- Ostrem, J.M.; Shokat, K.M. Direct small-molecule inhibitors of KRAS: From structural insights to mechanism-based design. Nat. Rev. Drug Discov. 2016, 15, 771–785. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Huang, X.; Shi, X.; Jiang, M.; Liu, H.; Zhao, L. LAMTOR1 decreased exosomal PD-L1 to enhance immunotherapy efficacy in non-small cell lung cancer. Mol. Cancer 2024, 23, 184. [Google Scholar] [CrossRef] [PubMed]
- Abe, T.; Umeki, I.; Kanno, S.I.; Inoue, S.I.; Niihori, T.; Aoki, Y. LZTR1 facilitates polyubiquitination and degradation of RAS-GTPases. Cell Death Differ. 2020, 27, 1023–1035. [Google Scholar] [CrossRef] [PubMed]
- Bie, J.; Li, Y.; Song, C.; Weng, Q.; Zhao, L.; Su, L.; Zhao, Z.; Ye, Y.; Shen, Z.; Ji, J.; et al. LAMTOR1 ablation impedes cGAS degradation caused by chemotherapy and promotes antitumor immunity. Proc. Natl. Acad. Sci. USA 2024, 121, e2320591121. [Google Scholar] [CrossRef]
- de Araujo, M.E.G.; Naschberger, A.; Fürnrohr, B.G.; Stasyk, T.; Dunzendorfer-Matt, T.; Lechner, S.; Welti, S.; Kremser, L.; Shivalingaiah, G.; Offterdinger, M.; et al. Crystal structure of the human lysosomal mTORC1 scaffold complex and its impact on signaling. Science 2017, 358, 377–381. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; García-Seisdedos, D.; Hewapathirana, S.; Kamatchinathan, S.; Kundu, D.J.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M.; et al. The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022, 50, D543–D552. [Google Scholar] [CrossRef]

| Type | Sequence (5′-3′) |
|---|---|
| Forward 1 | AAAGACAATACTGTGCCTCTGAA |
| Reverse 1 | GTTTATATTCAGTCATCTTTTCGGCAGACCGCA |
| Forward 2 | ATGACTGAATATAAACTTGTGGTAGTTGG |
| Reverse 2 | TTATCTAGAGTCGCGGGATCCTTACATTATAATGCATTTTTTAATTTTCACACAG |
| Subtype | Mutated Base | Sequence (5′-3′) |
|---|---|---|
| G12C | GGT-TGT | F: TTGGAGCTTGTGGCGTAGGCAAGAGTGCCTTG |
| R: TACGCCACAAGCTCCAACTACCACAAGTTTTATTCA | ||
| G12D | GGT-GAT | F: TTGGAGCTGATGGCGTAGGCAAGAGTGCCTTG |
| R: TACGCCATCAGCTCCAACTACCACAAGTTTATATT | ||
| G12V | GGT-GTT | F: TTGGAGCTGTTGGCGTAGGCAAGAGTGCCTTG |
| R: TACGCCAACAGCTCCAACTACCACAAGTTTATATT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, J.; Wang, B.; Zou, M.; Zhou, H.; Ding, Y.; Ren, W.; Fang, L.; Zhang, J. Mapping the Interactome of KRAS and Its G12C/D/V Mutants by Integrating TurboID Proximity Labeling with Quantitative Proteomics. Biology 2025, 14, 477. https://doi.org/10.3390/biology14050477
Song J, Wang B, Zou M, Zhou H, Ding Y, Ren W, Fang L, Zhang J. Mapping the Interactome of KRAS and Its G12C/D/V Mutants by Integrating TurboID Proximity Labeling with Quantitative Proteomics. Biology. 2025; 14(5):477. https://doi.org/10.3390/biology14050477
Chicago/Turabian StyleSong, Jiangwei, Busong Wang, Mingjie Zou, Haiyuan Zhou, Yibing Ding, Wei Ren, Lei Fang, and Jingzi Zhang. 2025. "Mapping the Interactome of KRAS and Its G12C/D/V Mutants by Integrating TurboID Proximity Labeling with Quantitative Proteomics" Biology 14, no. 5: 477. https://doi.org/10.3390/biology14050477
APA StyleSong, J., Wang, B., Zou, M., Zhou, H., Ding, Y., Ren, W., Fang, L., & Zhang, J. (2025). Mapping the Interactome of KRAS and Its G12C/D/V Mutants by Integrating TurboID Proximity Labeling with Quantitative Proteomics. Biology, 14(5), 477. https://doi.org/10.3390/biology14050477







