Fine-Mapping of OvANS: A Novel Gene Controlling White Flowers in Orychophragmus violaceus
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Statistical Analysis
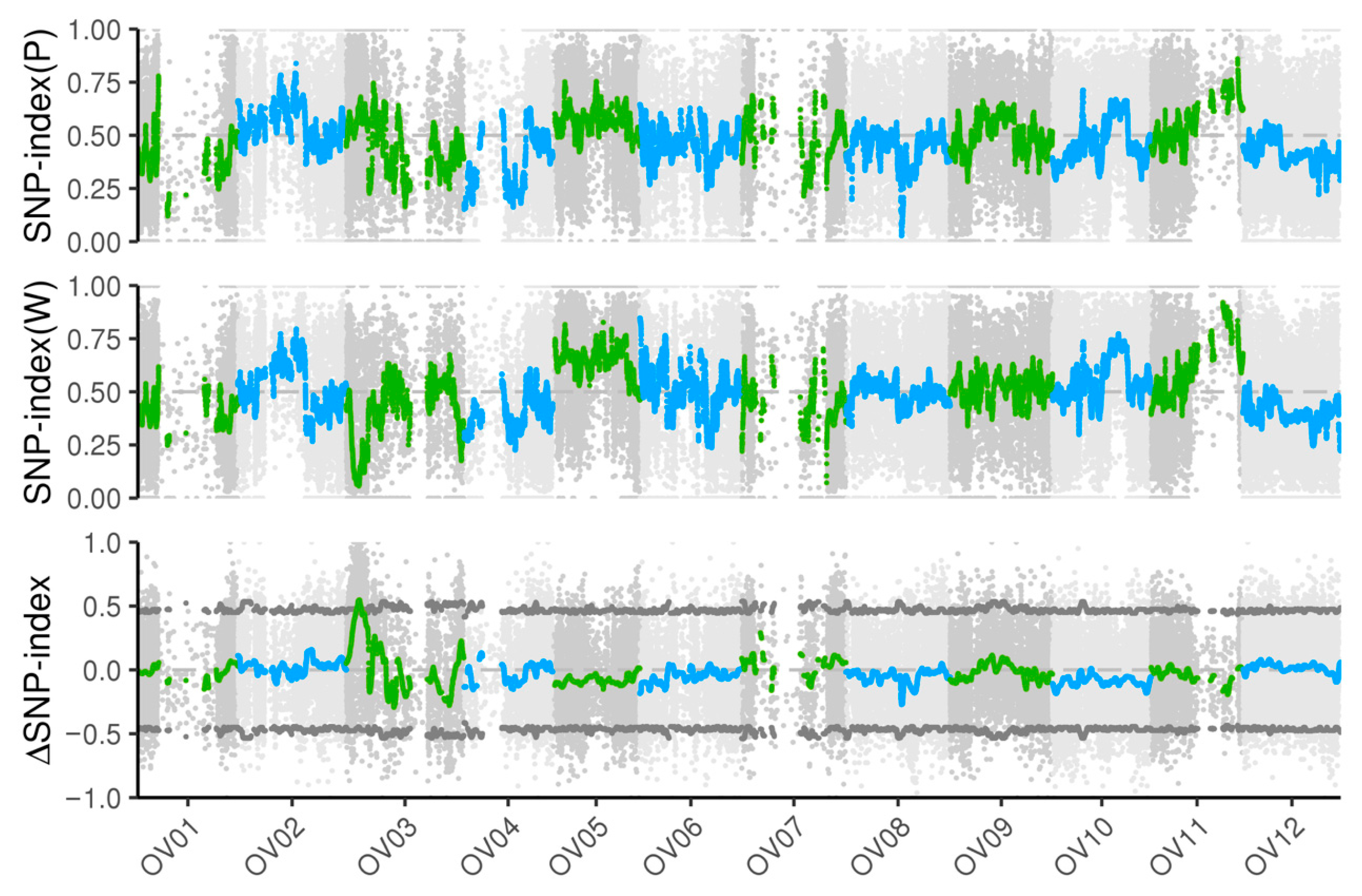
2.3. BSA-Seq Analysis
2.4. Fine-Mapping of Ovwf Locus
2.5. RNA Sequencing Data Analysis
3. Results
3.1. The Phenotype of O. violaceus
3.2. A Single Recessive Locus wf Controls the White Flower Phenotype in O. violaceus
3.3. The wf Locus Is Located on Chromosome Ov03 by BSA-Seq Analysis
3.4. The Ovwf Locus Was Fine-Mapped to a 1.37 Mb Region
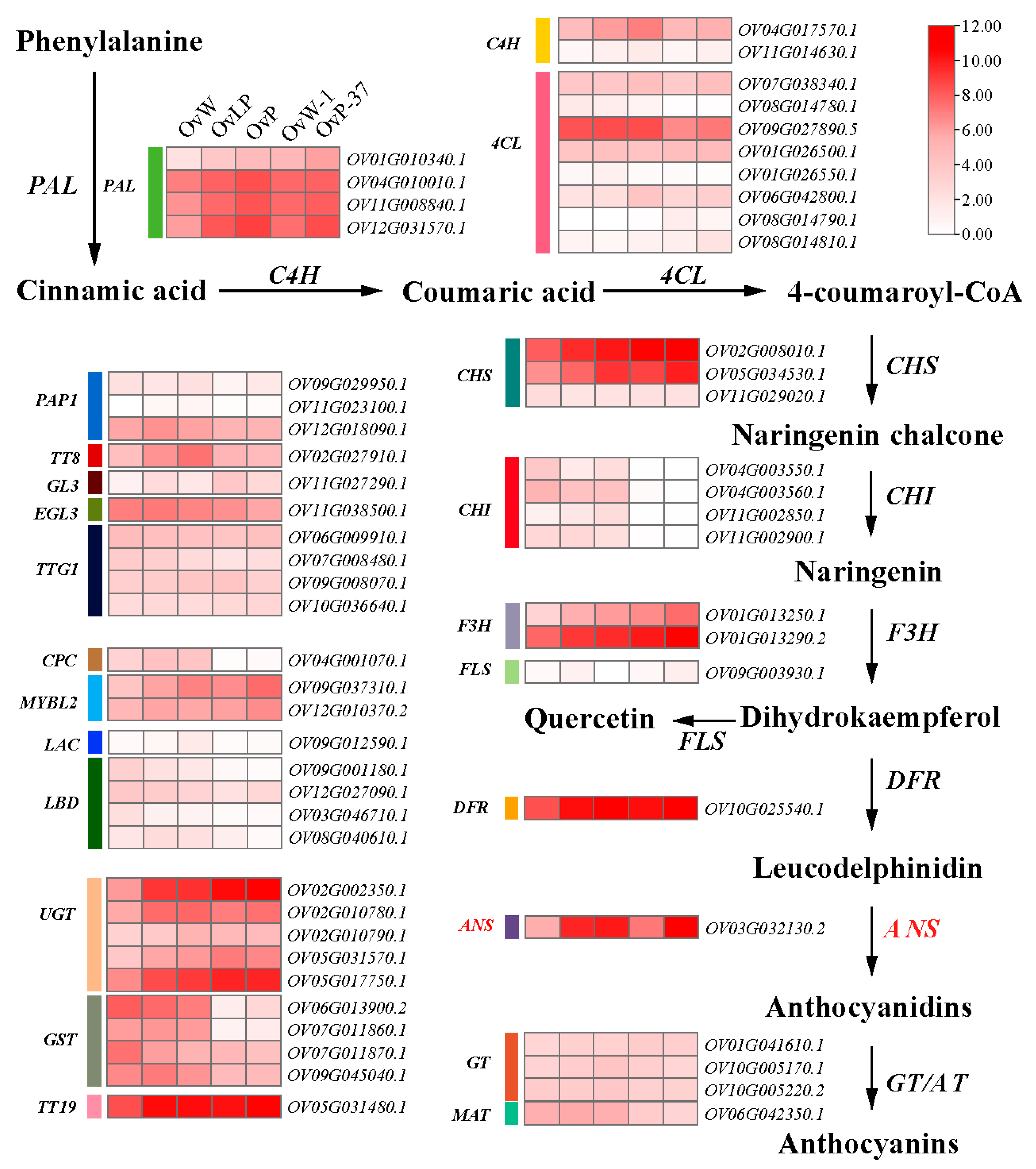
3.5. OvANS Was Predicted to Be the Candidate Gene for the Ovwf Locus
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Pan, Q.; Zeng, P.; Li, Z. Unraveling Large and Polyploidy Genome of the Crucifer Orychophragmus violaceus in China, a Potential Oil Crop. Plants 2023, 12, 374. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Teitgen, A.M.; Shirani, A.; Ling, J.; Busta, L.; Cahoon, R.E.; Zhang, W.; Li, Z.; Chapman, K.D.; Berman, D.; et al. Discontinuous fatty acid elongation yields hydroxylated seed oil with improved function. Nat. Plants 2018, 4, 711–720. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, G.; Cui, W.; Ge, C.; Li, B.; Li, M.; Liu, S.; Wang, L. Recent Advances in the Nutritional Value, Chemical Compositions, Pharmacological Activity, and Application Value of Orychophragmus violaceus: A Comprehensive Review. Molecules 2024, 29, 1314. [Google Scholar] [CrossRef]
- Tanaka, Y.; Sasaki, N.; Ohmiya, A. Biosynthesis of plant pigments: Anthocyanins, betalains and carotenoids. Plant J. 2008, 54, 733–749. [Google Scholar] [CrossRef]
- Honda, T.; Tatsuzawa, F.; Kobayashi, N.; Kasai, H.; Nagumo, S.; Shigihara, A.; Saito, N. Acylated anthocyanins from the violet-blue flowers of Orychophragonus violaceus. Phytochemistry 2005, 66, 1844–1851. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, C.C.; Chen, H.D.; Shen, W.J.; Chen, L.L.; Tan, C.; Chen, D.Z. Identification and characterization of the gene OvANS associated with purple flower in Orychophragmus violaceus. BMC Plant Biol. 2025, 25, 1349. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, Z.; Yan, Z.; Liu, J.; Zhang, J.; Liu, G. Integrated transcriptomic and metabolomic analyses reveal the molecular mechanism of flower color differentiation in Orychophragmus violaceus. Front. Plant Sci. 2025, 16, 1509120. [Google Scholar] [CrossRef]
- Shi, M.Z.; Xie, D.Y. Biosynthesis and metabolic engineering of anthocyanins in Arabidopsis thaliana. Recent. Pat. Biotechnol. 2014, 8, 47–60. [Google Scholar] [CrossRef]
- Li, X.; Bu, F.; Zhang, M.; Li, Z.; Zhang, Y.; Chen, H.; Xue, W.; Guo, R.; Qi, J.; Kim, C.; et al. Enhancing nature’s palette through the epigenetic breeding of flower color in chrysanthemum. New Phytol. 2025, 245, 2117–2132. [Google Scholar] [CrossRef]
- Sun, Y.; Zheng, Y.; Wang, W.; Yao, H.; Ali, Z.; Xiao, M.; Ma, Z.; Li, J.; Zhou, W.; Cui, J.; et al. VvFHY3 links auxin and endoplasmic reticulum stress to regulate grape anthocyanin biosynthesis at high temperatures. Plant Cell 2024, 37, koae303. [Google Scholar] [CrossRef] [PubMed]
- Li, S. Transcriptional control of flavonoid biosynthesis: Fine-tuning of the MYB-bHLH-WD40 (MBW) complex. Plant Signal Behav. 2014, 9, e27522. [Google Scholar] [CrossRef]
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 2015, 20, 176–185. [Google Scholar] [CrossRef]
- Yan, C.; An, G.; Zhu, T.; Zhang, W.; Zhang, L.; Peng, L.; Chen, J.; Kuang, H. Independent activation of the BoMYB2 gene leading to purple traits in Brassica oleracea. Theor. Appl. Genet. 2019, 132, 895–906. [Google Scholar] [CrossRef]
- Chen, D.; Liu, Y.; Yin, S.; Qiu, J.; Jin, Q.; King, G.J.; Wang, J.; Ge, X.; Li, Z. Alternatively Spliced BnaPAP2.A7 Isoforms Play Opposing Roles in Anthocyanin Biosynthesis of Brassica napus L. Front. Plant Sci. 2020, 11, 983. [Google Scholar] [CrossRef]
- He, Q.; Wu, J.Q.; Xue, Y.H.; Zhao, W.B.; Li, R.; Zhang, L.G. The novel gene, located on chromosome A07, with a short intron 1 controls the purple-head trait of Chinese cabbage (L.). Hortic. Res. 2020, 7, 97. [Google Scholar] [CrossRef] [PubMed]
- Heng, S.; Cheng, Q.; Zhang, T.; Liu, X.; Huang, H.; Yao, P.; Liu, Z.; Wan, Z.; Fu, T. Fine-mapping of the BjPur gene for purple leaf color in Brassica juncea. Theor. Appl. Genet. 2020, 133, 2989–3000. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Chen, P.; Tang, X.; Zhong, T.; Yang, T.; Nwafor, C.C.; Yang, C.; Ge, X.; An, H.; Li, Z.; et al. Genome assembly of the Brassicaceae diploid Orychophragmus violaceus reveals complex whole-genome duplication and evolution of dihydroxy fatty acid metabolism. Plant Commun. 2023, 4, 100432. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Yang, Y.; Zhang, X.; Zhang, L.; Fu, Y.; Guo, Z.; Chen, S.; Wu, J.; Schnable, J.C.; Yi, K.; et al. The genome of Orychophragmus violaceus provides genomic insights into the evolution of Brassicaceae polyploidization and its distinct traits. Plant Commun. 2023, 4, 100431. [Google Scholar] [CrossRef]
- Cruz, C.D. GENES—A software package for analysis in experimental statistics and quantitative genetics. Acta Sci.-Agron. 2013, 35, 271–276. [Google Scholar] [CrossRef]
- Matuszczak, M.; Spasibionek, S.; Gacek, K.; Bartkowiak-Broda, I. Cleaved amplified polymorphic sequences (CAPS) marker for identification of two mutant alleles of the rapeseed BnaA.FAD2 gene. Mol. Biol. Rep. 2020, 47, 7607–7621. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef] [PubMed]
- Van der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; Del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.1–11.10.33. [Google Scholar] [CrossRef]
- Mansfeld, B.N.; Grumet, R. QTLseqr: An R Package for Bulk Segregant Analysis with Next-Generation Sequencing. Plant Genome 2018, 11, 180006. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Chen, C.J.; Wu, Y.; Li, J.W.; Wang, X.; Zeng, Z.H.; Xu, J.; Liu, Y.L.; Feng, J.T.; Chen, H.; He, Y.H.; et al. TBtools-II: A “one for all, all for one”bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Tan, C.; Chen, H.; Dai, G.; Liu, Y.; Shen, W.; Wang, C.; Liu, D.; Liu, S.; Xu, S.; Zhu, B.; et al. Identification and characterization of the gene BraANS.A03 associated with purple leaf color in pak choi (Brassica rapa L. ssp. chinensis). Planta 2023, 258, 19. [Google Scholar] [CrossRef]
- Si, J.; Zhou, X.; Chen, X.; Ming, H.; Liu, H.; Hui, M. Identification and characterization of a key gene controlling purple leaf coloration in non-heading Chinese cabbage (Brassica rapa). Planta 2025, 261, 80. [Google Scholar] [CrossRef]
- Li, H.; Zhu, L.; Yuan, G.; Heng, S.; Yi, B.; Ma, C.; Shen, J.; Tu, J.; Fu, T.; Wen, J. Fine mapping and candidate gene analysis of an anthocyanin-rich gene, BnaA.PL1, conferring purple leaves in Brassica napus L. Mol. Genet. Genom. 2016, 291, 1523–1534. [Google Scholar] [CrossRef]
- Chen, D.; Jin, Q.; Pan, J.; Liu, Y.; Tang, Y.; E, Y.; Xu, L.; Yang, T.; Qiu, J.; Chen, X.; et al. Fine mapping of genes controlling pigment accumulation in oilseed rape (Brassica napus L.). Mol. Breed. 2023, 43, 19. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Zhou, H.; Zhou, D.; Wu, J.; Liu, L.; Guo, Y.; Wang, T.; Tan, C.; Chen, D.; Ge, X.; et al. The introgression of BjMYB113 from Brassica juncea leads to purple leaf trait in Brassica napus. BMC Plant Biol. 2024, 24, 735. [Google Scholar] [CrossRef]
- Zhang, B.; Liu, C.; Wang, Y.Q.; Yao, X.; Wang, F.; Wu, J.S.; King, G.J.; Liu, K.D. Disruption of a gene converts flower colour from white to yellow in species. New Phytol. 2015, 206, 1513–1526. [Google Scholar] [CrossRef]
- Ye, S.; Hua, S.; Ma, T.; Ma, X.; Chen, Y.; Wu, L.; Zhao, L.; Yi, B.; Ma, C.; Tu, J.; et al. Genetic and multi-omics analyses reveal BnaA07.PAP2In-184-317 as the key gene conferring anthocyanin-based color in Brassica napus flowers. J. Exp. Bot. 2022, 73, 6630–6645. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.; Zhang, K.; Chai, L.; Zheng, B.; Zhang, J.; Jiang, J.; Tan, C.; Li, H.; Chen, D.; Jiang, L. Unraveling the mechanism of flower color variation in Brassica napus by integrated metabolome and transcriptome analyses. Front. Plant Sci. 2024, 15, 1419508. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.; Zhang, Q.; Shen, W.; Liu, Y.; Zhang, D.; Chen, L.; Chen, D. Expression profiles of microRNA-mRNA and their potential impact on anthocyanin accumulation in purple petals of Brassica napus. BMC Plant Biol. 2024, 24, 1223. [Google Scholar] [CrossRef]
- Zheng, Y.; Shi, R.; Chen, W.; Wang, X.; Dun, X.; Wang, H.; Deng, J. Precise pigment biosynthesis for flower color design in Brassica napus. Hortic. Res. 2025, 12, uhaf193. [Google Scholar] [CrossRef]
- Hu, Q.; Hansen, N.; Laursen, J.; Dixelius, C.; Andersen, B. Intergeneric hybrids between Brassica napus and Orychophragmus violaceus containing traits of agronomic importance for oilseed rape breeding. Theor. Appl. Genet. 2002, 105, 834–840. [Google Scholar] [CrossRef]
- Ma, N.; Li, Z.Y.; Cartagena, J.A.; Fukui, K. GISH and AFLP analyses of novel Brassica napus lines derived from one hybrid between B. napus and Orychophragmus violaceus. Plant Cell Rep. 2006, 25, 1089–1093. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.G.; Hu, T.T.; Ge, X.H.; Du, X.Z.; Ding, L.; Li, Z.Y. Production and characterization of intergeneric somatic hybrids between Brassica napus and Orychophragmus violaceus and their backcrossing progenies. Plant Cell Rep. 2008, 27, 1611–1621. [Google Scholar] [CrossRef] [PubMed]
- Fu, W.; Chen, D.; Pan, Q.; Li, F.; Zhao, Z.; Ge, X.; Li, Z. Production of red-flowered oilseed rape via the ectopic expression of Orychophragmus violaceus OvPAP2. Plant Biotechnol. J. 2018, 16, 367–380. [Google Scholar] [CrossRef] [PubMed]


| Type | Population | Purple | White | Theoretical Ratio | Actual Ratio | χ2 |
|---|---|---|---|---|---|---|
| F2 | 1224 | 929 | 295 | 03:01 | 3.15:1 | 0.48 |
| BC1F1 | 508 | 261 | 247 | 01:01 | 1.06:1 | 0.33 |
| Note: χ2 (0.05, 1) = 3.84. | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Xie, L.; Zhu, Z.; Tan, C.; Gao, L.; Shen, W.; Wan, S.; Ge, X.; Chen, D.; Zhu, B. Fine-Mapping of OvANS: A Novel Gene Controlling White Flowers in Orychophragmus violaceus. Biology 2025, 14, 1669. https://doi.org/10.3390/biology14121669
Liu Y, Xie L, Zhu Z, Tan C, Gao L, Shen W, Wan S, Ge X, Chen D, Zhu B. Fine-Mapping of OvANS: A Novel Gene Controlling White Flowers in Orychophragmus violaceus. Biology. 2025; 14(12):1669. https://doi.org/10.3390/biology14121669
Chicago/Turabian StyleLiu, Yi, Liwen Xie, Zichen Zhu, Chen Tan, Liwei Gao, Wenjie Shen, Shubei Wan, Xianhong Ge, Daozong Chen, and Bin Zhu. 2025. "Fine-Mapping of OvANS: A Novel Gene Controlling White Flowers in Orychophragmus violaceus" Biology 14, no. 12: 1669. https://doi.org/10.3390/biology14121669
APA StyleLiu, Y., Xie, L., Zhu, Z., Tan, C., Gao, L., Shen, W., Wan, S., Ge, X., Chen, D., & Zhu, B. (2025). Fine-Mapping of OvANS: A Novel Gene Controlling White Flowers in Orychophragmus violaceus. Biology, 14(12), 1669. https://doi.org/10.3390/biology14121669







