Lipidomic and Transcriptomic Reveals Variations in Lipid Deposition During Goose Fatty Liver Formation
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Sample Collection
2.2. Liver Histology Analysis
2.3. Liver Chemical Composition Determination
2.4. Transcriptomic Analysis
2.5. Screening of Differentially Expressed Genes and Functional Enrichment Analysis
2.6. Lipidomic Analysis
2.7. Identification of Differential Lipid Molecules and Functional Enrichment Analysis
2.8. Statistical Analysis
3. Results
3.1. Overfeeding Dramatically Changed the Global Appearance, Hepatic Histology and Chemical Composition of Goose Livers
3.2. Analysis of Gene Expression Levels in Goose Livers
3.3. Identifying Differential Expression Genes and Key Pathways at Different Overfeeding Stages
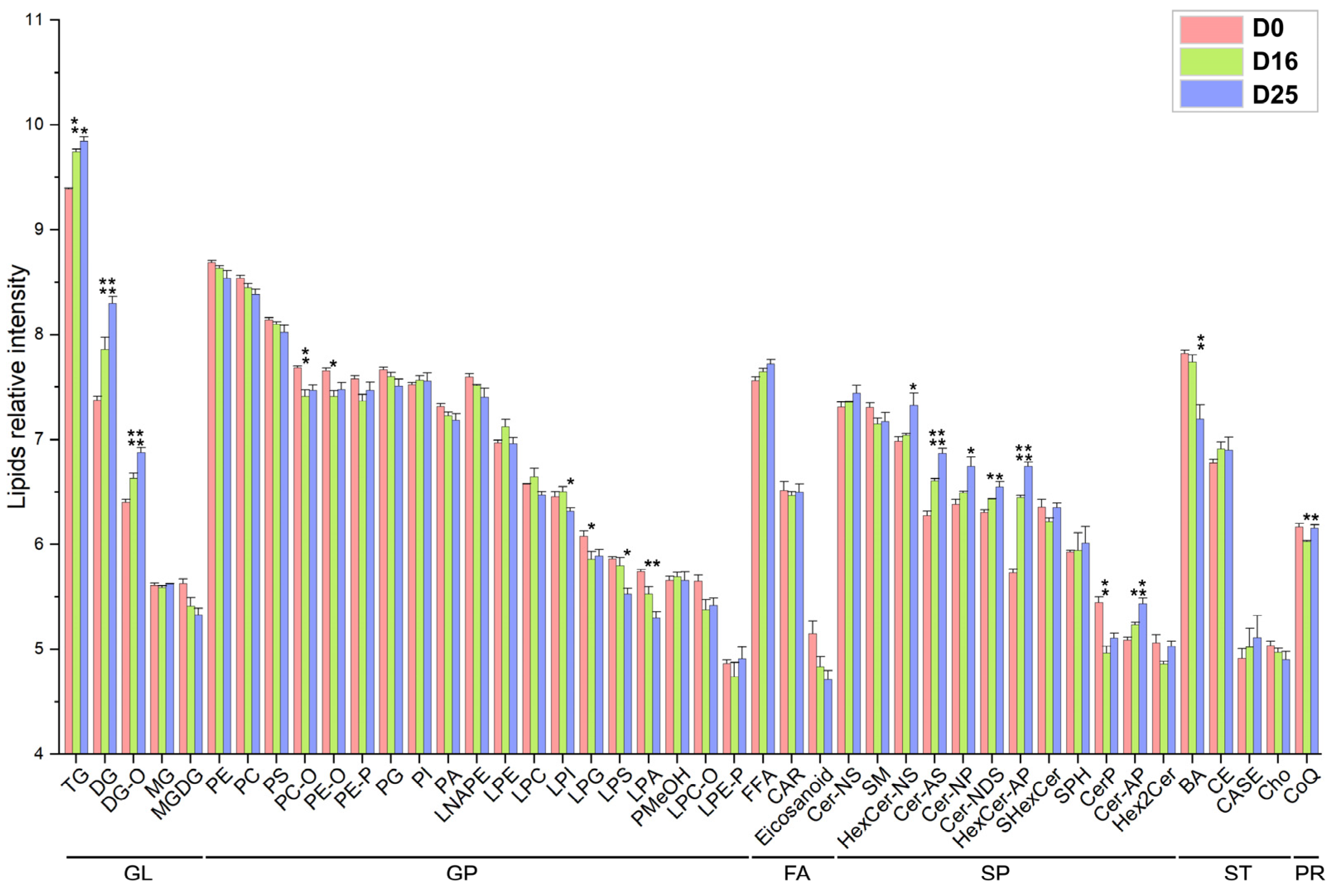
3.4. Analysis of Lipid Composition in Goose Livers
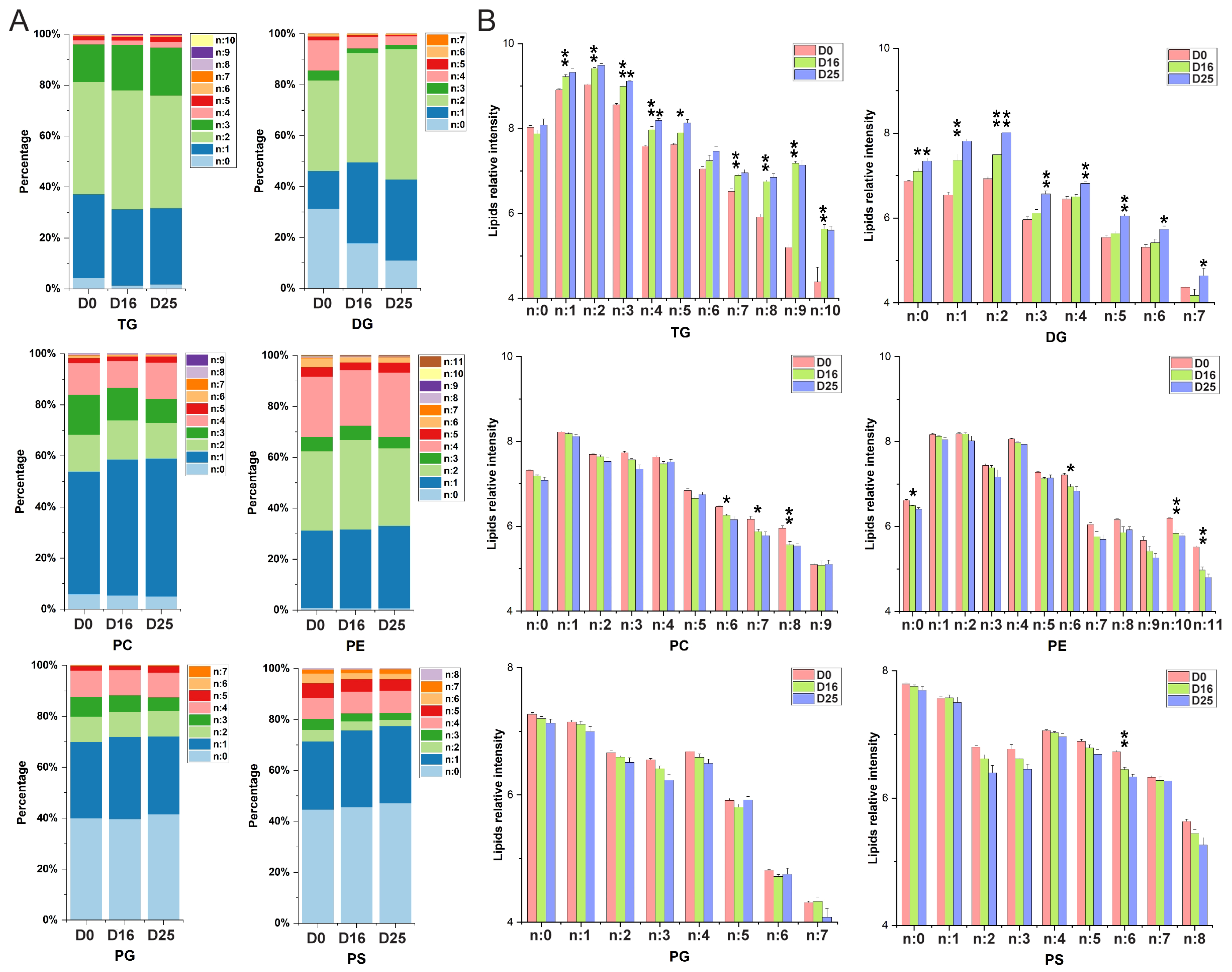
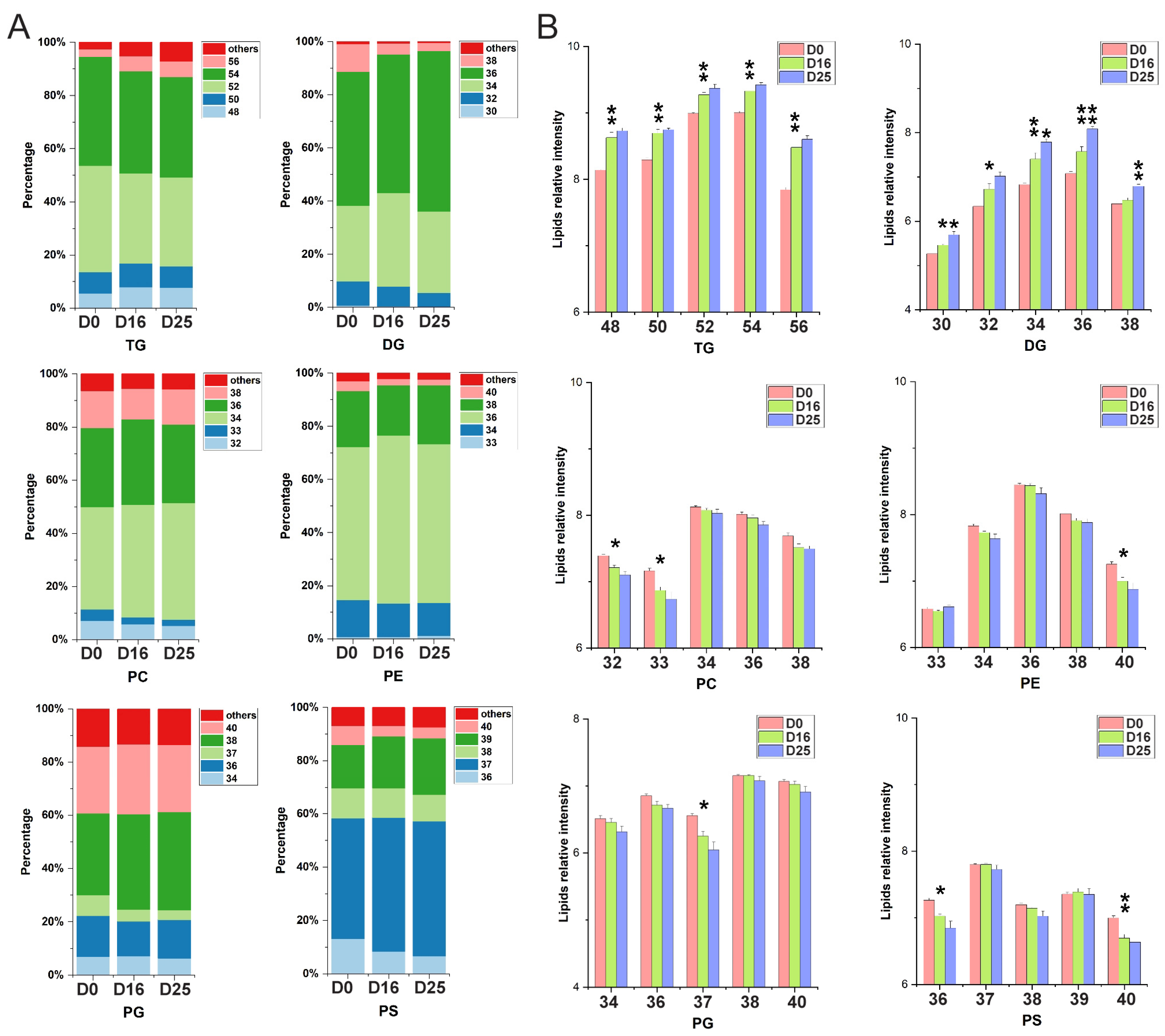
3.5. Dynamic Changes of Fatty Acid Composition in Each Lipid Subclass with Respect to the Carbon Chain Length and Saturation
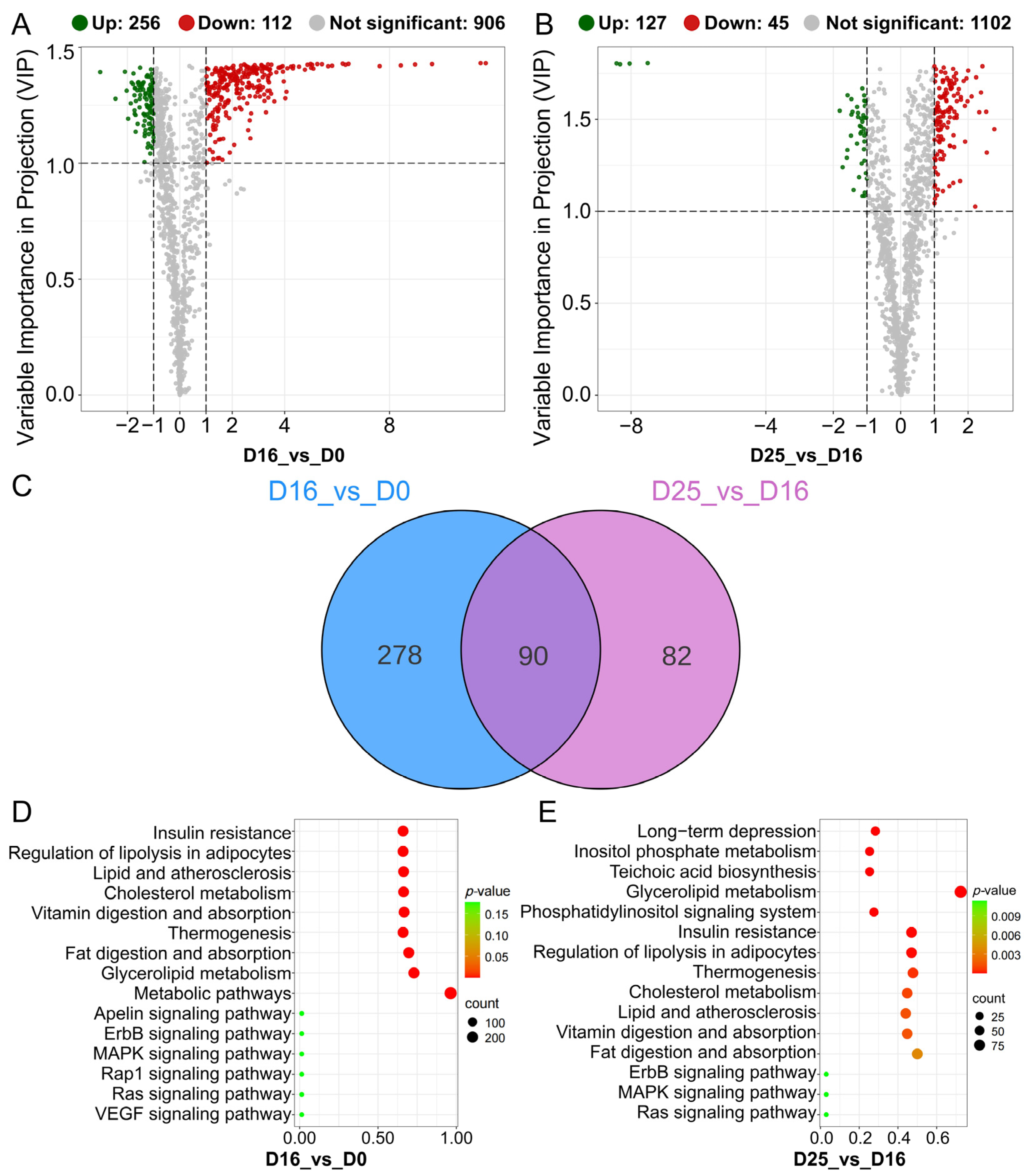
3.6. Identifying Differential Lipid Molecules and Key Pathways at Different Overfeeding Stages
3.7. Integrated Lipidomic and Transcriptomic Analysis in Goose Fatty Livers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DEGs | differentially expressed genes |
| DLMs | differential lipid molecules |
| NAFLD | non-alcoholic fatty liver disease |
| CA | crude ash |
| CP | crude protein |
| CF | crude fat |
| padj | adjusted p-value |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| MTBE | methyl tert-butyl ether |
| LC-MS/MS | Liquid Chromatography–Tandem Mass Spectrometry |
| LC-ESI-MS | Liquid Chromatography–Electrospray Ionization–Tandem Mass Spectrometry |
| LIT | linear ion trap |
| QQQ | triple quadrupole |
| IS | ion spray |
| GS1 | Gas 1 |
| GS2 | Gas 2 |
| CUR | curtain gas |
| CAD | collision gas |
| MRM | multiple reaction monitoring |
| DP | declustering potential |
| CE | collision energy |
| QC | quality control |
| BPCs | base peak ion chromatograms |
| RSD | relative standard deviation |
| RT | retention time |
| OPLS-DA | orthogonal partial least squares–discriminate analysis |
| VIP | variable importance in projection |
| ANOVA | analysis of variance |
| HSD | honestly significant difference |
| PCA | Principal Component Analysis |
| GL | glycerolipids |
| GP | glycerophospholipids |
| FA | fatty acyls |
| SP | sphingolipids |
| ST | sterol lipids |
| PR | prenol lipids |
| TG | triacylglycerols |
| PE | phosphatidylethanolamines |
| PC | phosphatidylcholine |
| DG | diglycerides |
| HexCer-NS | non-hydroxy sphingosine hexosylceramides |
| Cer-AS | alpha-hydroxy sphingosine ceramide |
| Cer-NP | non-hydroxy phytosphingosine ceramide |
| Cer-NDS | non-hydroxy dihydrosphingosine ceramide |
| HexCer-AP | alpha-hydroxy phytosphingosine hexosylceramide |
| LPA | lysophosphatidic acid |
| LPS | lysophosphatidylserine |
| BA | bile acids |
| PS | phosphatidylserine |
| PG | phosphatidylglycerol |
| PA | phosphatidic acid |
| LNA | lysophosphatidic acid |
| Diglyceride | 1,2-Diacyl-sn-glycerol |
| SFAs | saturated fatty acids |
| HDL | high-density lipoprotein |
| FFA | free fatty acids |
References
- Gong, Y.; Lyu, W.; Shi, X.; Zou, X.; Lu, L.; Yang, H.; Xiao, Y. A Serum Metabolic Profiling Analysis During the Formation of Fatty Liver in Landes Geese via GC-TOF/MS. Front. Physiol. 2020, 11, 581699. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.; Han, C.; Deng, D.; Ye, F.; Gan, X.; Liu, H.; Li, L.; Xu, H.; Wei, S. Research progress into the physiological changes in metabolic pathways in waterfowl with hepatic steatosis. Br. Poult. Sci. 2021, 62, 118–124. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.; Ning, R.; Han, C.; Wei, S.; Teng, Y.; Li, L.; Liu, H.; Hu, S.; Kang, B.; Xu, H. Lipidomics analysis reveals new insights into the goose fatty liver formation. Poult. Sci. 2023, 102, 102428. [Google Scholar] [CrossRef]
- Osman, R.H.; Liu, L.; Xia, L.L.; Zhao, X.; Wang, Q.Q.; Sun, X.X.; Zhang, Y.H.; Yang, B.; Zheng, Y.; Gong, D.Q.; et al. Fads1 and 2 are promoted to meet instant need for long-chain polyunsaturated fatty acids in goose fatty liver. Mol. Cell. Biochem. 2016, 418, 103–117. [Google Scholar] [CrossRef]
- Hermier, D.; Rousselot-Pailley, D.; Peresson, R.; Sellier, N. Influence of orotic acid and estrogen on hepatic lipid storage and secretion in the goose susceptible to liver steatosis. Biochim. Biophys. Acta 1994, 1211, 97–106. [Google Scholar] [CrossRef]
- Geng, T.; Xia, L.; Li, F.; Xia, J.; Zhang, Y.; Wang, Q.; Yang, B.; Montgomery, S.; Cui, H.; Gong, D. The role of endoplasmic reticulum stress and insulin resistance in the occurrence of goose fatty liver. Biochem. Biophys. Res. Commun. 2015, 465, 83–87. [Google Scholar] [CrossRef]
- Liu, D.D.; Han, C.C.; Wan, H.F.; He, F.; Xu, H.Y.; Wei, S.H.; Du, X.H.; Xu, F. Effects of inhibiting PI3K-Akt-mTOR pathway on lipid metabolism homeostasis in goose primary hepatocytes. Animal 2016, 10, 1319–1327. [Google Scholar] [CrossRef]
- Lu, L.; Chen, Y.; Wang, Z.; Li, X.; Chen, W.; Tao, Z.; Shen, J.; Tian, Y.; Wang, D.; Li, G.; et al. The goose genome sequence leads to insights into the evolution of waterfowl and susceptibility to fatty liver. Genome Biol. 2015, 16, 89. [Google Scholar] [CrossRef]
- Liu, L.; Zhao, X.; Wang, Q.; Sun, X.; Xia, L.; Wang, Q.; Yang, B.; Zhang, Y.; Montgomery, S.; Meng, H.; et al. Prosteatotic and Protective Components in a Unique Model of Fatty Liver: Gut Microbiota and Suppressed Complement System. Sci. Rep. 2016, 6, 31763. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Jiang, S.; Zhang, Y.; Zhang, R.; Gong, D. Detection of miR-33 Expression and the Verification of Its Target Genes in the Fatty Liver of Geese. Int. J. Mol. Sci. 2015, 16, 12737–12752. [Google Scholar] [CrossRef]
- Wei, R.; Teng, Y.; Han, C.; Wei, S.; Li, L.; Liu, H.; Hu, S.; Kang, B.; Xu, H. Multi-omics reveals goose fatty liver formation from metabolic reprogramming. Front. Vet. Sci. 2024, 11, 1122904. [Google Scholar] [CrossRef]
- Hermier, D.; Saadoun, A.; Salichon, M.R.; Sellier, N.; Rousselot-Paillet, D.; Chapman, M.J. Plasma lipoproteins and liver lipids in two breeds of geese with different susceptibility to hepatic steatosis: Changes induced by development and force-feeding. Lipids 1991, 26, 331–339. [Google Scholar] [CrossRef]
- Yang, J.S.; Park, Y. Insecticide Exposure and Development of Nonalcoholic Fatty Liver Disease. J. Agric. Food Chem. 2018, 66, 10132–10138. [Google Scholar] [CrossRef]
- Hong, L.; Sun, Z.; Xu, D.; Li, W.; Cao, N.; Fu, X.; Huang, Y.; Tian, Y.; Li, B. Transcriptome and lipidome integration unveils mechanisms of fatty liver formation in Shitou geese. Poult. Sci. 2024, 103, 103280. [Google Scholar] [CrossRef]
- Thompson, W.L.; Takebe, T. Human liver model systems in a dish. Dev. Growth Differ. 2021, 63, 47–58. [Google Scholar] [CrossRef]
- Igarashi, R.; Oda, M.; Okada, R.; Yano, T.; Takahashi, S.; Pastuhov, S.; Matano, M.; Masuda, N.; Togasaki, K.; Ohta, Y.; et al. Generation of human adult hepatocyte organoids with metabolic functions. Nature 2025, 641, 1248–1257. [Google Scholar] [CrossRef]
- Clark, E.L.; Archibald, A.L.; Daetwyler, H.D.; Groenen, M.A.M.; Harrison, P.W.; Houston, R.D.; Kuhn, C.; Lien, S.; Macqueen, D.J.; Reecy, J.M.; et al. From FAANG to fork: Application of highly annotated genomes to improve farmed animal production. Genome Biol. 2020, 21, 285. [Google Scholar] [CrossRef]
- Jones, H.E.; Wilson, P.B. Progress and opportunities through use of genomics in animal production. Trends Genet. 2022, 38, 1228–1252. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.; Zhang, J.; Wang, X.; Xing, G.; Guo, Z.; Zhang, Q.; Huang, W.; Xie, M.; Hou, S. Selective analysis of resistance and susceptibility to duck hepatitis A virus genotype 3 in Pekin duck. Anim. Res. One Health 2023, 1, 146–155. [Google Scholar] [CrossRef]
- Tan, X.; He, Z.; Fahey, A.G.; Zhao, G.; Liu, R. Research progress and applications of genome-wide association study in farm animals. Anim. Res. One Health 2023, 1, 56–77. [Google Scholar] [CrossRef]
- Fan, W.; Hou, S.; Zhou, Z. The Duck 1000 Genomes Project: Achievements and perspectives. Anim. Res. One Health 2024, 2, 366–376. [Google Scholar] [CrossRef]
- Wei, R.; Deng, D.; Teng, Y.; Lu, C.; Luo, Z.; Abdulai, M.; Liu, H.; Xu, H.; Li, L.; Hu, S.; et al. Study on the effect of different types of sugar on lipid deposition in goose fatty liver. Poult. Sci. 2022, 101, 101729. [Google Scholar] [CrossRef]
- Li, X.; Hu, S.; Wang, W.; Tang, B.; Zheng, C.; Hu, J.; Hu, B.; Li, L.; Liu, H.; Wang, J. Effects of cage versus floor rearing system on goose intestinal histomorphology and cecal microbial composition. Poult. Sci. 2022, 101, 101931. [Google Scholar] [CrossRef]
- Geng, T.; Zhao, X.; Xia, L.; Liu, L.; Li, F.; Yang, B.; Wang, Q.; Montgomery, S.; Cui, H.; Gong, D. Supplementing dietary sugar promotes endoplasmic reticulum stress-independent insulin resistance and fatty liver in goose. Biochem. Biophys. Res. Commun. 2016, 476, 665–669. [Google Scholar] [CrossRef]
- Roy, B.; Gupta, M.; Goswami, B.K. Serial section histopathology image dataset of colon and pancreas tissue captured through light microscope. Data Brief 2024, 54, 110429. [Google Scholar] [CrossRef]
- Rio, D.C.; Ares, M., Jr.; Hannon, G.J.; Nilsen, T.W. Purification of RNA using TRIzol (TRI reagent). Cold Spring Harb. Protoc. 2010, 2010, pdb.prot5439. [Google Scholar] [CrossRef]
- Senst, A.; Bonsiepe, H.; Kron, S.; Schulz, I. Application of the Agilent 2100 Bioanalyzer instrument as quality control for next-generation sequencing. J. Forensic. Sci. 2024, 69, 2192–2196. [Google Scholar] [CrossRef] [PubMed]
- Withanage, M.H.H.; Liang, H.; Zeng, E. RNA-Seq Experiment and Data Analysis. Methods Mol. Biol. 2022, 2418, 405–424. [Google Scholar]
- Dillies, M.A.; Rau, A.; Aubert, J.; Hennequet-Antier, C.; Jeanmougin, M.; Servant, N.; Keime, C.; Marot, G.; Castel, D.; Estelle, J.; et al. A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief. Bioinform. 2013, 14, 671–683. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Dennis, G., Jr.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003, 4, R60. [Google Scholar] [CrossRef]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef]
- Matyash, V.; Liebisch, G.; Kurzchalia, T.V.; Shevchenko, A.; Schwudke, D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J. Lipid Res. 2008, 49, 1137–1146. [Google Scholar] [CrossRef]
- Faul, F.; Erdfelder, E.; Lang, A.G.; Buchner, A. G*Power 3: A flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav. Res. Methods 2007, 39, 175–191. [Google Scholar] [CrossRef]
- Hong, S.E.; Mun, S.J.; Lee, Y.J.; Yoo, T.; Suh, K.S.; Kang, K.W.; Son, M.J.; Kim, W.; Choi, M. Single-cell eQTL analysis identifies genetic variation underlying metabolic dysfunction-associated steatohepatitis. Nat. Genet. 2025, 57, 1638–1648. [Google Scholar] [CrossRef]
- Fan, W.; Liu, W.; Liu, H.; Meng, Q.; Xu, Y.; Guo, Y.; Wang, B.; Zhou, Z.; Hou, S. Dynamic accumulation of fatty acids in duck (Anas platyrhynchos) breast muscle and its correlations with gene expression. BMC Genom. 2020, 21, 58. [Google Scholar] [CrossRef]
- Liu, R.; Kong, F.; Xing, S.; He, Z.; Bai, L.; Sun, J.; Tan, X.; Zhao, D.; Zhao, G.; Wen, J. Dominant changes in the breast muscle lipid profiles of broiler chickens with wooden breast syndrome revealed by lipidomics analyses. J. Anim. Sci. Biotechnol. 2022, 13, 93. [Google Scholar] [CrossRef]
- Yu, Y.; Lyu, W.; Fu, Z.; Fan, Q.; Xiao, Y.; Ren, Y.; Yang, H. Metabolic Profiling Analysis of Liver in Landes Geese During the Formation of Fatty Liver via GC-TOF/MS. Front. Physiol. 2021, 12, 783498. [Google Scholar] [CrossRef]
- Fernandez, X.; Lazzarotto, V.; Bernadet, M.D.; Manse, H. Comparison of the composition and sensory characteristics of goose fatty liver obtained by overfeeding and spontaneous fattening1. Poult. Sci. 2019, 98, 6149–6160. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.; Han, C.; Wei, S.; Teng, Y.; Li, L.; Liu, H.; Hu, S.; Kang, B.; Xu, H. Integrative analysis of transcriptome and lipidome reveals fructose pro-steatosis mechanism in goose fatty liver. Front. Nutr. 2022, 9, 1052600. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.H.; Meng, H.; Duan, X.J.; Xu, G.Q.; Zhang, J.; Gong, D.Q. Gene expression profile in the liver tissue of geese after overfeeding. Poult. Sci. 2011, 90, 107–117. [Google Scholar] [CrossRef]
- Chartrin, P.; Bernadet, M.D.; Guy, G.; Mourot, J.; Hocquette, J.F.; Rideau, N.; Duclos, M.J.; Baeza, E. Does overfeeding enhance genotype effects on energy metabolism and lipid deposition in breast muscle of ducks? Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2006, 145, 413–418. [Google Scholar] [CrossRef]
- Hermansson, M.; Hokynar, K.; Somerharju, P. Mechanisms of glycerophospholipid homeostasis in mammalian cells. Prog. Lipid Res. 2011, 50, 240–257. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Jin, L.; Li, Y.; Tang, Q.; Hu, S.; Xu, H.; Gill, C.A.; Li, M.; Wang, J. Transcriptomic analysis between Normal and high-intake feeding geese provides insight into adipose deposition and susceptibility to fatty liver in migratory birds. BMC Genom. 2019, 20, 372. [Google Scholar] [CrossRef]
- Gandemer, G. Lipids in muscles and adipose tissues, changes during processing and sensory properties of meat products. Meat Sci. 2002, 62, 309–321. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.A.; Kersten, S.; Qi, L. Lipoprotein Lipase and Its Regulators: An Unfolding Story. Trends Endocrinol. Metab. 2021, 32, 48–61. [Google Scholar] [CrossRef]
- Wang, Z.; Yao, T.; Song, Z. Involvement and mechanism of DGAT2 upregulation in the pathogenesis of alcoholic fatty liver disease. J. Lipid Res. 2010, 51, 3158–3165. [Google Scholar] [CrossRef]
- Amin, N.B.; Saxena, A.R.; Somayaji, V.; Dullea, R. Inhibition of Diacylglycerol Acyltransferase 2 Versus Diacylglycerol Acyltransferase 1: Potential Therapeutic Implications of Pharmacology. Clin. Ther. 2023, 45, 55–70. [Google Scholar] [CrossRef]
- McCoy, M.G.; Sun, G.S.; Marchadier, D.; Maugeais, C.; Glick, J.M.; Rader, D.J. Characterization of the lipolytic activity of endothelial lipase. J. Lipid Res. 2002, 43, 921–929. [Google Scholar] [CrossRef]
- Chen, S.; Subbaiah, P.V. Phospholipid and fatty acid specificity of endothelial lipase: Potential role of the enzyme in the delivery of docosahexaenoic acid (DHA) to tissues. Biochim. Biophys. Acta 2007, 1771, 1319–1328. [Google Scholar] [CrossRef]
- Barchuk, M.; Miksztowicz, V.; Zago, V.; Cevey, A.; Lopez, G.; Goren, N.; Friedman, S.; Gelpi, R.J.; Morales, C.; Fernandez Tome, M.D.C.; et al. Endothelial Lipase Is an Alternative Pathway for Fatty Acid Release from Lipoproteins: Evidence from a High Fat Diet Model of Obesity in Rats. Lipids 2018, 53, 993–1003. [Google Scholar] [CrossRef]
- Li, X.; Ge, J.; Li, Y.; Cai, Y.; Zheng, Q.; Huang, N.; Gu, Y.; Han, Q.; Li, Y.; Sun, R.; et al. Integrative lipidomic and transcriptomic study unravels the therapeutic effects of saikosaponins A and D on non-alcoholic fatty liver disease. Acta Pharm. Sin. B 2021, 11, 3527–3541. [Google Scholar] [CrossRef]
- Morrow, M.R.; Batchuluun, B.; Wu, J.; Ahmadi, E.; Leroux, J.M.; Mohammadi-Shemirani, P.; Desjardins, E.M.; Wang, Z.; Tsakiridis, E.E.; Lavoie, D.C.T.; et al. Inhibition of ATP-citrate lyase improves NASH, liver fibrosis, and dyslipidemia. Cell Metab. 2022, 34, 919–936.e918. [Google Scholar] [CrossRef] [PubMed]
- Lewis, G.F.; Carpentier, A.; Adeli, K.; Giacca, A. Disordered fat storage and mobilization in the pathogenesis of insulin resistance and type 2 diabetes. Endocr. Rev. 2002, 23, 201–229. [Google Scholar] [CrossRef] [PubMed]
- Carpentier, A.C.; Frisch, F.; Cyr, D.; Genereux, P.; Patterson, B.W.; Giguere, R.; Baillargeon, J.P. On the suppression of plasma nonesterified fatty acids by insulin during enhanced intravascular lipolysis in humans. Am. J. Physiol.-Endocrinol. Metab. 2005, 289, E849–E856. [Google Scholar] [CrossRef] [PubMed]
- Carpentier, A.C. Postprandial fatty acid metabolism in the development of lipotoxicity and type 2 diabetes. Diabetes Metab. 2008, 34, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Bessone, F.; Razori, M.V.; Roma, M.G. Molecular pathways of nonalcoholic fatty liver disease development and progression. Cell. Mol. Life Sci. 2019, 76, 99–128. [Google Scholar] [CrossRef] [PubMed]
- Ferre, P.; Foufelle, F. Hepatic steatosis: A role for de novo lipogenesis and the transcription factor SREBP-1c. Diabetes Obes. Metab. 2010, 12, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Cao, G.; Lin, M.; Gu, W.; Su, Z.; Duan, Y.; Song, W.; Liu, H.; Zhang, F. The rules and regulatory mechanisms of FOXO3 on inflammation, metabolism, cell death and aging in hosts. Life Sci. 2023, 328, 121877. [Google Scholar] [CrossRef]
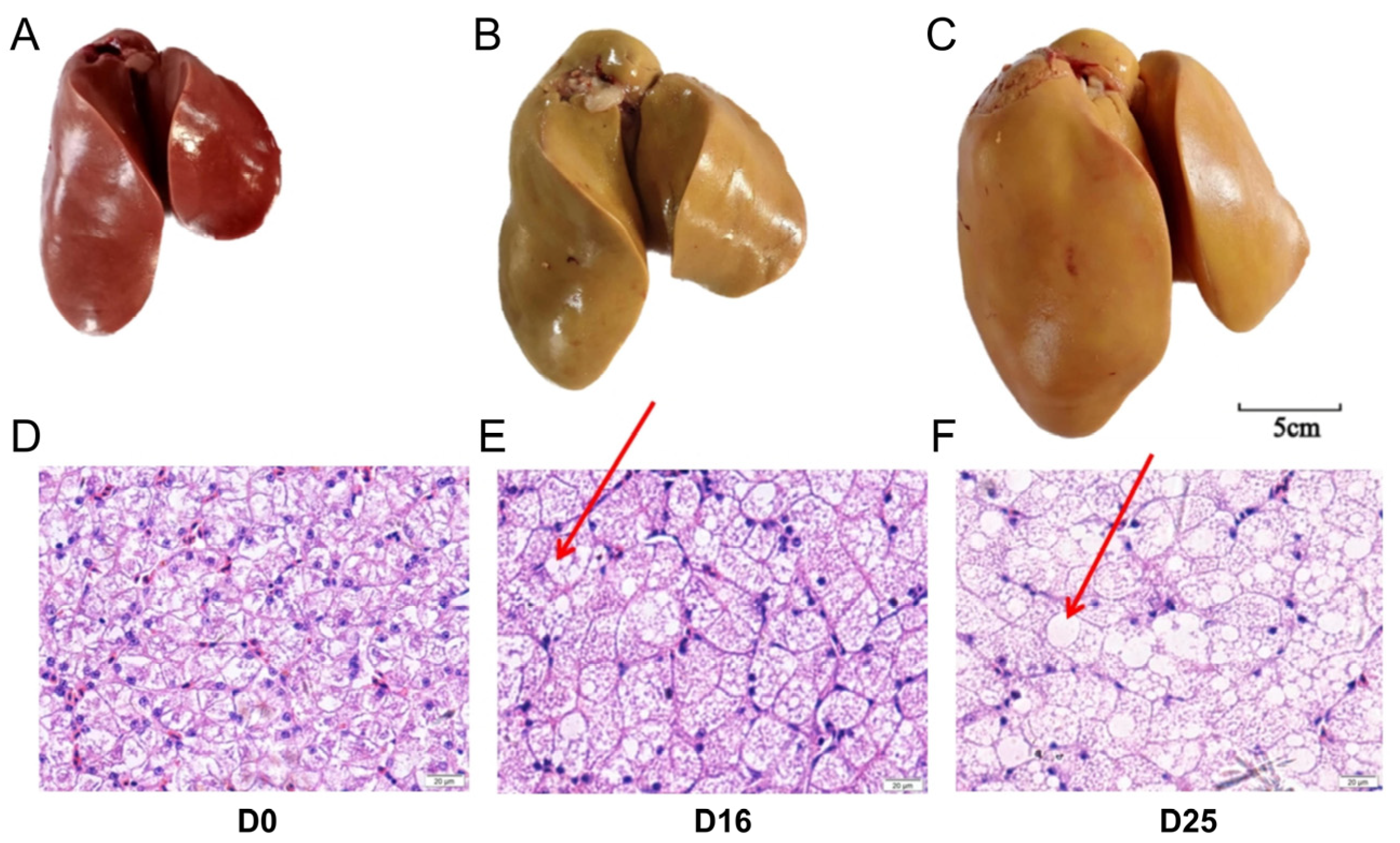
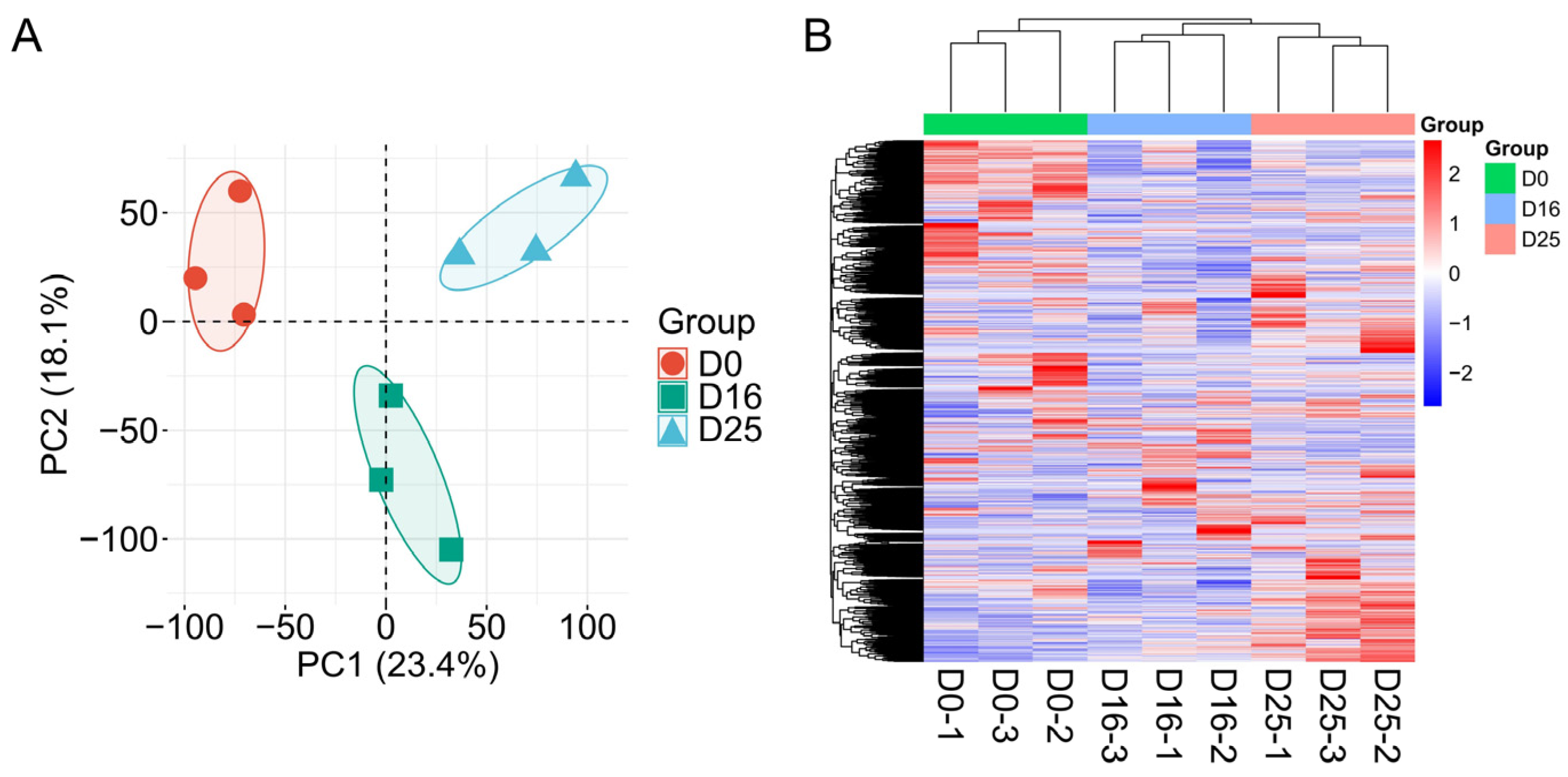
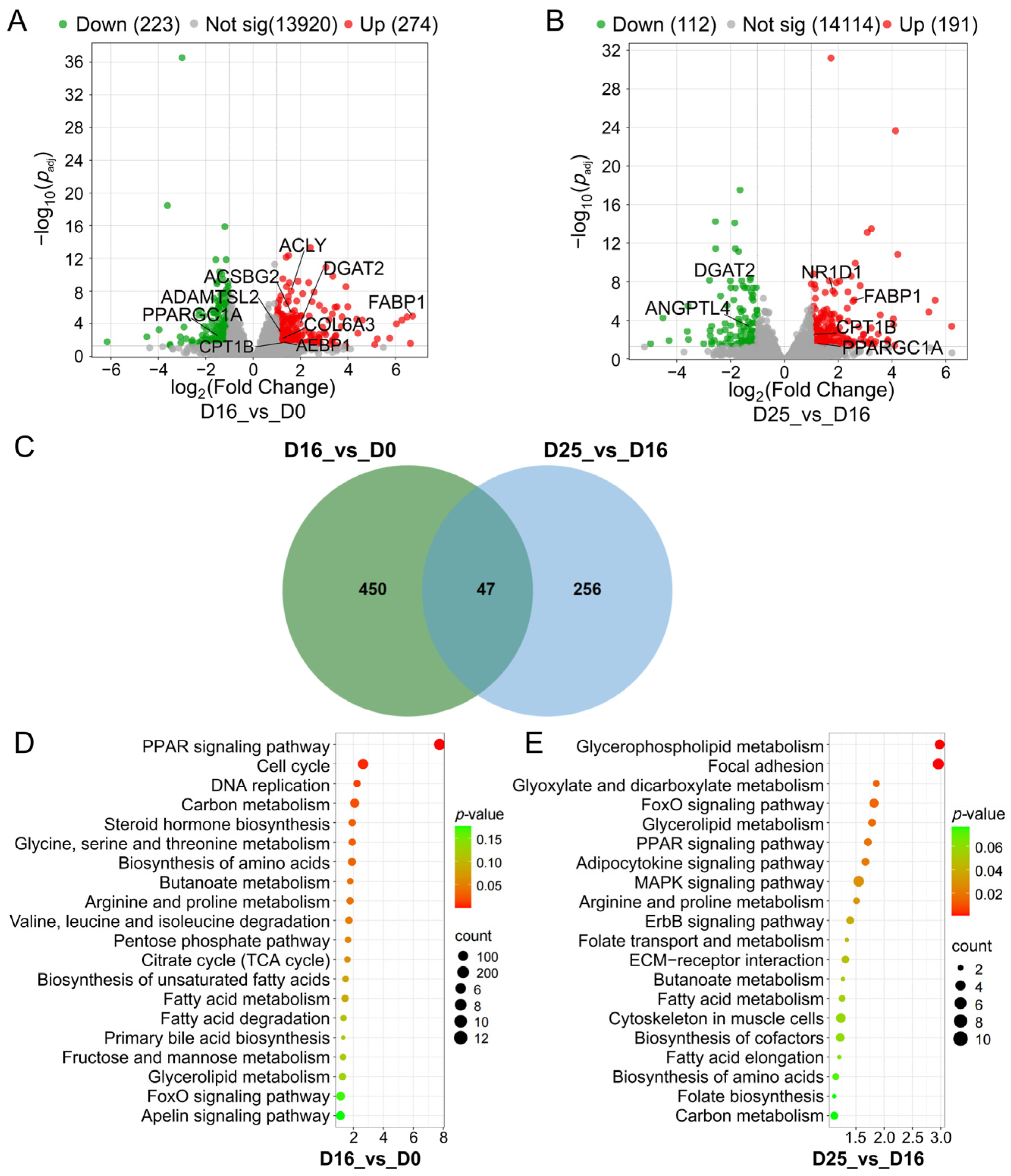
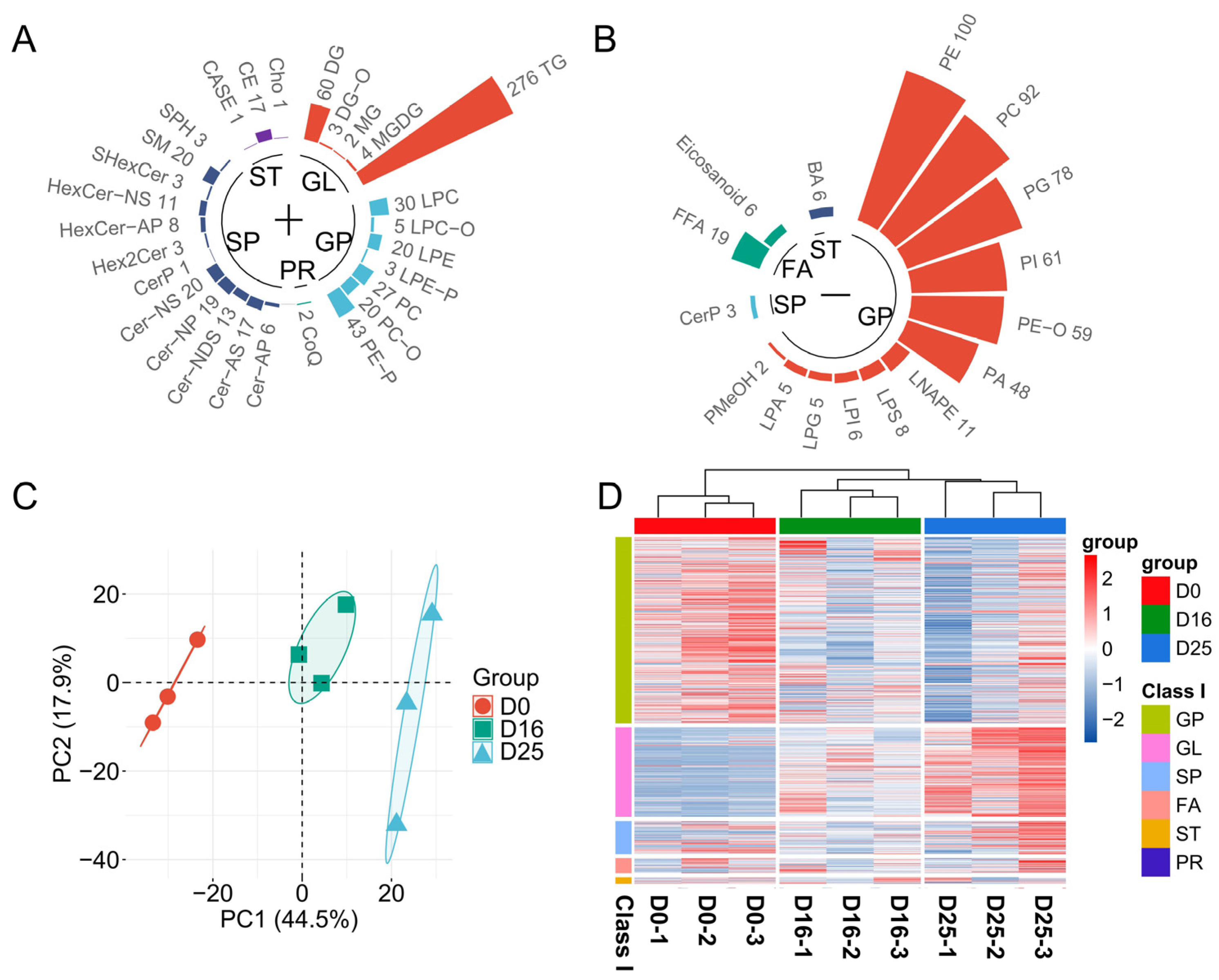
| Item | D0 | D16 | D25 | SEM | p-Value |
|---|---|---|---|---|---|
| Body weight (kg) | 4.09 b | 5.83 a | 5.51 a | 0.289 | <0.01 |
| Liver weight (g) | 183.83 c | 353.60 b | 502.45 a | 47.784 | <0.01 |
| Liver index (%) | 3.87 c | 6.08 b | 9.36 a | 0.873 | <0.01 |
| Moisture (%) | 64.58 a | 52.38 b | 39.85 c | 3.581 | <0.01 |
| Crude ash (%) | 1.28 a | 0.81 b | 0.52 c | 0.114 | <0.01 |
| Crude fat (%) | 5.52 c | 25.21 b | 43.85 a | 5.537 | <0.01 |
| Crude protein (%) | 15.96 a | 11.84 b | 9.35 b | 0.972 | <0.01 |
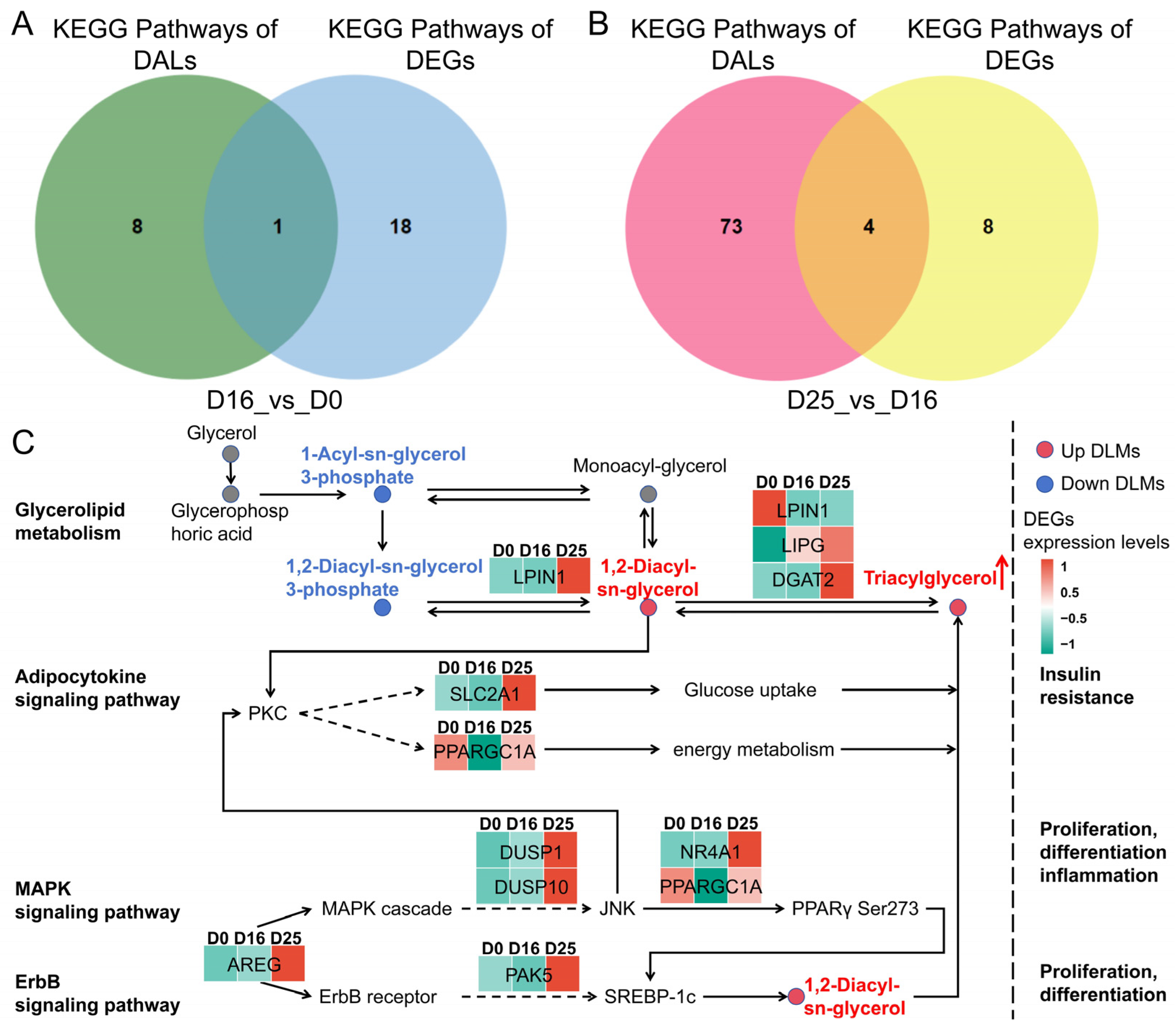
| Comparison | KEG ID | KEGG Term | KEGG Term | DLMs |
|---|---|---|---|---|
| D16 vs. D0 | ko00561 | ko00561 | ALDH7A1, DGAT2, LIPG, LPL ALDH7A1, DGAT2, LIPG, LPL | 1,2-Diacyl-sn-glycerol 3-phosphate, 1,2-Diacyl-sn-glycerol (Diglyceride), Triacylglycerol |
| D25 vs. D16 | ko00561 | Glycerolipid metabolism | DGAT2, DGKI, LPIN1, LPIN2 | 1-Acyl-sn-glycerol3-phosphate, 1,2-Diacyl-sn-glycerol (Diglyceride), Triacylglycerol |
| ko04920 | Adipocytokine signaling pathway | ACSBG2, NFKBIA, PPARGC1A, SLC2A1 | Diglyceride | |
| ko04010 | MAPK signaling pathway | AREG, DUSP1, DUSP10, DUSP16, EPHA2, FOS, IGF1, NR4A1, PGF | Diglyceride | |
| ko04012 | ErbB signaling pathway | AREG, CDKN1A, HBEGF, PAK5 | Diglyceride |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Q.; Bai, C.; Zhang, M.; Yue, B.; Zhang, J.; Kong, M.; Wang, B.; Wang, B.; Fan, W. Lipidomic and Transcriptomic Reveals Variations in Lipid Deposition During Goose Fatty Liver Formation. Biology 2025, 14, 1617. https://doi.org/10.3390/biology14111617
Zhang Q, Bai C, Zhang M, Yue B, Zhang J, Kong M, Wang B, Wang B, Fan W. Lipidomic and Transcriptomic Reveals Variations in Lipid Deposition During Goose Fatty Liver Formation. Biology. 2025; 14(11):1617. https://doi.org/10.3390/biology14111617
Chicago/Turabian StyleZhang, Qi, Chuning Bai, Mingai Zhang, Bin Yue, Jing Zhang, Min Kong, Binghan Wang, Baowei Wang, and Wenlei Fan. 2025. "Lipidomic and Transcriptomic Reveals Variations in Lipid Deposition During Goose Fatty Liver Formation" Biology 14, no. 11: 1617. https://doi.org/10.3390/biology14111617
APA StyleZhang, Q., Bai, C., Zhang, M., Yue, B., Zhang, J., Kong, M., Wang, B., Wang, B., & Fan, W. (2025). Lipidomic and Transcriptomic Reveals Variations in Lipid Deposition During Goose Fatty Liver Formation. Biology, 14(11), 1617. https://doi.org/10.3390/biology14111617






