Telomere-to-Telomere Gap-Free Genome Assembly and Comparative Analysis of the Opsariichthys bidens (Cypriniformes: Xenocyprididae)
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. DNA Extraction, Library Construction and Sequencing
2.3. Genome Assembly and Evaluation
2.4. Gene Prediction and Annotation
2.5. Genome Comparison and Identification of Newly Assembled Genes
2.6. Gene Family and Phylogenomic Analysis
2.7. Gene Positive Selection Analysis
3. Results and Discussion
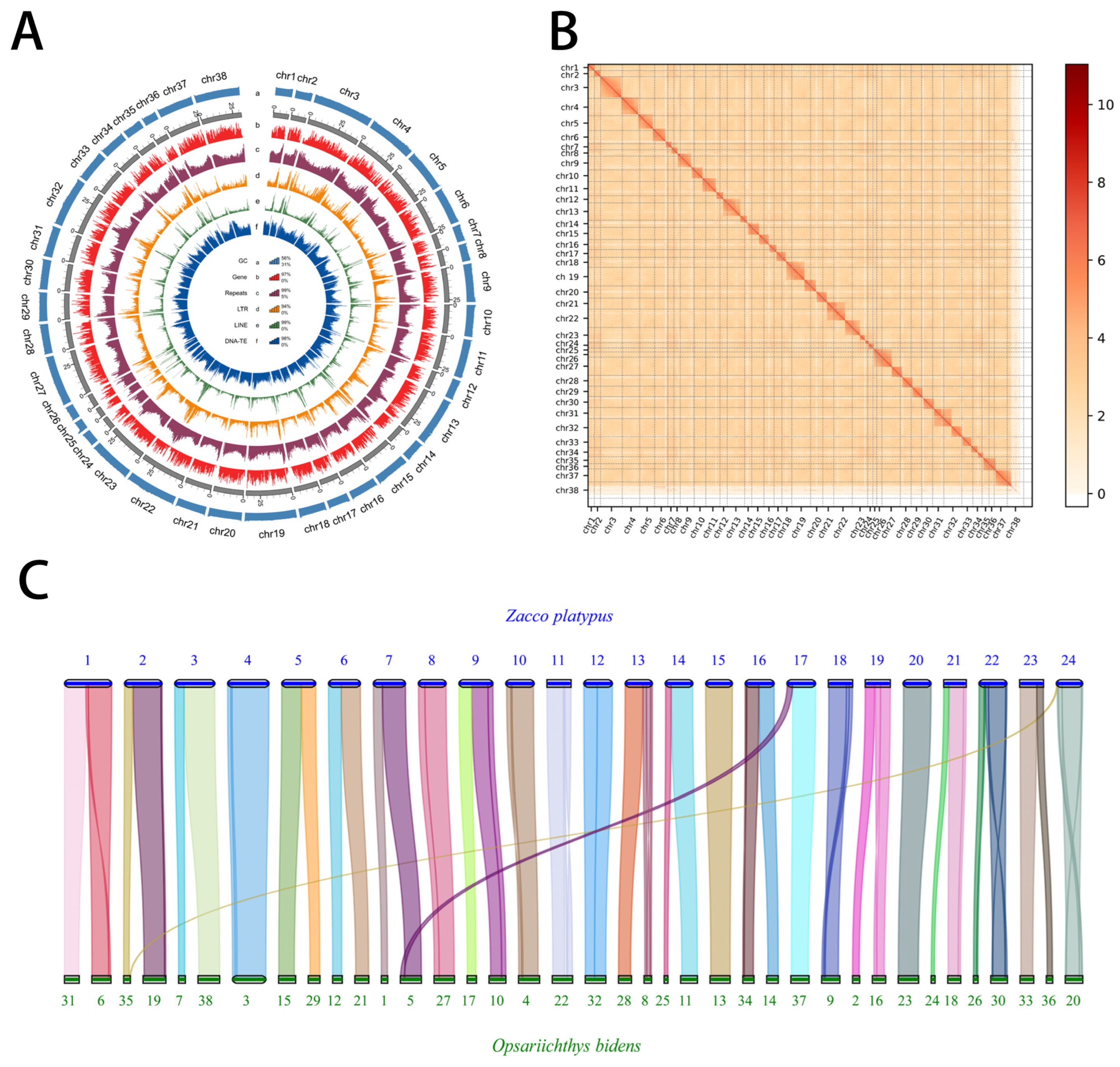
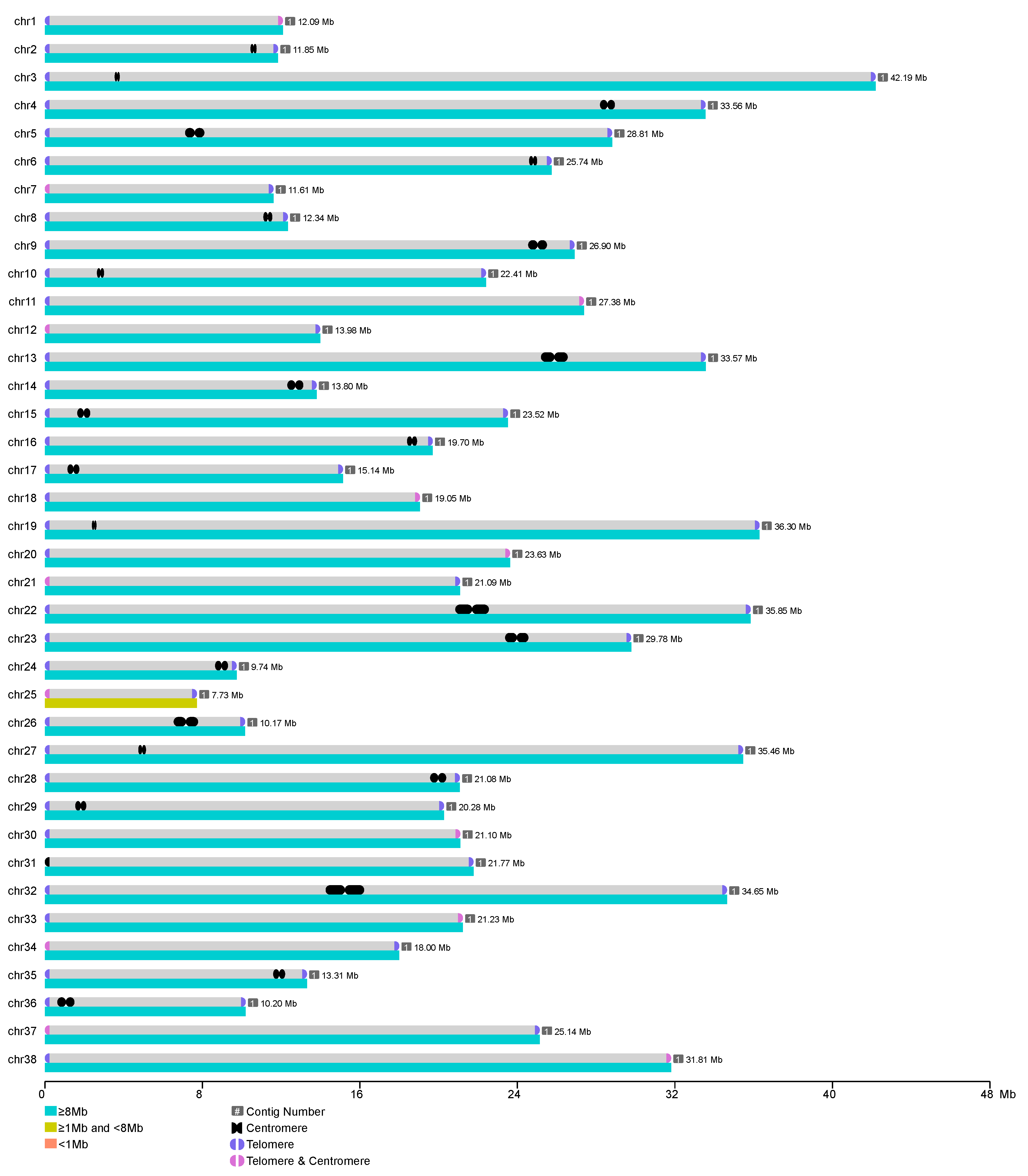
3.1. Genome Sequencing and Gap-Free Assembly
3.2. Genome Annotation and Repetitive Element Analysis
3.3. Comparative Genomics Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- He, S.P.; Chen, Y.Y.; Nakajima, T. Sequencing and Phylogenetic Analysis of Cytochrome b Gene in Low-Grade Cyprinidae Fishes from East Asia. Chin. Sci. Bull. 2000, 45, 2297–2302. [Google Scholar] [CrossRef]
- Betancur-R, R.; Wiley, E.O.; Arratia, G.; Acero, A.; Bailly, N.; Miya, M.; Lecointre, G.; Orti, G. Phylogenetic classification of bony fishes. BMC Evol. Biol. 2017, 17, 162. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.; Zhu, C.D. Taihu Lake Fish Fauna; Shanghai Science and Technology Press: Shanghai, China, 2005; ISBN 7-5323-7995-7. [Google Scholar]
- Poff, N.L. Landscape filters and species traits: Towards mechanistic understanding and prediction in stream ecology. J. N. Am. Benthol. Soc. 1997, 16, 391–409. [Google Scholar] [CrossRef]
- Murphy, M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2009, 417, 1–13. [Google Scholar] [CrossRef]
- Wilhelm, F.D. Reactive oxygen species, antioxidants and fish mitochondria. Front. Biosci. 2007, 1, 1229–1237. [Google Scholar] [CrossRef]
- Oufiero, C.E.; Whitlow, K.R. The evolution of phenotypic plasticity in fish swimming. Curr. Zool. 2016, 62, 475–488. [Google Scholar] [CrossRef]
- McGuigan, K.; Franklin, C.E.; Moritz, C.; Blows, M.W. Adaptation of rainbow fish to lake and stream habitats. Evolution 2003, 57, 104–118. [Google Scholar] [CrossRef]
- Liu, X.Y.; Chen, C.; Gong, W.Q.; Chen, K.J.; Li, Q.S.; Liu, P.; He, Y.L. Biological characteristics of Opsariichthys bidens in Anhua. J. Beijing Agric. Univ. 2015, 30, 49–52. [Google Scholar] [CrossRef]
- Su, J.X.; Qiu, F.X.; Zhou, W.X.; Xie, C.X.; Wang, W.M.; Chen, C.F. Age, growth, reproduction and feeding habits of Opsariichthys bidens in Sandaohe Reservoir. Fish. Water Conserv. 1993, 1, 15–18. [Google Scholar] [CrossRef]
- Jin, B.; Jin, K.W. Preliminary study on the biology of Opsariichthys bidens in Tiejia Reservoir. Fish Sci. 1985, 4, 8–12. [Google Scholar] [CrossRef]
- Perdices, A.; Sayanda, D.; Coelho, M.M. Mitochondrial diversity of Opsariichthys bidens (teleostei, cyprinidae) in three Chinese drainages. Mol. Phylogenet. Evol. 2005, 37, 920–927. [Google Scholar] [CrossRef]
- Deng, Y.; Wang, X.; Lin, P.C.; Liu, H.Z.; Wang, X.Z. Genetic divergence and population differentiation analysis of Opsariichthys bidens from Yiluo He. Acta Hydrobiol. Sin. 2020, 44, 339–345. [Google Scholar] [CrossRef]
- Chen, W.; Schmidt, B.V.; He, S. The potential colonization histories of Opsariichthys bidens (Cyprinidae) in China using bayesian binary MCMC analysis. Gene 2018, 15, 1–8. [Google Scholar] [CrossRef]
- Xu, X.J.; Guan, W.Z.; Niu, B.L.; Guo, D.D.; Xie, Q.P.; Zhan, W.; Yi, S.K.; Lou, B. Chromosome-level assembly of the Chinese hooksnout carp (Opsariichthys bidens) genome using PacBio sequencing and hi-C technology. Front. Genet. 2022, 12, 788547. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Gui, L.; Zhu, Y.F.; Xu, C.; Zhou, W.Z.; Li, M.Y. Chromosome-level assembly of male Opsariichthys bidens genome provides insights into the regulation of the GnRH signaling pathway and genome evolution. Biology 2022, 11, 1500. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.J.; Yu, J.Y.; Ge, J.H.; Yi, S.K.; Weng, X.D.; Guan, W.Z.; Niu, B.L.; Zhang, Z.H.; Lou, B. A male-specific insert of Opsariichthys bidens identified based on genome-wide association analyses and comparative genomics. Aquac. Rep. 2024, 35, 101982. [Google Scholar] [CrossRef]
- Xu, X.J.; Zhang, X.H.; Guan, W.Z.; Yu, J.Y.; Niu, B.L.; Yi, S.K.; Lou, B. Three genome assemblies of Opsariichthys bidens from yangzte river, pearl river and qiantang river basins. Sci. Data 2024, 11, 1110. [Google Scholar] [CrossRef]
- Chen, S.F.; Zhou, Y.Q.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Cheng, H.Y.; Concepcion, G.T.; Feng, X.W.; Zhang, H.W.; Li, H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods 2021, 18, 170–175. [Google Scholar] [CrossRef]
- Wingett, S.W.; Ewels, P.; Furlan-Magaril, M.; Nagano, T.; Schoenfelder, S.; Fraser, P.; Andrews, S. HiCUP: Pipeline for mapping and processing hi-C data. F1000Research 2015, 20, 1310. [Google Scholar] [CrossRef]
- Roach, M.J.; Schmidt, S.A.; Borneman, A.R. Purge haplotigs: Allelic contig reassignment for third-gen diploid genome assemblies. BMC Bioinform. 2018, 19, 460. [Google Scholar] [CrossRef]
- Dudchenko, O.; Batra, S.S.; Omer, A.D.; Nyquist, S.K.; Hoeger, M.; Durand, N.C.; Shamim, M.S.; Machol, I.; Lander, E.S.; Aiden, A.P.; et al. De novo assembly of the aedes aegypti genome using hi-C yields chromosome-length scaffolds. Science 2017, 356, 92–95. [Google Scholar] [CrossRef]
- Xu, M.Y.; Guo, L.D.; Gu, S.Q.; Wang, O.; Zhang, R.; Peters, B.A.; Fan, G.Y.; Liu, X.; Xu, X.; Deng, L.; et al. TGS-GapCloser: A fast and accurate gap closer for large genomes with low coverage of error-prone long reads. GigaScience 2020, 9, giaa094. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.Z.; Ye, C.; Li, X.Z.; Chen, Q.Y.; Wu, Y.; Zhang, F.; Pan, R.; Zhang, S.J.; Chen, S.X.; Wang, X.; et al. quarTeT: A telomere-to-telomere toolkit for gap-free genome assembly and centromeric repeat identification. Hortic. Res. 2023, 10, 127. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- Rhie, A.; Walenz, B.P.; Koren, S.; Phillippy, A.M. Merqury: Reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biol. 2020, 21, 245. [Google Scholar] [CrossRef]
- Manni, M.; Berkeley, M.R.; Seppey, M.; Simao, F.A.; Zdobnov, E.M. BUSCO update: Novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol. Biol. Evol. 2021, 38, 4647–4654. [Google Scholar] [CrossRef]
- Huang, N.; Li, H. miniBUSCO: A faster and more accurate reimplementation of BUSCO. bioRxiv 2023. [Google Scholar] [CrossRef]
- Tarailo-Graovac, M.; Chen, N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinform. 2009, 25, 4.10.1–4.10.14. [Google Scholar] [CrossRef]
- Jurka, J.; Kapitonov, V.V.; Pavlicek, A.; Klonowski, P.; Kohany, O.; Walichiewicz, J. Repbase update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005, 110, 462–467. [Google Scholar] [CrossRef] [PubMed]
- Ou, S.; Jiang, N. LTR_FINDER_parallel: Parallelization of LTR_FINDER enabling rapid identification of long terminal repeat retrotransposons. Mob. DNA 2019, 10, 48. [Google Scholar] [CrossRef] [PubMed]
- Flynn, J.M.; Hubley, R.; Goubert, C.; Rosen, J.; Clark, A.G.; Feschotte, C.; Smit, A.F. RepeatModeler2 for automated genomic discovery of transposable element families. Proc. Natl. Acad. Sci. USA 2020, 117, 9451–9457. [Google Scholar] [CrossRef] [PubMed]
- Stanke, M.; Keller, O.; Gunduz, I.; Hayes, A.; Waack, S.; Morgenstern, B. AUGUSTUS: Ab initio prediction of alternative transcripts. Nucleic Acids Res. 2006, 34, W435–W439. [Google Scholar] [CrossRef]
- Burge, C.; Karlin, S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 1997, 268, 78–94. [Google Scholar] [CrossRef]
- Birney, E.; Clamp, M.; Durbin, R. GeneWise and genomewise. Genome Res. 2004, 14, 988–995. [Google Scholar] [CrossRef]
- Shumate, A.; Salzberg, S.L. Liftoff: Accurate mapping of gene annotations. Bioinformatics 2021, 37, 1639–1643. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Holt, C.; Yandell, M. MAKER2: An annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinform. 2011, 12, 491. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Lu, S.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; DeWeese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; et al. CDD: A conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 2011, 39, D225–D229. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Reuter, K.; Drost, H.G. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods 2021, 18, 366–368. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.; Binns, D.; Chang, H.Y.; Fraser, M.; Li, W.Z.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res3 2013, 41, e121. [Google Scholar] [CrossRef]
- Griffiths-Jones, S. Rfam: Annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2004, 33, D121–D124. [Google Scholar] [CrossRef]
- Griffiths-Jones, S. miRBase: MicroRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006, 34, D140–D144. [Google Scholar] [CrossRef]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Goel, M.; Sun, H.; Jiao, W.B.; Schneeberger, K. SyRI: Finding genomic rearrangements and local sequence differences from whole—Genome assemblies. Genome Biol. 2019, 20, 277. [Google Scholar] [CrossRef]
- Li, L.; Stoeckert, C.J.; Roos, D.S. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003, 13, 2178–2189. [Google Scholar] [CrossRef]
- Nakamura, T.; Yamada, K.D.; Tomii, K.; Katoh, K. Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics 2018, 34, 2490–2492. [Google Scholar] [CrossRef]
- Wu, H.; Luo, L.Y.; Zhang, Y.H.; Zhang, C.Y.; Huang, J.H.; Mo, D.X.; Zhao, L.M.; Wang, Z.X.; Wang, Y.C.; He-Hua, E.; et al. Telomere-to-telomere genome assembly of a male goat reveals variants associated with cashmere traits. Nat. Commun. 2024, 15, 10041. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Mendes, F.K.; Vanderpool, D.; Fulton, B.; Hahn, M.W. CAFE 5 models variation in evolutionary rates among gene families. Bioinformatics 2021, 36, 5516–5518. [Google Scholar] [CrossRef] [PubMed]
- De, B.T.; Cristianini, N.; Demuth, J.P.; Hahn, M.W. CAFE: A computational tool for the study of gene family evolution. Bioinformatics 2006, 22, 1269–1271. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef] [PubMed]
- Nath, S.; Shaw, D.E.; White, M.A. Improved contiguity of the threespine stickleback genome using long-read sequencing. G3 Genes Genomes Genet. 2021, 11, jkab007. [Google Scholar] [CrossRef]
- Harvey, W.T.; Ebert, P.; Ebler, J.; Audano, P.A.; Munson, K.M.; Hoekzema, K.; Porubsky, D.; Beck, C.R.; Marschall, T.; Garimella, K.; et al. Whole-genome long-read sequencing downsampling and its effect on variant calling precision and recall. Genome Res. 2023, 33, 2029–2040. [Google Scholar] [CrossRef]
- Lu, L. Genome Assembly and Population History of Siniperca chuatsi and Siniperca knerii. Master’s Dissertation, Shanghai Ocean University, Shanghai, China, 2023. [Google Scholar] [CrossRef]
- Marshall, H.D.; Coulson, M.W.; Carr, S.M. Near neutrality, rate heterogeneity, and linkage govern mitochondrial genome evolution in atlantic cod (gadus morhua) and other gadine fish. Mol. Biol. Evol. 2008, 26, 579–589. [Google Scholar] [CrossRef][Green Version]
- Terekhanova, N.V.; Logacheva, M.D.; Penin, A.A.; Neretina, T.V.; Barmintseva, A.E.; Bazykin, G.A.; Kondrashov, A.S.; Mugue, N.S. Fast evolution from precast bricks: Genomics of young freshwater populations of threespine stickleback gasterosteus aculeatus. PLoS Genet. 2014, 10, e1004696. [Google Scholar] [CrossRef]
- Davidson, W.S.; Koop, B.F.; Jones, S.J.; Iturra, P.; Vidal, R.; Maass, A.; Jonassen, I.; Lien, S.; Omholt, S.W. Sequencing the genome of the Atlantic salmon (Salmo salar). Genome Biol. 2010, 11, 403. [Google Scholar] [CrossRef]
- Hemmer-Hansen, J.; Therkildsen, N.O.; Pujolar, J.M. Population genomics of marine fishes: Next-generation prospects and challenges. Biol. Bull. 2014, 227, 117–132. [Google Scholar] [CrossRef]
- Fox, J. Counter-gradient variation in gene expression between fish populations facilitates colonization of low-dissolved oxygen environments. Mol. Ecol. 2024, 33, e17419. [Google Scholar] [CrossRef]
- Zhang, J.J.; Wang, Y.B.; Wang, Q.; Zhang, X.H.; Liu, B.L. Transcriptomic analysis of muscle tissues of Larimichthys crocea with different flow resistance. Oceanol. Limnol. Sin. 2023, 54, 1507–1516. [Google Scholar] [CrossRef]
- Takle, H.; Castro, V. Swimming Physiology of Fish; Springer: Berlin/Heidelberg, Germany, 2013; pp. 257–274. ISBN 978-3-64-231048-5. [Google Scholar]
- Ma, Q.; Luo, Y.; Zhong, J.; Limbu, S.M.; Li, L.Y.; Chen, L.Q.; Qiao, F.; Zhang, M.L.; Lin, Q.; Du, Z.Y. Hypoxia tolerance in fish depends on catabolic preference between lipids and carbohydrates. Zool. Res. 2023, 44, 954–966. [Google Scholar] [CrossRef]
- Zhao, N.; Guo, H.B.; Jia, L.; Deng, Q.X.; Zhu, C.H.; Zhang, B. High-quality chromosome-level genome assembly of redlip mullet (Planiliza haematocheila). Zool. Res. 2022, 42, 796–799. [Google Scholar] [CrossRef]




| Contig Length (bp) | Contig Number | Scaffold Length (bp) | Scaffold Number | |
|---|---|---|---|---|
| N90 | 12,343,832 | 31 | 12,343,832 | 31 |
| N80 | 19,053,543 | 25 | 19,053,543 | 25 |
| N70 | 21,089,684 | 21 | 21,089,684 | 21 |
| N60 | 22,405,681 | 17 | 22,405,681 | 17 |
| N50 | 25,739,500 | 13 | 25,739,500 | 13 |
| Total length | 841,960,764 | NA | 841,960,764 | NA |
| Number (≥100bp) | NA | 38 | NA | 38 |
| Number (≥2kb) | NA | 38 | NA | 38 |
| Max length | 42,194,854 | NA | 42,194,854 | NA |
| Chromosome | Length (bp) | Number of Contigs | Number of Gaps | Number of Start Repeat Unit | Number of End Repeat Unit |
|---|---|---|---|---|---|
| chr1 | 12,090,187 | 1 | 0 | 1878 | 2521 |
| chr2 | 11,846,864 | 1 | 0 | 2173 | 478 |
| chr3 | 42,194,854 | 1 | 0 | 2308 | 1440 |
| chr4 | 33,560,156 | 1 | 0 | 2598 | 2236 |
| chr5 | 28,811,627 | 1 | 0 | 725 | 1437 |
| chr6 | 25,739,500 | 1 | 0 | 6251 | 23 |
| chr7 | 11,611,510 | 1 | 0 | 4712 | 1556 |
| chr8 | 12,343,832 | 1 | 0 | 1890 | 2057 |
| chr9 | 26,899,604 | 1 | 0 | 1771 | 1723 |
| chr10 | 22,405,681 | 1 | 0 | 1898 | 1442 |
| chr11 | 27,380,574 | 1 | 0 | 1375 | 2676 |
| chr12 | 13,984,826 | 1 | 0 | 1318 | 1118 |
| chr13 | 33,569,505 | 1 | 0 | 1953 | 1989 |
| chr14 | 13,802,948 | 1 | 0 | 1689 | 2244 |
| chr15 | 23,521,344 | 1 | 0 | 4962 | 1668 |
| chr16 | 19,700,785 | 1 | 0 | 3074 | 32 |
| chr17 | 15,140,455 | 1 | 0 | 1221 | 674 |
| chr18 | 19,053,543 | 1 | 0 | 1940 | 1682 |
| chr19 | 36,299,173 | 1 | 0 | 3630 | 1463 |
| chr20 | 23,626,312 | 1 | 0 | 2530 | 41 |
| chr21 | 21,089,684 | 1 | 0 | 33 | 1642 |
| chr22 | 35,847,689 | 1 | 0 | 1872 | 2997 |
| chr23 | 29,779,289 | 1 | 0 | 2608 | 3403 |
| chr24 | 9,738,410 | 1 | 0 | 1417 | 1527 |
| chr25 | 7,725,830 | 1 | 0 | 2161 | 1856 |
| chr26 | 10,171,860 | 1 | 0 | 963 | 1397 |
| chr27 | 35,461,429 | 1 | 0 | 2234 | 1577 |
| chr28 | 21,077,629 | 1 | 0 | 1723 | 1847 |
| chr29 | 20,275,876 | 1 | 0 | 45 | 2229 |
| chr30 | 21,102,698 | 1 | 0 | 3138 | 7 |
| chr31 | 21,769,779 | 1 | 0 | 0 | 1951 |
| chr32 | 34,646,360 | 1 | 0 | 1195 | 2618 |
| chr33 | 21,231,445 | 1 | 0 | 3229 | 1936 |
| chr34 | 18,000,321 | 1 | 0 | 9 | 3372 |
| chr35 | 13,313,169 | 1 | 0 | 1688 | 3629 |
| chr36 | 10,202,237 | 1 | 0 | 1736 | 1113 |
| chr37 | 25,135,100 | 1 | 0 | 78 | 1344 |
| chr38 | 31,808,679 | 1 | 0 | 1458 | 2 |
| Statistical Indicators | GCA_037194365.1 | GCA_037194315.1 | GCA_037194245.1 | GWHBEIO00000000-1 | GWHBEIO00000000-2 | This Study |
|---|---|---|---|---|---|---|
| Sex | male | female | female | female | male | female |
| Total size of assembled genome (Mb) | 852.41 | 843.11 | 840.94 | 818.78 Mb | 992.9 | 841.96 |
| Contig N50 | 9.01 | 2.9 | 5.27 | 4.66 | 5.2 | 25.74 |
| Contig N90 | NA | NA | NA | NA | NA | 12.34 |
| Number of Contigs | NA | NA | NA | 403 | 1373 | 38 |
| Scaffold N50 (Mb) | 21.01 | 23.62 | 24.75 | 25.29 | 19.44 | 25.74 |
| Scaffold N90 (Mb) | NA | NA | NA | NA | NA | 12.34 |
| Scaffold number | 228 | 450 | 98 | 39 | 38 | 38 |
| chromosome number | 37 | 38 | 39 | 39 | 38 | 38 |
| Number of gap-free chromosome | NA | NA | NA | NA | NA | 38 |
| Number of gaps | NA | NA | NA | NA | NA | 0 |
| Number of telomeres(pairs/single) | NA | NA | NA | NA | NA | 37/1 |
| TE size | 360.05 | 356.29 | 356.64 | 347.06 | 357.31 | 407.46 |
| GC content | 38.3 | 38.3 | 38.1 | NA | 37.9 | 38.4 |
| Chromosome anchoring ratio (%) | 90.39 | 95.67 | 99.01 | 95.66% | 89.31 | 98.09 |
| Total chromosome length (Mb) | 770.52 | 806.65 | 832.70 | 814.71 | 886.81 | 841.96 |
| Gene number | 26,556 | 25,036 | 26,283 | 23,992 | 36,738 | 29,816 |
| Functional proteins | 25,383 (95.58%) | 23,139 (92.42%) | 24,493 (93.19%) | 22,869 (95.4%) | 30,922 (84.17%) | 27,169 (91.12%) |
| BUSCO completeness genome (%) | 97.2 | 96.6 | 96.8 | 96.6 | 97.5 | 99.34 |
| BUSCO completeness (protein) | 91.7 | 91.8 | 94.5 | 93.5 | NA | 97.31 |
| DNA matching rate (%) | 98.78 | 99.05 | 98.98 | 98.76 | 99.5 | 99.94 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Liu, Q.; Yin, D.; Wang, P.; Jiang, M.; Liu, J.; Sun, N.; Yan, Y.; Liu, K. Telomere-to-Telomere Gap-Free Genome Assembly and Comparative Analysis of the Opsariichthys bidens (Cypriniformes: Xenocyprididae). Biology 2025, 14, 1544. https://doi.org/10.3390/biology14111544
Wang X, Liu Q, Yin D, Wang P, Jiang M, Liu J, Sun N, Yan Y, Liu K. Telomere-to-Telomere Gap-Free Genome Assembly and Comparative Analysis of the Opsariichthys bidens (Cypriniformes: Xenocyprididae). Biology. 2025; 14(11):1544. https://doi.org/10.3390/biology14111544
Chicago/Turabian StyleWang, Xinyue, Qi Liu, Denghua Yin, Pan Wang, Min Jiang, Jie Liu, Ning Sun, Yunzhi Yan, and Kai Liu. 2025. "Telomere-to-Telomere Gap-Free Genome Assembly and Comparative Analysis of the Opsariichthys bidens (Cypriniformes: Xenocyprididae)" Biology 14, no. 11: 1544. https://doi.org/10.3390/biology14111544
APA StyleWang, X., Liu, Q., Yin, D., Wang, P., Jiang, M., Liu, J., Sun, N., Yan, Y., & Liu, K. (2025). Telomere-to-Telomere Gap-Free Genome Assembly and Comparative Analysis of the Opsariichthys bidens (Cypriniformes: Xenocyprididae). Biology, 14(11), 1544. https://doi.org/10.3390/biology14111544






