Genome-Wide Identification and Analysis of Fruit Expression Patterns of the TCP Gene Family in Three Genera of Juglandaceae
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. Identification of the TCP Gene Family
2.2.2. Physicochemical Property Analysis of TCP Proteins
2.2.3. Phylogenetic Tree Construction and Visualization
2.2.4. Conserved Motif, Gene Structure and Promoter Analysis
2.2.5. Chromosomal Localization, Gene Duplication and Syntenic Analysis
2.2.6. Expression Pattern Analysis of TCP Genes
2.2.7. RNA Extraction and qRT-PCR Analysis
3. Results
3.1. Identification and Physicochemical Properties of TCP Gene Family
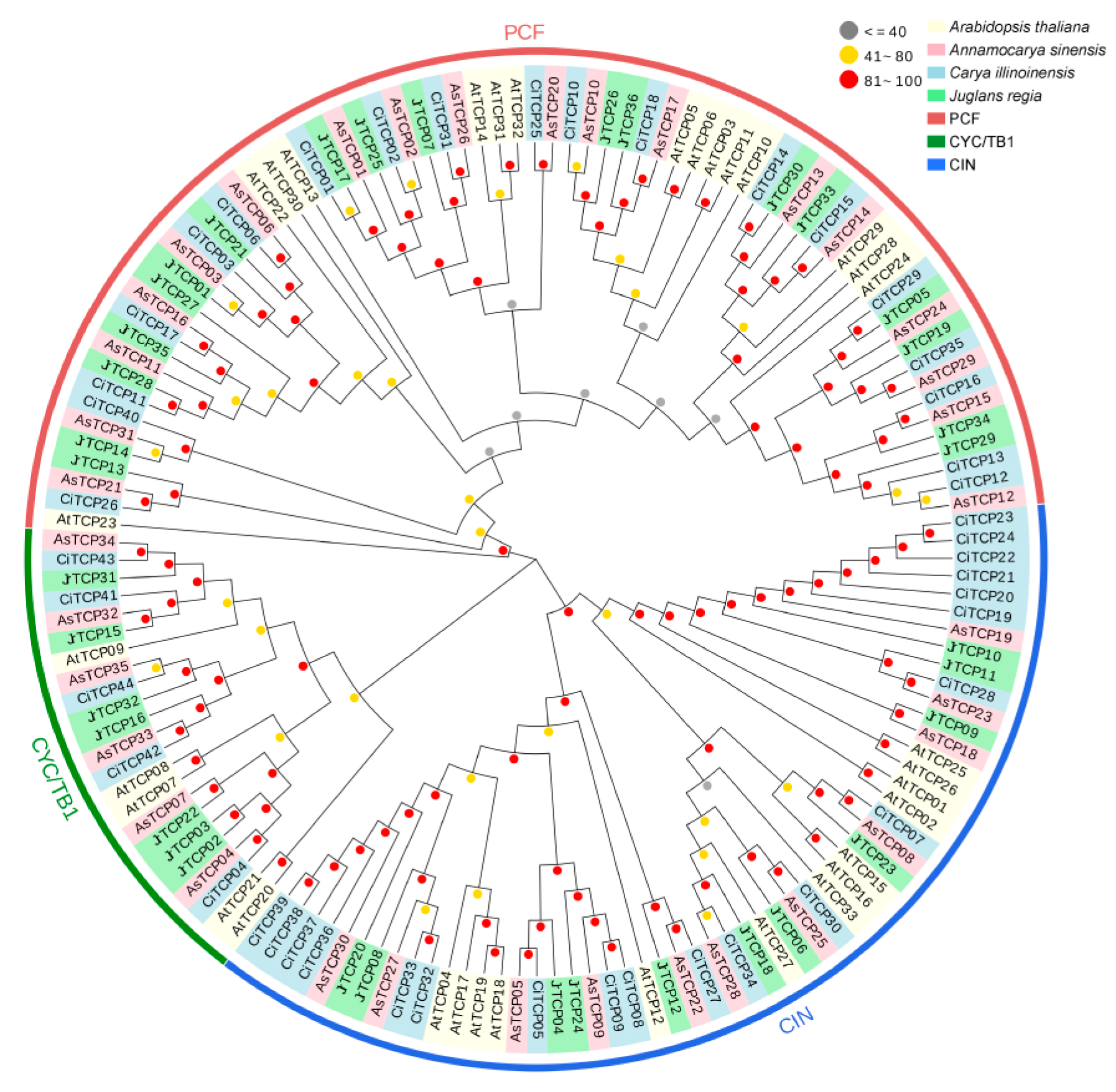
3.2. Phylogenetic Analysis of the TCP Gene Family
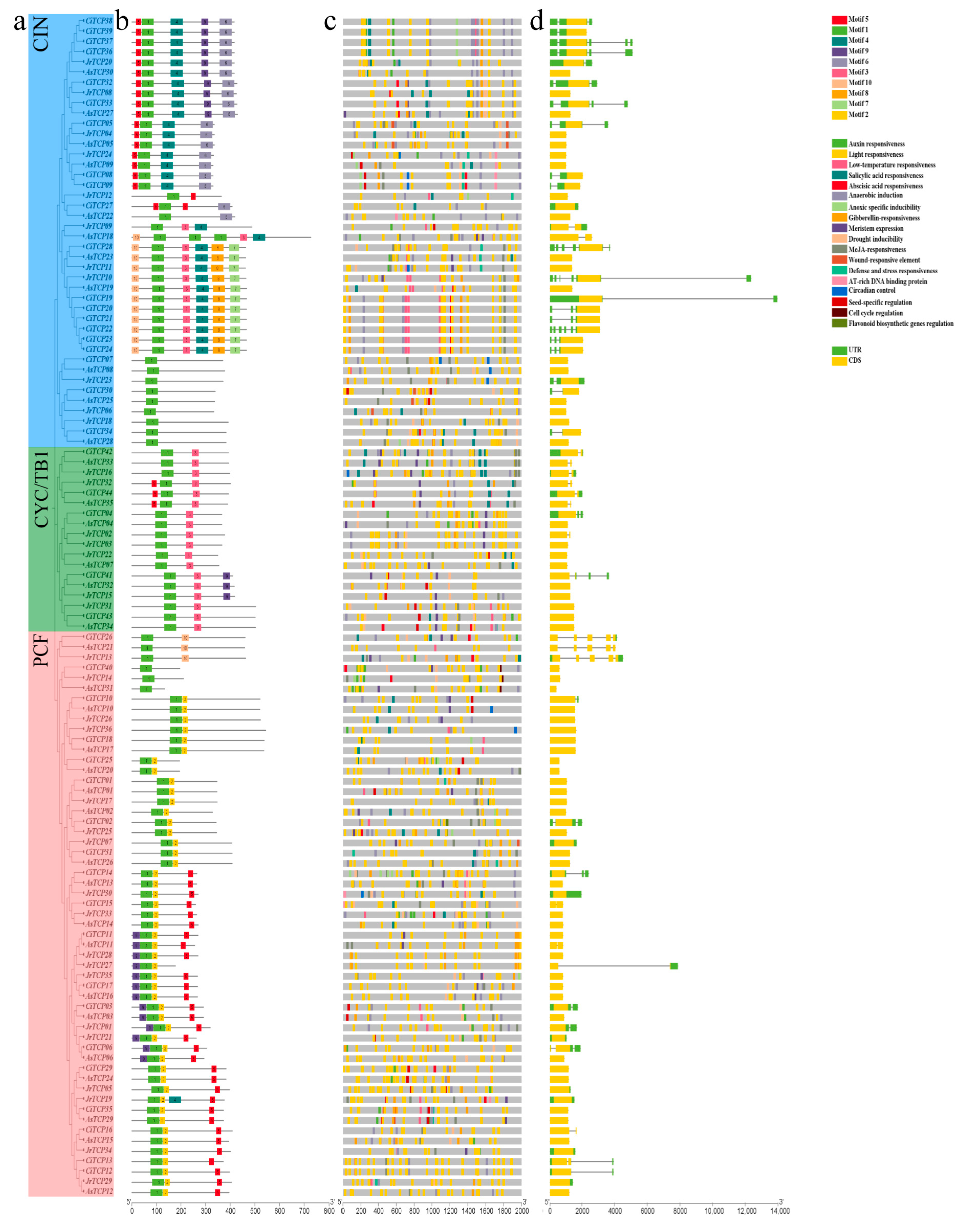
3.3. Conserved Motifs, Gene Structure, and Promoter Analysis of TCP Gene Family
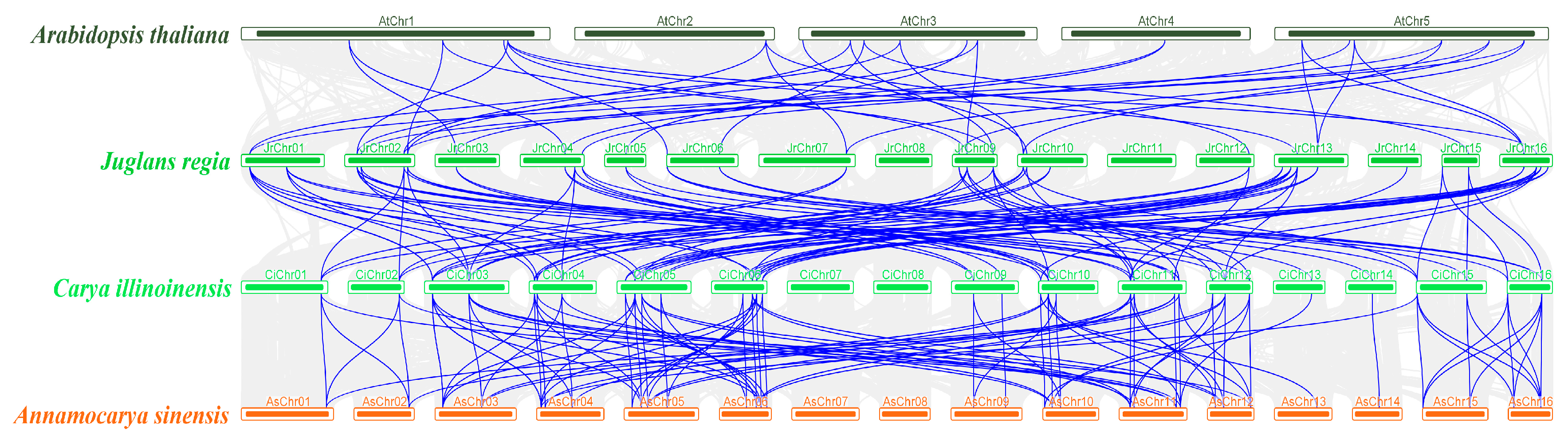
3.4. Chromosomal Localization and Gene Collinearity Analysis of TCP Genes
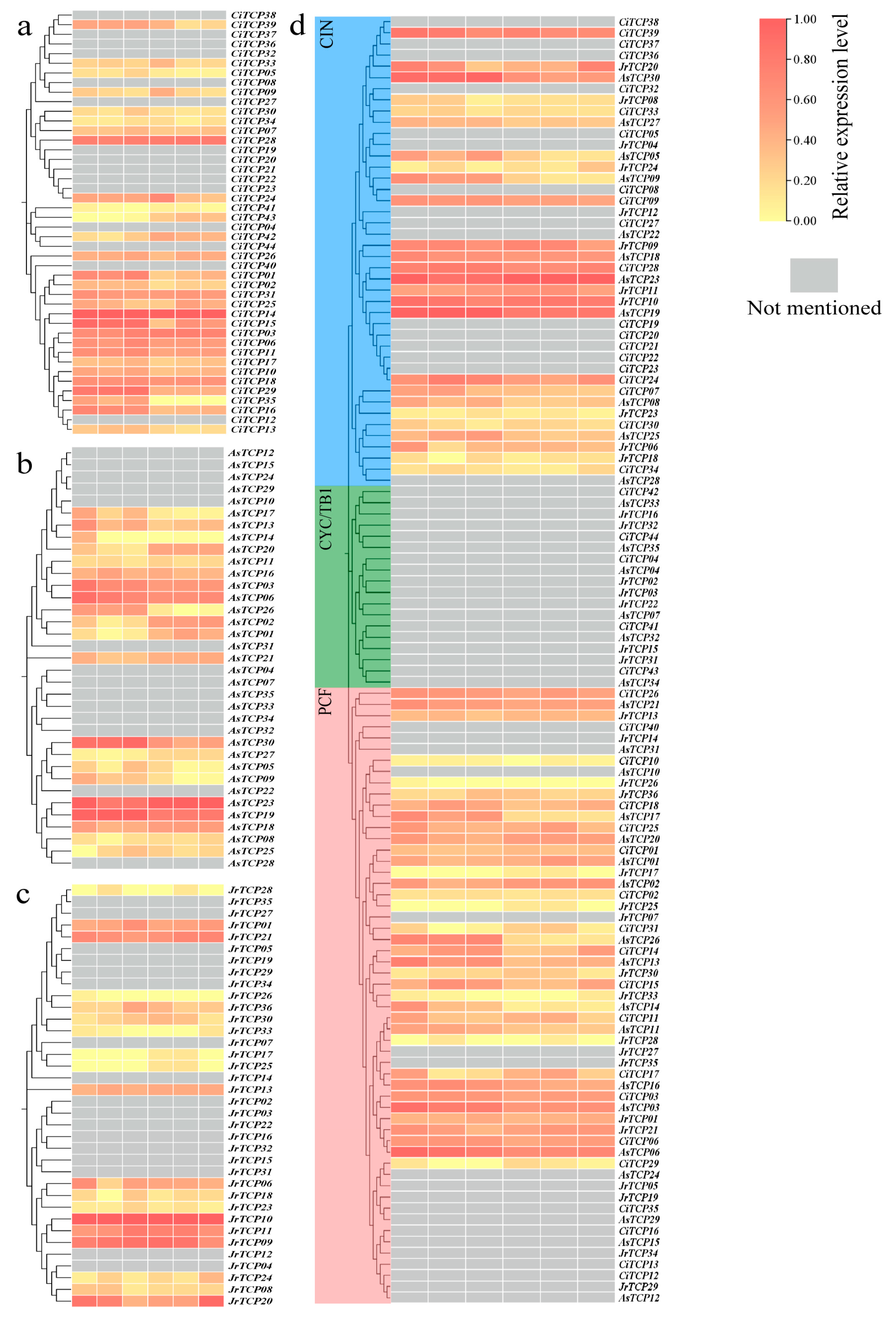
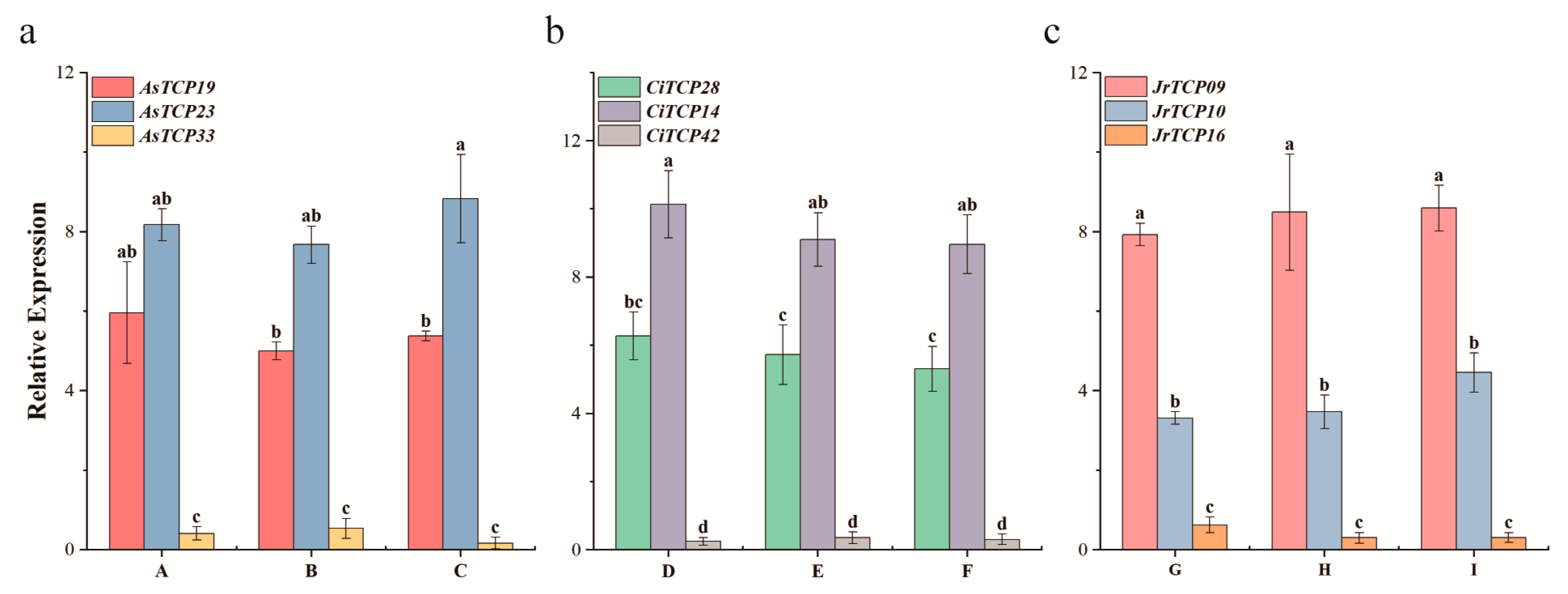
3.5. Expression Patterns of TCP Genes in Fruit
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| As | Annamocarya sinensis |
| Batch CD-search | Batch Conserved Domain Search |
| bp | Base Pair |
| cDNA | Complementary DNA |
| CDS | Coding Sequence |
| Chr | Chromosome |
| Ci | Carya illinoinensis |
| CIN | CINCINNATA |
| CYC/TB1 | Cycloidea/Teosinte Branched1 |
| Jr | Juglans regia |
| PCF | Proliferating Cell Nuclear Antigen Factors |
| qRT-PCR | Quantitative Real-Time PCR |
| MEME | Motif-BasedSequenceAnalysis Tools |
References
- Liu, Q.; Zhang, G.; Chen, S. The structure and regulatory role of plant transcription factors. Chin. Sci. Bull. 2000, 45, 1465–1474. [Google Scholar]
- Liu, Q.; Zhao, N.; Yamaguch-Shinozaki, K.; Shinozaki, K. The role of DREB transcription factors in improving the stress resistance of plants. Chin. Sci. Bull. 2000, 45, 11–16. [Google Scholar]
- Yang, Z.; Wang, X.; Li, X.; Yang, C. Research Progress of Transcription Factors in Higher Plants. Hereditas 2004, 26, 403–408. [Google Scholar]
- Da, L.; Carpenter, R.; Copsey, L.; Vincent, C.; Clark, J.; Coen, E. Control of Organ Asymmetry in Flowers of Antirrhinum. Cell 1999, 99, 367–376. [Google Scholar] [CrossRef]
- Doebley, J.; Stec, A.; Hubbard, L. The evolution of apical dominance in maize. Nature 1997, 386, 485–488. [Google Scholar] [CrossRef]
- Kosugi, S.; Ohashi, Y. PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell 1997, 9, 1607–1619. [Google Scholar]
- Liu, L.; Gao, H. Research Progress on the Family of TCP Genes. Biotechnol. Bull. 2016, 32, 14–22. [Google Scholar]
- Cubas, P.; Lauter, N.; Doebley, J.; Coen, E. The TCP domain: A motif found in proteins regulating plant growth and development. Plant J. 1999, 18, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Martín-Trillo, M.; Cubas, P. TCP genes: A family snapshot ten years later. Trends Plant Sci. 2010, 15, 31–39. [Google Scholar] [CrossRef]
- Zhou, Q.; Han, Y.; Zhu, Y.; Chen, S.; Li, X.; Peng, B.; Yuan, H. Genome-Wide Identification, Classification and Expression Analysis of TCP Gene Family in Tea Plant. Acta Hortic. Sin. 2019, 46, 2021–2036. [Google Scholar]
- Howarth, D.G.; Donoghue, M.J. Phylogenetic analysis of the “ECE” (CYC/TB1) clade reveals duplications predating the core eudicots. Proc. Natl. Acad. Sci. USA 2006, 103, 9101–9106. [Google Scholar] [CrossRef]
- Fenn, M.A.; Giovannoni, J.J. Phytohormones in fruit development and maturation. Plant J. 2021, 105, 446–458. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Martínez, J.A.; Poza-Carrión, C.; Cubas, P. Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 2007, 19, 458–472. [Google Scholar] [PubMed]
- Leng, X.; Wei, H.; Xu, X.; Ghuge, S.A.; Jia, D.; Liu, G.; Wang, Y.; Yuan, Y. Genome-wide identification and transcript analysis of TCP transcription factors in grapevine. BMC Genom. 2019, 20, 786. [Google Scholar] [CrossRef]
- Chen, C.; Zhang, Y.; Chen, Y.; Chen, H.; Gong, R. Sweet cherry TCP gene family analysis reveals potential functions of PavTCP1, PavTCP2 and PavTCP3 in fruit light responses. BMC Genom. 2024, 25, 3. [Google Scholar] [CrossRef] [PubMed]
- Gramzow, L.; Klupsch, K.; Fernández-Pozo, N.; Hölzer, M.; Marz, M.; Rensing, S.A.; Theißen, G. Comparative transcriptomics identifies candidate genes involved in the evolutionary transition from dehiscent to indehiscent fruits in Lepidium (Brassicaceae). BMC Plant Biol. 2022, 22, 340. [Google Scholar] [CrossRef]
- Zhou, H.; Hu, Y.; Ebrahimi, A.; Liu, P.; Woeste, K.; Zhao, P.; Zhang, S. Whole genome based insights into the phylogeny and evolution of the Juglandaceae. BMC Ecol. Evol. 2021, 21, 191. [Google Scholar] [CrossRef]
- Ma, H.; Zhang, W.E.; Pan, X.; Wei, X.; Huang, W. Walnut allelopathy and its application prospects: A review. Jiangsu Agric. Sci. 2019, 47, 57–63. [Google Scholar]
- Lin, R.; Li, R.; Lu, A.; Zhu, J.; Chen, Z. Comparative flower development of Juglans regia, Cyclocarya paliurus and Engelhardia spicata: Homology of floral envelopes in Juglandaceae. Bot. J. Linn. Soc. 2016, 181, 279–293. [Google Scholar] [CrossRef]
- Mu, X.; Tong, L.; Sun, M.; Zhu, Y.-X.; Wen, J.; Lin, Q.; Liu, B. Phylogeny and divergence time estimation of the walnut family (Juglandaceae) based on nuclear RAD-Seq and chloroplast genome data. Mol. Phylogenet. Evol. 2020, 147, 106802. [Google Scholar]
- Wan, W.; He, X.; Deng, L. Advances in breeding of Carya Nutt. South. Hortic. 2023, 34, 62–66. [Google Scholar]
- Ma, M.; Li, D. Research Progress on Chemical Constitutents and Pharmacological Actions of Plants of Genus Juglans. Guangzhou Chem. Ind. 2021, 49, 29–32. [Google Scholar]
- Tan, F.; Shi, Y.; Zou, R.; Jiang, Y.; Xiong, Z. Comparative Analysis of Nutritional Components of Annamocarya Sinensis and Juglans Regia. Food Sci. Technol. 2020, 45, 93–97. [Google Scholar]
- Xu, T.; Geng, S. Comprehensive evaluation and analysis of oil quality of 9 Pecan varieties. China Oils Fats 2023, 1–11. [Google Scholar] [CrossRef]
- Morales-de la Peña, M.; Rábago-Panduro, L.M.; Martín-Belloso, O.; Welti-Chanes, J. Challenges and Benefits of Using Pecan Kernels, Derivatives, and Byproducts as Alternative Ingredients in Food Product Development. Food Rev. Int. 2023, 39, 2530–2542. [Google Scholar] [CrossRef]
- Li, M.; Liu, Y.; Sun, C.; Meng, Y.; Yang, K.; Hou, L.; Wang, J. Research Advance about Nutrients and Medicinal Value of Walnut. J. Chin. Cereals Oils 2009, 24, 166–170. [Google Scholar]
- Ji, Y.; Zhang, W.; Li, D.; Shen, L. The complete chloroplast genome sequence of Annamocarya sinensis (Juglandaceae), an Endangered species endemic to Yunnan Province, China. Mitochondrial DNA B Resour. 2020, 5, 2021–2023. [Google Scholar] [CrossRef]
- Sun, S.; Liu, J.; Wu, X.; Zhang, G.; Zong, D.; Li, D. Population dynamics and soil nutrient characteristics of the endangered Annamocarya sinensis in Yunnan, China. BMC Plant Biol. 2025, 25, 689. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Xiao, L.; Zhang, Z.; Zhang, R.; Wang, Z.; Huang, C.; Huang, R.; Luan, Y.; Fan, T.; Wang, J.; et al. The genomes of pecan and Chinese hickory provide insights into Carya evolution and nut nutrition. GigaScience 2019, 8, giz036. [Google Scholar] [CrossRef]
- Stevens, K.A.; Woeste, K.; Chakraborty, S.; Crepeau, M.W.; Leslie, C.A.; Martínez-García, P.J.; Puiu, D.; Romero-Severson, J.; Coggeshall, M.; Dandekar, A.M.; et al. Genomic Variation Among and Within Six Juglans Species. G3 Genes|Genomes|Genet. 2018, 8, 2153–2165. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; He, W.; Wang, L.; Lan, Y.; Wu, M. TCP transcription factor identification in pecan (Carya illinoensis) and salt tolerance function analysis of CiTCP8. Sci. Hortic. 2024, 330, 113051. [Google Scholar] [CrossRef]
- Shad, M.A.; Wu, S.; Rao, M.J.; Luo, X.; Huang, X.; Wu, Y.; Zhou, Y.; Wang, L.; Ma, C.; Hu, L. Evolution and Functional Dynamics of TCP Transcription Factor Gene Family in Passion Fruit (Passiflora edulis). Plants 2024, 13, 2568. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A.; et al. The conserved domain database in 2023. Nucleic Acids Res. 2023, 51, D384–D388. [Google Scholar] [CrossRef]
- Subramanian, B.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.-H. Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 2019, 47, W270–W275. [Google Scholar] [CrossRef] [PubMed]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.-Z.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis Transcription Factors: Genome-Wide Comparative Analysis Among Eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef] [PubMed]
- Gull, S.; Uddin, S.; Hussain, H.A.; Wang, S.; Bayar, J.; Liu, J. Genome-wide analysis reveals the TCP-miR159-miR319 module is crucial for rice (Oryza sativa L.) growth and response to drought and salinity. Plant Stress 2023, 10, 100215. [Google Scholar] [CrossRef]
- Jiu, S.; Xu, Y.; Wang, J.; Wang, L.; Wang, S.; Ma, C.; Guan, L.; Abdullah, M.; Zhao, M.; Xu, W.; et al. Genome-Wide Identification, Characterization, and Transcript Analysis of the TCP Transcription Factors in Vitis vinifera. Front. Genet. 2019, 10, 1276. [Google Scholar] [CrossRef]
- Ma, X.; Ma, J.; Fan, D.; Li, C.; Jiang, Y.; Luo, K. Genome-wide Identification of TCP Family Transcription Factors from Populus euphratica and Their Involvement in Leaf Shape Regulation. Sci. Rep. 2016, 6, 32795. [Google Scholar] [CrossRef]
- Xu, H.; Wang, N.; Xiang, Y.; Sheng, Q.; Xu, Y.; Pei, D.; Wang, H. Genome assembly and comparative analysis reveal the imbalanced subgenomes divergence and evolutionary history of Juglans cathayensis. Plant J. 2025, 122, e70252. [Google Scholar] [CrossRef]
- Ji, F.; Ma, Q.; Zhang, W.; Liu, J.; Feng, Y.; Zhao, P.; Song, X.; Chen, J.; Zhang, J.; Wei, X.; et al. A genome variation map provides insights into the genetics of walnut adaptation and agronomic traits. Genome Biol. 2021, 22, 300. [Google Scholar] [CrossRef]
- Zhou, H.; Hwarari, D.; Ma, H.; Xu, H.; Yang, L.; Luo, Y. Genomic survey of TCP transcription factors in plants: Phylogenomics, evolution and their biology. Front. Genet. 2022, 13, 1060546. [Google Scholar] [CrossRef] [PubMed]
- Mi, Y.; Tian, Y.; Yang, H.; Deng, Y.; Sun, Y.; Li, Y. Identification and Expression Analysis of TCP Gene Family in Robinia pseudoacacia. Mol. Plant Breed. 2023, 1–13. Available online: https://kns.cnki.net/kcms/detail/46.1068.S.20230505.1238.008.html (accessed on 15 May 2025).
- Chen, Z.; Qiao, Z.; Li, J.; Zhang, X.; Ma, S.; He, C.; Zong, D. Genome-wide Identification and Analysis of the TCP Gene Family in Populus yunnanensis. Biotechnol. Bull. 2024, 40, 214–226. [Google Scholar]
- Li, X.; Li, S.; Yao, L. Bioinformatics and expression analysis of TCP gene family in Punica granatum L. Jiangsu Agric. Sci. 2024, 52, 41–48. [Google Scholar]
- Zhang, B.; Liu, G.; Tian, J.; Wang, D. Genome-wide identification and expression analysis of TCP gene family in Docynia delavayi(Franch.) Schneid. Chin. J. Biotechnol. 2025, 41, 809–824. [Google Scholar]
- González-Grandío, E.; Cubas, P. Chapter 9—TCP Transcription Factors: Evolution, Structure, and Biochemical Function. In Plant Transcription Factors; Gonzalez, D.H., Ed.; Academic Press: Boston, MA, USA, 2016; pp. 139–151. [Google Scholar]
- Chen, Y.-T.; Su, W.-Y.; Wu, W.; Zhu, Q.-G.; Yin, X.-R.; Sun, N.-J.; Liu, X.-F. Transcriptional responses of postharvest ABA treatment in Persimmon fruit. Postharvest Biol. Technol. 2024, 217, 113097. [Google Scholar] [CrossRef]
- Lan, J.; Qin, G. The molecular function and regulation of Class II TCP transcription factors. Sci. Sin. Vitae 2021, 51, 1542–1557. [Google Scholar] [CrossRef]
- Danisman, S. TCP Transcription Factors at the Interface between Environmental Challenges and the Plant’s Growth Responses. Front. Plant Sci. 2016, 7, 1930. [Google Scholar] [CrossRef]

| Species | Gene Number | Number of Amino Acid | Molecular Weight (kDa) | Theoretical pI | Instability Index | Grand Average of Hydropathicity | Transmembrane- Domain Number |
|---|---|---|---|---|---|---|---|
| Carya illinoinensis | 44 | 537~193 | 56.2~20.9 | 9.8~5.9 | 71.8~38.3 | −0.17~−0.87 | 0 |
| Juglans regia | 36 | 543~177 | 57.0~18.8 | 10.1~5.5 | 67.0~38.5 | −0.20~−0.88 | 0~2 |
| Annamocarya sinensis | 35 | 727~133 | 79.4~14.0 | 10.4~5.8 | 77.5~39.7 | −0.16~−0.86 | 0~1 |
| Total | 115 | 727~133 | 79.4~14.0 | 10.4~5.5 | 77.5~38.3 | −0.16~−0.88 | 0~3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, S.; Wu, X.; Liu, J.; Zhang, Y.; Shi, R.; Li, D. Genome-Wide Identification and Analysis of Fruit Expression Patterns of the TCP Gene Family in Three Genera of Juglandaceae. Biology 2025, 14, 1529. https://doi.org/10.3390/biology14111529
Sun S, Wu X, Liu J, Zhang Y, Shi R, Li D. Genome-Wide Identification and Analysis of Fruit Expression Patterns of the TCP Gene Family in Three Genera of Juglandaceae. Biology. 2025; 14(11):1529. https://doi.org/10.3390/biology14111529
Chicago/Turabian StyleSun, Shengjie, Xiaodong Wu, Jiaole Liu, Yinlong Zhang, Rui Shi, and Dan Li. 2025. "Genome-Wide Identification and Analysis of Fruit Expression Patterns of the TCP Gene Family in Three Genera of Juglandaceae" Biology 14, no. 11: 1529. https://doi.org/10.3390/biology14111529
APA StyleSun, S., Wu, X., Liu, J., Zhang, Y., Shi, R., & Li, D. (2025). Genome-Wide Identification and Analysis of Fruit Expression Patterns of the TCP Gene Family in Three Genera of Juglandaceae. Biology, 14(11), 1529. https://doi.org/10.3390/biology14111529






