Genome-Wide SNP Analysis Reveals the Unique Genetic Diversity Represented by Fat-Tailed Coarse-Wooled Sheep Breeds of Kazakhstan
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Sampling and Data Collection
2.2. DNA Isolation, Genome-Wide SNP Genotyping and SNP Data Filtering
2.3. Genetic Diversity and Phylogenetic Analyses
2.4. Population Structure and Demographic Analyses
3. Results
3.1. Genome-Wide SNP Genotyping Data Assessment
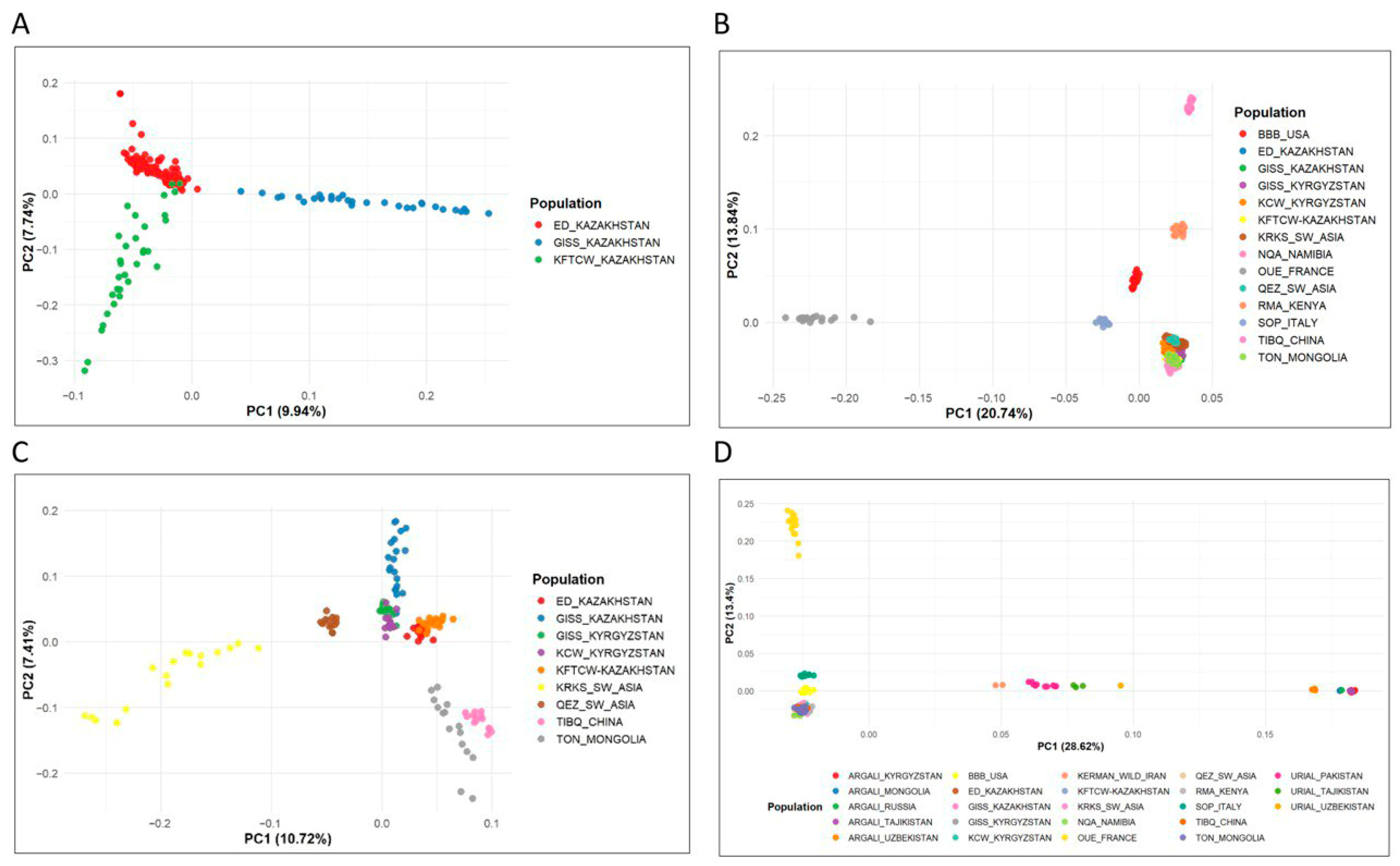
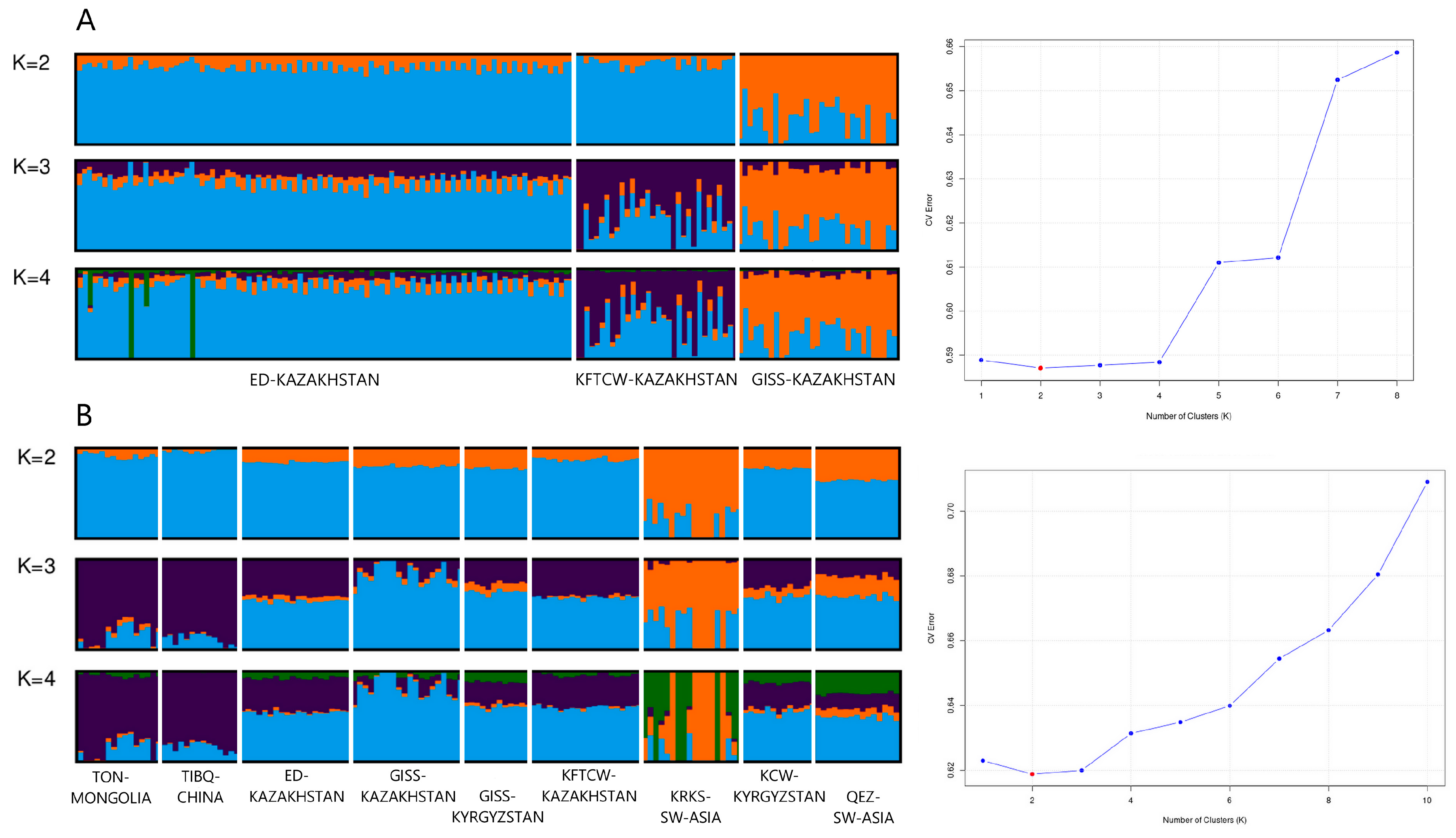
3.2. Genetic Diversity and Population Structure Analyses
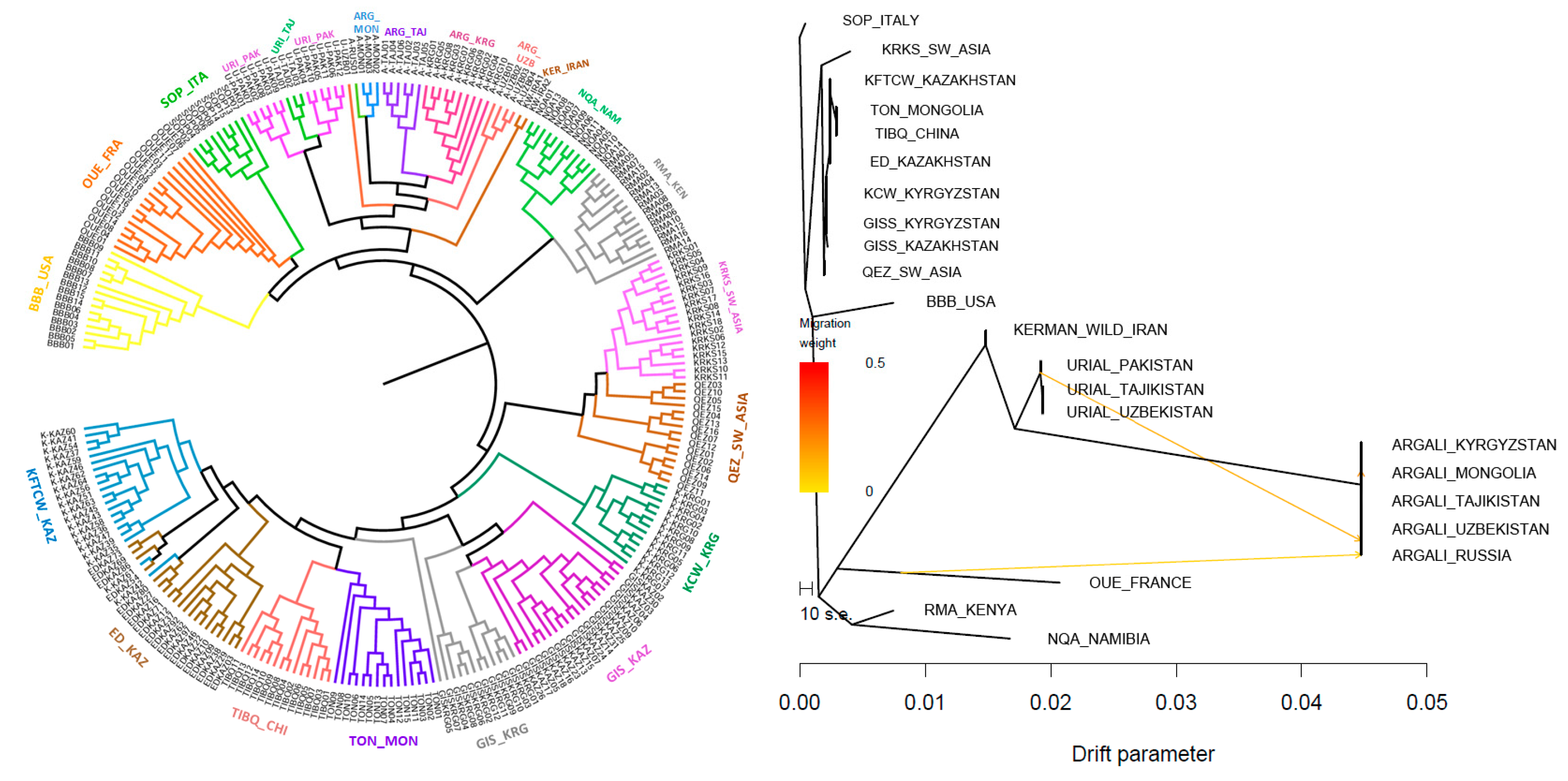
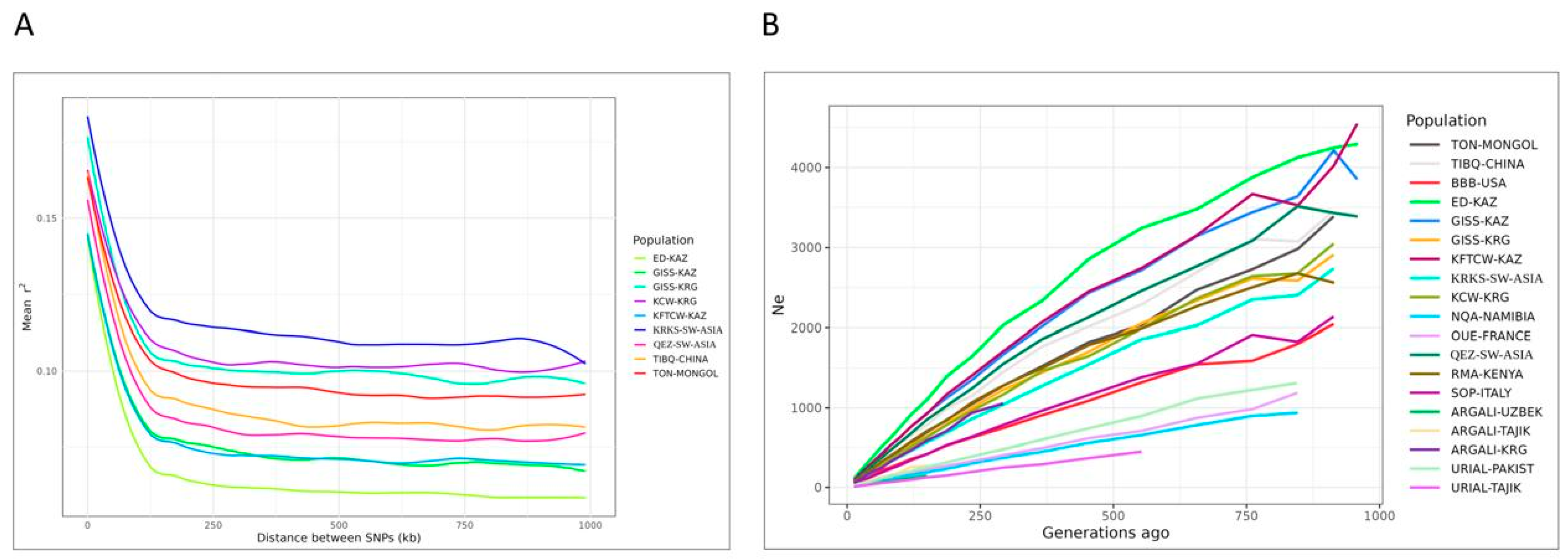
3.3. Phylogenetics and Demographic Analyses, and Breed Differentiation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| SNP | Single Nucleotide Polymorphism |
| RSE | Republican State Enterprise |
| EDTA | Ethylenediaminetetraacetic acid |
| PCA | Principal Component Analysis |
| LD | Linkage Disequilibrium |
References
- Kassenbayev, G.; Kerimova, U.; Rakhimzhanova, G.; Shalgimbayeva, K. Animal husbandry market in Kazakhstan: Dynamics and prognosis. Sci. Horiz. 2024, 27, 176–188. [Google Scholar] [CrossRef]
- Byuro Natsional’noy Statistiki (Office of National Statistics). Available online: https://stat.gov.kz/ru/industries/business-statistics/stat-forrest-village-hunt-fish/publications/287928/ (accessed on 22 July 2025).
- Zhumadillayev, N.K. Dissertation on the Topic “Scientific and Practical Justification for Improving Productive Traits and Creating New Genotypes of Fat-Tailed Sheep in Various Regions of Kazakhstan” (Nauchno-Prakticheskoe Obosnovanie Povysheniya Produktivnykh Kachestv i Sozdaniya Novykh Genotipov Kurdyuchnykh Ovets v Razlichnykh Regionakh Kazakhstana). 2025. Available online: https://mbook.kz/media/360/items/58/index.html (accessed on 23 July 2025).
- Isaeva, D.; Korotkevich, O. Characteristics of the Edilbay sheep breed of the Republic of Kazakhstan. Bull. NSAU (Novosib. State Agrar. Univ.) 2022, 157–163. [Google Scholar] [CrossRef]
- Katasheva, A.; Iskakov, K.; Kulataev, B.; Abdramanov, A.; Sattorov, S. Improving the efficiency of the production of mutton of kazakh fat-tailed coarse-wool sheep. Izdenister Natigeler 2024, 2, 55–62. [Google Scholar] [CrossRef]
- Zhumadillaev, N. Introductory crossing in the meat-fat sheep production of western Kazakhstan. Sheep Goats Woolen Bus. 2021, 4, 7–12. [Google Scholar] [CrossRef]
- Rákossy, Z.; Petrescu-Mag, I.V.; Kovacs, E.; Gavriloaie, C.; Coroian, C.O.; Păpuc, T.; Ungureanu, T.; Botha, M. Black wooled sheep breeds of the world, I. ABAH Bioflux 2019, 11, 69–78. [Google Scholar]
- Deniskova, T.; Dotsev, A.; Lushihina, E.; Shakhin, A.; Kunz, E.; Medugorac, I.; Reyer, H.; Wimmers, K.; Khayatzadeh, N.; Sölkner, J.; et al. Population Structure and Genetic Diversity of Sheep Breeds in the Kyrgyzstan. Front. Genet. 2019, 10, 1311. [Google Scholar] [CrossRef]
- Hiendleder, S.; Kaupe, B.; Wassmuth, R.; Janke, A. Molecular analysis of wild and domestic sheep questions current nomenclature and provides evidence for domestication from two different subspecies. Proc. Biol. Sci. 2002, 269, 893–904. [Google Scholar] [CrossRef]
- Zeder, M.A. Domestication and early agriculture in the Mediterranean Basin: Origins, diffusion, and impact. Proc. Natl. Acad. Sci. USA 2008, 105, 11597–11604. [Google Scholar] [CrossRef]
- Peters, J.; Helmer, D.; Von Den Driesch, A.; Saña Segui, M. Early animal husbandry in the Northern Levant. Paléorient 1999, 25, 27–48. [Google Scholar] [CrossRef]
- Hiendleder, S.; Mainz, K.; Plante, Y.; Lewalski, H. Analysis of mitochondrial DNA indicates that domestic sheep are derived from two different ancestral maternal sources: No evidence for contributions from urial and argali sheep. J. Hered. 1998, 89, 113–120. [Google Scholar] [CrossRef]
- Larsen, P.F.; Demontis, D.; Nielsen, V.H.; Thirstrup, J.P.; Loeschcke, V.; Pertoldi, C. The Transferability of Illumina Canine BeadChip Single-Nucleotide Polymorphisms (SNPs) to American Mink (Neovison vison). Biochem. Genet. 2012, 50, 717–721. [Google Scholar] [CrossRef]
- Haynes, G.D.; Latch, E.K. Identification of novel single nucleotide polymorphisms (SNPs) in deer (Odocoileus spp.) using the BovineSNP50 BeadChip. PLoS ONE 2012, 7, e36536. [Google Scholar] [CrossRef]
- More, M.; Gutiérrez, G.; Rothschild, M.; Bertolini, F.; Abel, F. Evaluation of SNP Genotyping in Alpacas Using the Bovine HD Genotyping Beadchip. Front. Genet. 2019, 10, 433129. [Google Scholar] [CrossRef]
- Kijas, J.W.; Lenstra, J.A.; Hayes, B.; Boitard, S.; Porto Neto, L.R.; San Cristobal, M.; Servin, B.; McCulloch, R.; Whan, V.; Gietzen, K.; et al. Genome-Wide Analysis of the World’s Sheep Breeds Reveals High Levels of Historic Mixture and Strong Recent Selection. PLoS Biol. 2012, 10, e1001258. [Google Scholar] [CrossRef]
- Dossybayev, K.; Orazymbetova, Z.; Mussayeva, A.; Saitou, N.; Zhapbasov, R.; Makhatov, B.; Bekmanov, B. Genetic diversity of different breeds of Kazakh sheep using microsatellite analysis. Arch. Anim. Breed. 2019, 62, 305–312. [Google Scholar] [CrossRef]
- Mukhametzharova, I.; Islamov, Y.; Shauyenov, S.; Ibrayev, D.; Atavliyeva, S.; Tarlykov, P. Genetic Characterization of Kazakh Native Sheep Breeds Using Mitochondrial DNA. OnLine J. Biol. Sci. 2018, 18, 341–348. [Google Scholar] [CrossRef]
- Amandykova, M.; Akhatayeva, Z.; Kozhakhmet, A.; Kapassuly, T.; Orazymbetova, Z.; Yergali, K.; Khamzin, K.; Iskakov, K.; Dossybayev, K. Distribution of Runs of Homozygosity and Their Relationship with Candidate Genes for Productivity in Kazakh Meat–Wool Sheep Breed. Genes 2023, 14, 1988. [Google Scholar] [CrossRef] [PubMed]
- Zhumadillayev, N.; Dossybayev, K.; Khamzina, A.; Kapasuly, T.; Khamzina, Z.; Tlevlesov, N. SNP Genotyping Characterizes the Genome Composition of the New Baisary Fat-Tailed Sheep Breed. Animals 2022, 12, 1468. [Google Scholar] [CrossRef] [PubMed]
- Dossybayev, K.; Amandykova, M.; Orakbayeva, A.; Adylkanova, S.; Kozhakhmet, A.; Yergali, K.; Kulboldin, T.; Kulataev, B.; Torekhanov, A. Genome-Wide Association Studies Revealed Several Candidate Genes of Meat Productivity in Saryarka Fat- Tailed Coarse-Wool Sheep Breed. Genes 2024, 15, 1549. [Google Scholar] [CrossRef] [PubMed]
- Kijas, J.W.; Townley, D.; Dalrymple, B.P.; Heaton, M.P.; Maddox, J.F.; McGrath, A.; Wilson, P.; Ingersoll, R.G.; McCulloch, R.; McWilliam, S.; et al. International Sheep Genomics Consortium. A genome wide survey of SNP variation reveals the genetic structure of sheep breeds. PLoS ONE 2009, 4, e4668. [Google Scholar] [CrossRef]
- Lv, F.H.; Cao, Y.H.; Liu, G.J.; Luo, L.Y.; Lu, R.; Liu, M.J.; Li, W.R.; Zhou, P.; Wang, X.H.; Shen, M.; et al. Whole-Genome Resequencing of Worldwide Wild and Domestic Sheep Elucidates Genetic Diversity, Introgression, and Agronomically Important Loci. Mol. Biol. Evol. 2022, 39, msab353. [Google Scholar] [CrossRef]
- FAO. Developing the Institutional Framework for the Management of Animal Genetic Resources. FAO Animal Production and Health Guidelines. No. 6. Rome. 2011. Available online: http://www.fao.org/docrep/014/ba0054e/ba0054e00.pdf (accessed on 22 July 2025).
- WIDDE. Available online: http://widde.toulouse.inra.fr/widde/widde/main.do?module=sheep (accessed on 22 July 2025).
- DRYAD. Available online: https://datadryad.org/ (accessed on 22 July 2025).
- Figshare. Available online: https://figshare.com/ (accessed on 22 July 2025).
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Weir, B.S.; Cockerham, C.C. Estimating F-Statistics for the Analysis of Population Structure. Evolution 1984, 38, 1358–1370. [Google Scholar] [CrossRef]
- Szpiech, Z.A.; Jakobsson, M.; Rosenberg, N.A. ADZE: A rarefaction approach for counting alleles private to combinations of populations. Bioinformatics 2008, 24, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Molecular Evolution, Phylogenetics and Epidemiology. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 22 July 2025).
- Van Rossum, G.; Drake, F.L. Python 3 Reference Manual; CreateSpace: Scotts Valley, CA, USA, 2009. [Google Scholar]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef] [PubMed]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. CLUMPAK: A program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 2015, 15, 1179–1191. [Google Scholar] [CrossRef]
- Pickrell, J.K.; Pritchard, J.K. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 2012, 8, e1002967. [Google Scholar] [CrossRef]
- Barbato, M.; Orozco-terWengel, P.; Tapio, M.; Bruford, M.W. SNeP: A tool to estimate trends in recent effective population size trajectories using genome-wide SNP data. Front. Genet. 2015, 6, 109. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2009. [Google Scholar]
- Pozharskiy, A.; Khamzina, A.; Gritsenko, D.; Khamzina, Z.; Kassymbekova, S.; Karimov, N.; Karymsakov, T.; Tlevlesov, N. SNP genotyping and population analysis of five indigenous Kazakh sheep breeds. Livest. Sci. 2020, 241, 104252. [Google Scholar] [CrossRef]
- Deniskova, T.E.; Dotsev, A.V.; Selionova, M.I.; Kunz, E.; Medugorac, I.; Reyer, H.; Wimmers, K.; Barbato, M.; Traspov, A.A.; Brem, G.; et al. Population structure and genetic diversity of 25 Russian sheep breeds based on whole-genome genotyping. Genet. Sel. Evol. 2018, 50, 29. [Google Scholar] [CrossRef]
- Popov, I.N. Edilbay Sheep (Edilbayevskaya Ovtsa); Bastau: Almaty, Kazakhstan, 1951; p. 62. [Google Scholar]
- Kanapin, K.K. Edilbay Sheep (Edilbayevskaya Ovtsa); Bastau: Almaty, Kazakhstan, 2009; pp. 25–30. [Google Scholar]
- Kiyatkin, P.F. Fat-Tailed Sheep and Breeding Work with Them (Kurduychnyie Ovtsi i Plemennaya Rabota s Nimi); Fan: Tashkent, Uzbekistan, 1968; pp. 4–120. [Google Scholar]
- Lus, Y.Y.; Medvedev, N.N. Fat-tailed sheep of Semipalatinsk province (Kurduychnaya ovtsa Semipalatinskoy gubernii). In Domestic aniMals of the Semipalatinsk Province: Mater. Commission of the Expeditionary Research; Publishing House of the USSR Academy of Sciences: Leningrad, Russia, 1928; pp. 176–265. [Google Scholar]
- Wei, C.; Wang, H.; Liu, G.; Wu, M.; Cao, J.; Liu, Z.; Liu, R.; Zhao, F.; Zhang, L.; Lu, J.; et al. Genome-wide analysis reveals population structure and selection in Chinese indigenous sheep breeds. BMC Genom. 2015, 16, 194. [Google Scholar] [CrossRef] [PubMed]
- Slow Food Foundation for Biodiversity. Available online: https://www.fondazioneslowfood.com/en/ark-of-taste-slow-food/gissar-sheep/ (accessed on 22 July 2025).
- Schillhorn van Veen, T.W. The Kyrgyz Sheep Herders at a Crossroads. Pastor. Dev. Netw. Ser. 1995, 38, 1–14. [Google Scholar]
- Kerven, C.; Steimann, B.; Ashley, L.; Dear, C.; Rahim, I. Pastoralism and Farming in Central Asia’s Mountains: A Research Review; University of Central Asia: Bishkek, Kyrgyzstan, 2011. [Google Scholar]
- van Marle-Köster, E.; Lashmar, S.F.; Retief, A.; Visser, C. Whole-Genome SNP Characterisation Provides Insight for Sustainable Use of Local South African Livestock Populations. Front. Genet. 2021, 12, 714194. [Google Scholar] [CrossRef] [PubMed]
- Eydivandi, S.; Sahana, G.; Momen, M.; Moradi, M.H.; Schönherz, A.A. Genetic diversity in Iranian indigenous sheep vis-à-vis selected exogenous sheep breeds and wild mouflon. Anim. Genet. 2020, 51, 772–787. [Google Scholar] [CrossRef]
- Bayraktar, M. Analysing the genetic diversity of three sheep breeds in Turkey and nearby countries using 50 K SNPs data. Anim. Biotechnol. 2024, 35, 2329106. [Google Scholar] [CrossRef]
- Liu, C.C.; Shringarpure, S.; Lange, K.; Novembre, J. Exploring Population Structure with Admixture Models and Principal Component Analysis. In Statistical Population Genomics. Methods in Molecular Biology; Dutheil, J.Y., Ed.; Humana: New York, NY, USA, 2020; Volume 2090. [Google Scholar]
- van Waaij, J.; Li, S.; Garcia-Erill, G.; Albrechtsen, A.; Wiuf, C. Evaluation of population structure inferred by principal component analysis or the admixture model. Genetics 2023, 225, iyad157. [Google Scholar] [CrossRef]
- Khabisi, M.M.; Foozi, M.A.; Lv, F.H.; Esmailizadeh, A. Genome-wide DNA arrays profiling unravels the genetic structure of Iranian sheep and pattern of admixture with worldwide coarse-wool sheep breeds. Genomics 2021, 113, 3501–3511. [Google Scholar] [CrossRef]
- Dotsev, A.V.; Moradi, M.H.; Deniskova, T.E.; Esmailizadeh, A.; Bakoev, N.F.; Koshkina, O.A.; Griffin, D.K.; Romanov, M.N.; Zinovieva, N.A. Molecular Genetic Assessment Aids in Clarifying Phylogenetic Status of Iranian Kerman Wild Sheep. Animals 2025, 15, 238. [Google Scholar] [CrossRef]
- Khamzina, A.; Smagulov, D.; Dossybayev, K.; Kantanen, J.; Khamzin, K. Assessing runs of homozygosity reveals production traits of Kazakh sheep breeds. Braz. J. Biol. 2025, 85, e292980. [Google Scholar] [CrossRef]
- Ali Awan, G.; Festa-Bianchet, M.; Ahmad, T. Poaching, recruitment and conservation of Punjab urial Ovis vignei punjabiensis. Wildl. Biol. 2006, 12, 443–449. [Google Scholar] [CrossRef]
- Stefan, M. Conservation of Tajik Markhor (Capra falconeri heptneri) and Urial (Ovis vignei) in Tajikistan and adjacent Afghanistan. Galemys Span. J. Mammal. 2010, 22, 407–419. [Google Scholar]
- Yuan, Z.; Liu, E.; Liu, Z.; Kijas, J.; Zhu, C.; Hu, S.; Ma, X.; Zhang, L.; Du, L.; Wang, H.; et al. Selection signature analysis reveals genes associated with tail type in Chinese indigenous sheep. Anim. Genet. 2016, 48, 55–66. [Google Scholar] [CrossRef]
- Ciani, E.; Lasagna, E.; D’Andrea, M.; Alloggio, I.; Marroni, F.; Ceccobelli, S.; Delgado Bermejo, J.V.; Sarti, F.M.; Kijas, J.; Lenstra, J.A.; et al. Merino and Merino-derived sheep breeds: A genome-wide intercontinental study. Genet. Sel. Evol. 2015, 47, 64. [Google Scholar] [CrossRef]
- Rochus, C.M.; Tortereau, F.; Plisson-Petit, F.; Restoux, G.; Moreno-Romieux, C.; Tosser-Klopp, G.; Servin, B. Revealing the selection history of adaptive loci using genome-wide scans for selection: An example from domestic sheep. BMC Genom. 2018, 19, 71. [Google Scholar] [CrossRef]
- Dotsev, A.; Koshkina, O.; Kharzinova, V.; Deniskova, T.; Reyer, H.; Kunz, E.; Mészáros, G.; Shakhin, A.; Petrov, S.; Medvedev, D.; et al. Genome-Wide Insights into Intraspecific Taxonomy and Genetic Diversity of Argali (Ovis ammon). Diversity 2023, 15, 627. [Google Scholar] [CrossRef]

| No. | Sheep Breed | Farm | Number of Animals | Overall |
|---|---|---|---|---|
| 1 | Edilbay (ED) | “Birlik Mal Zauyty” LLP, Birlik village, Zhangaly district, West Kazakhstan region | 33 | 97 |
| LLP “Suyindik asyl tukymdy koy zauyty”, Suyindik village, Kurmangazy district, Atyrau region | 32 | |||
| Peasant farm “Aigul”, Donenbay village, Ayagoz district, Abay region | 32 | |||
| 2 | Kazakh fat-tailed coarse-wooled (KFTCW) | Peasant farm “Kundyzdy”, Kundyzdy village, Ulken Naryn district, East Kazakhstan region | 32 | 32 |
| 3 | Gissar (GISS) | Peasant farm “Darbaza”, Darbaza village, Saryagash district, Turkestan region | 31 | 31 |
| Overall | 160 | |||
| Studied Sheep Groups | Total Number of Animals | Total Number of SNPs | Number of Animals After QC | Number of SNPs After QC | Number of Animals After LD Pruning | Number of SNPs After LD Pruning |
|---|---|---|---|---|---|---|
| Kazakhstani sheep breeds | 160 | 51,102 | 160 | 47,969 | 160 | 37,969 |
| Domestic sheep breeds | 225 | 39,273 | 182 | 36,798 | 182 | 30,230 |
| Domestic sheep breeds and wild sheep | 267 | 38,667 | 258 | 31,339 | 258 | 27,045 |
| No. | Population | N | He | Ho | F | FIS | Mean FST | Pn, % | Ar |
|---|---|---|---|---|---|---|---|---|---|
| 1 | ED-KAZ | 20 | 0.383901 | 0.396967 | −0.034014 | −0.008059360 | 0.126425371 | 98.90 | 1.90702 |
| 2 | KFTCW-KAZ | 20 | 0.379533 | 0.390952 | −0.030186 | −0.005761707 | 0.133995353 | 98.54 | 1.89864 |
| 3 | GISS-KAZ | 20 | 0.378127 | 0.394446 | −0.043173 | −0.016917877 | 0.135257999 | 98.35 | 1.89474 |
| 4 | TON-MONGOL | 15 | 0.376177 | 0.369286 | 0.018922 | 0.048046452 | 0.142830552 | 96.76 | 1.88441 |
| 5 | TIBQ-CHINA | 14 | 0.375390 | 0.362380 | 0.035135 | 0.065535223 | 0.146539513 | 96.21 | 1.88215 |
| 6 | KCW-KRG | 13 | 0.381521 | 0.405609 | −0.063152 | −0.023469213 | 0.136567911 | 97.62 | 1.90427 |
| 7 | GISS-KRG | 12 | 0.380844 | 0.399926 | −0.049898 | −0.008697962 | 0.133568616 | 97.60 | 1.90648 |
| 8 | KRKS-SW-ASIA | 18 | 0.372922 | 0.394711 | −0.058287 | −0.027327093 | 0.156338863 | 96.15 | 1.86975 |
| 9 | QEZ-SW-ASIA | 15 | 0.382087 | 0.391640 | −0.025053 | 0.007706401 | 0.131929368 | 98.02 | 1.90433 |
| 10 | SOP-ITALY | 9 | 0.382399 | 0.402810 | −0.053236 | −0.002107075 | 0.158924142 | 96.21 | 1.65776 |
| 11 | OUE-FRANCE | 18 | 0.316522 | 0.271522 | 0.142368 | 0.154365564 | 0.301822277 | 79.97 | 1.65567 |
| 12 | BBB-USA | 15 | 0.362649 | 0.366821 | −0.011258 | 0.019518782 | 0.191684706 | 93.19 | 1.83533 |
| 13 | RMA-KENYA | 15 | 0.352499 | 0.372089 | −0.055720 | −0.020860007 | 0.194441609 | 93.11 | 1.82236 |
| 14 | NQA-NAMIBIA | 12 | 0.352181 | 0.418734 | −0.189218 | −0.134063327 | 0.290036023 | 74.21 | 1.65979 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dossybayev, K.; Amandykova, M.; Ualiyeva, D.; Kapassuly, T.; Kozhakhmet, A.; Ciani, E.; Bekmanov, B.; Amzeyev, R. Genome-Wide SNP Analysis Reveals the Unique Genetic Diversity Represented by Fat-Tailed Coarse-Wooled Sheep Breeds of Kazakhstan. Biology 2025, 14, 1478. https://doi.org/10.3390/biology14111478
Dossybayev K, Amandykova M, Ualiyeva D, Kapassuly T, Kozhakhmet A, Ciani E, Bekmanov B, Amzeyev R. Genome-Wide SNP Analysis Reveals the Unique Genetic Diversity Represented by Fat-Tailed Coarse-Wooled Sheep Breeds of Kazakhstan. Biology. 2025; 14(11):1478. https://doi.org/10.3390/biology14111478
Chicago/Turabian StyleDossybayev, Kairat, Makpal Amandykova, Daniya Ualiyeva, Tilek Kapassuly, Altynay Kozhakhmet, Elena Ciani, Bakytzhan Bekmanov, and Rauan Amzeyev. 2025. "Genome-Wide SNP Analysis Reveals the Unique Genetic Diversity Represented by Fat-Tailed Coarse-Wooled Sheep Breeds of Kazakhstan" Biology 14, no. 11: 1478. https://doi.org/10.3390/biology14111478
APA StyleDossybayev, K., Amandykova, M., Ualiyeva, D., Kapassuly, T., Kozhakhmet, A., Ciani, E., Bekmanov, B., & Amzeyev, R. (2025). Genome-Wide SNP Analysis Reveals the Unique Genetic Diversity Represented by Fat-Tailed Coarse-Wooled Sheep Breeds of Kazakhstan. Biology, 14(11), 1478. https://doi.org/10.3390/biology14111478






