From Genome-Wide SNPs to Neuroimmune Crosstalk: Mapping the Genetic Landscape of IBD and Its Brain Overlap
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection and Preprocessing
2.1.1. GWAS Data
2.1.2. GTEx Data
2.1.3. Pathway Enrichment Analysis
2.1.4. Brain and Signaling-Related Pathway Analysis
2.1.5. Single Nucleotide Polymorphism (SNP) Analysis
2.1.6. SNP-to-Gene Mapping Strategy
2.1.7. Word Cloud Generation
2.1.8. Genetic Overlap Analysis with Brain Disorders
2.1.9. Overlap Among IBD Subtypes
2.2. Gene Expression Analysis
2.2.1. Average Gene Expression Calculation
2.2.2. Visualization
3. Results
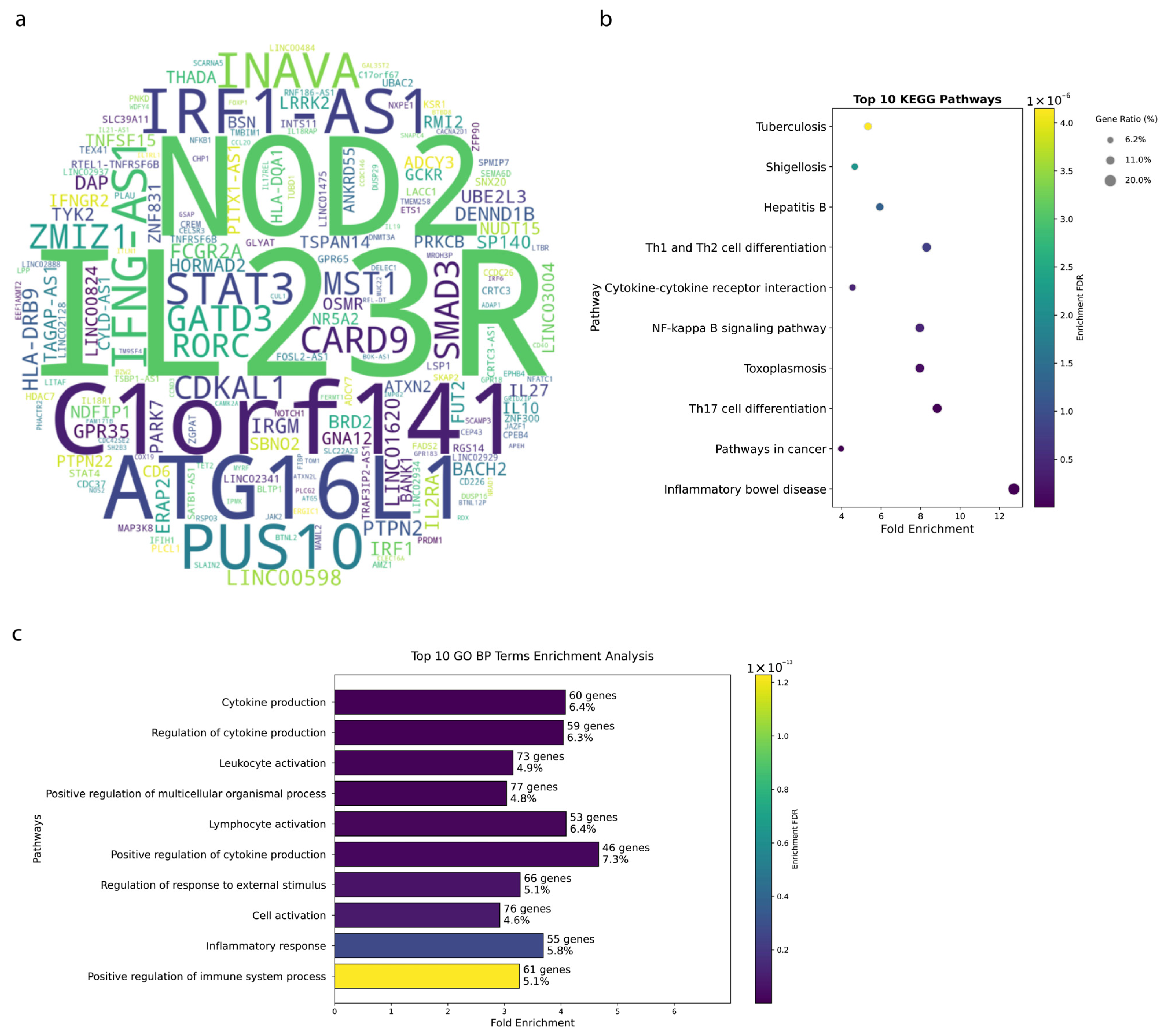
3.1. Genetic Architecture of IBD Reveals Key Susceptibility Loci
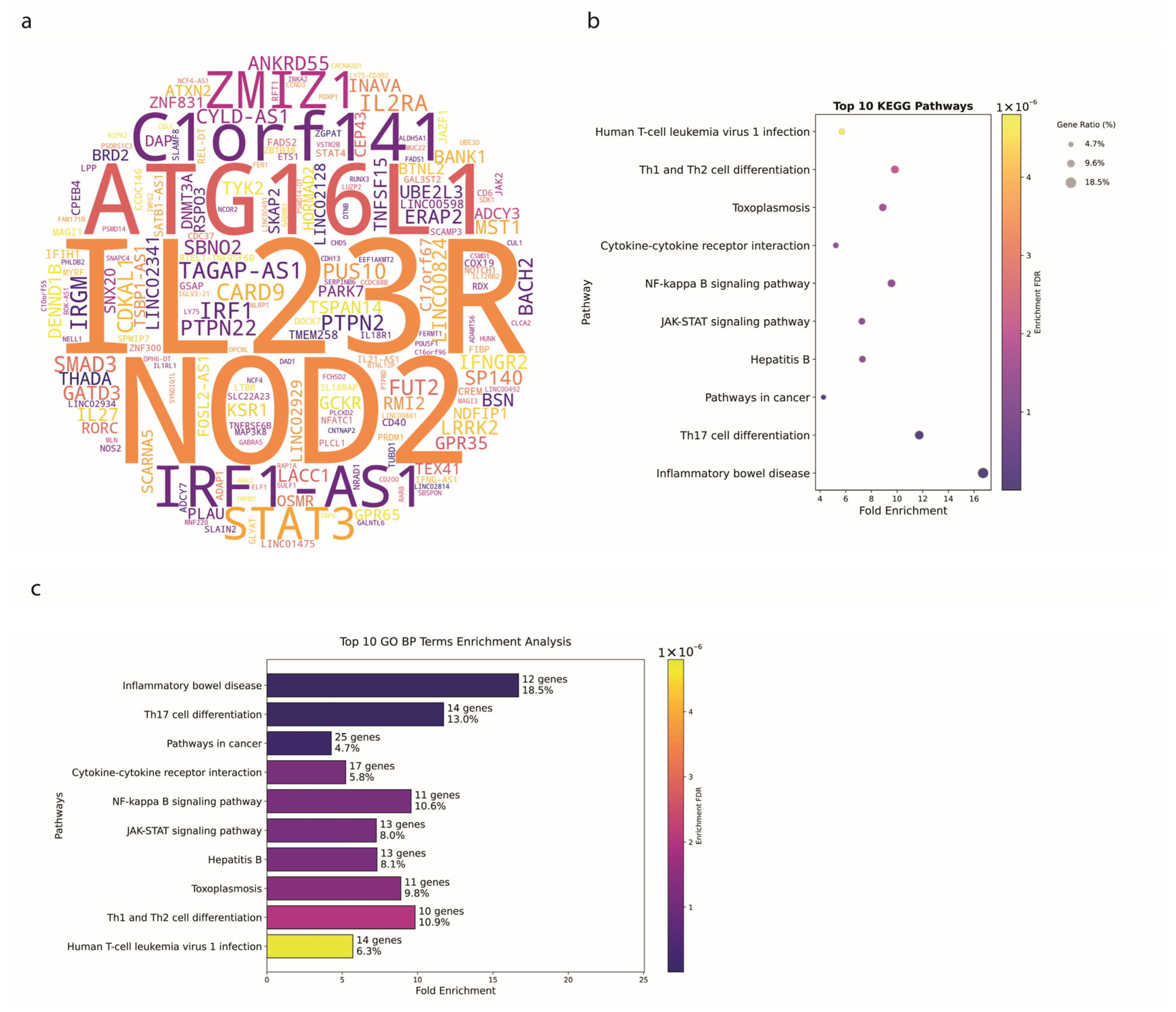
3.2. Crohn’s Disease Exhibits Distinct Genetic Signatures
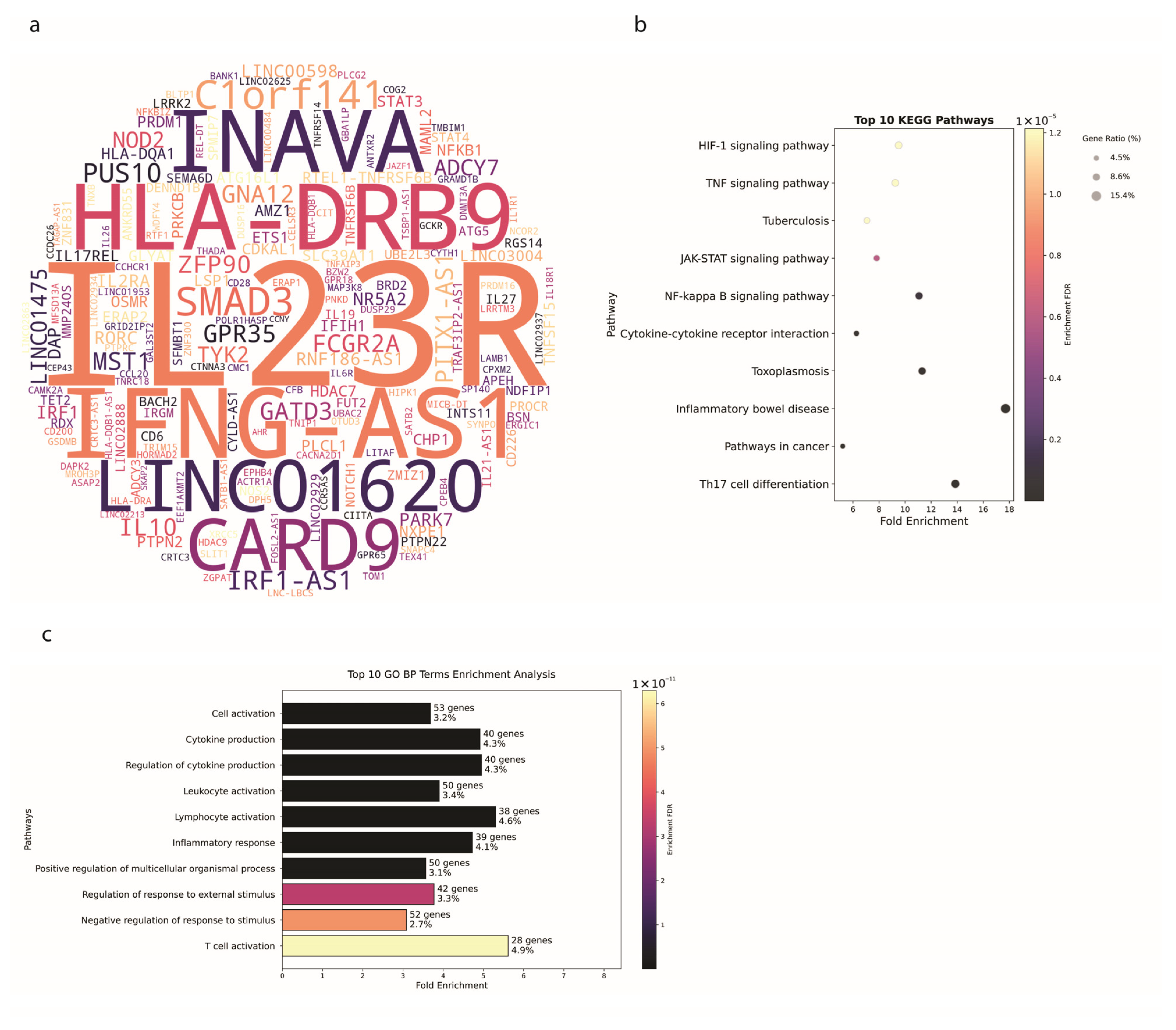
3.3. Ulcerative Colitis Shows Tissue-Specific Genetic Patterns
3.4. IBD-Associated Genes Show Differential Expression Patterns Along the Gut–Brain Axis
3.5. Genetic Variant Architecture Reveals Disease-Specific and Shared Susceptibility Patterns
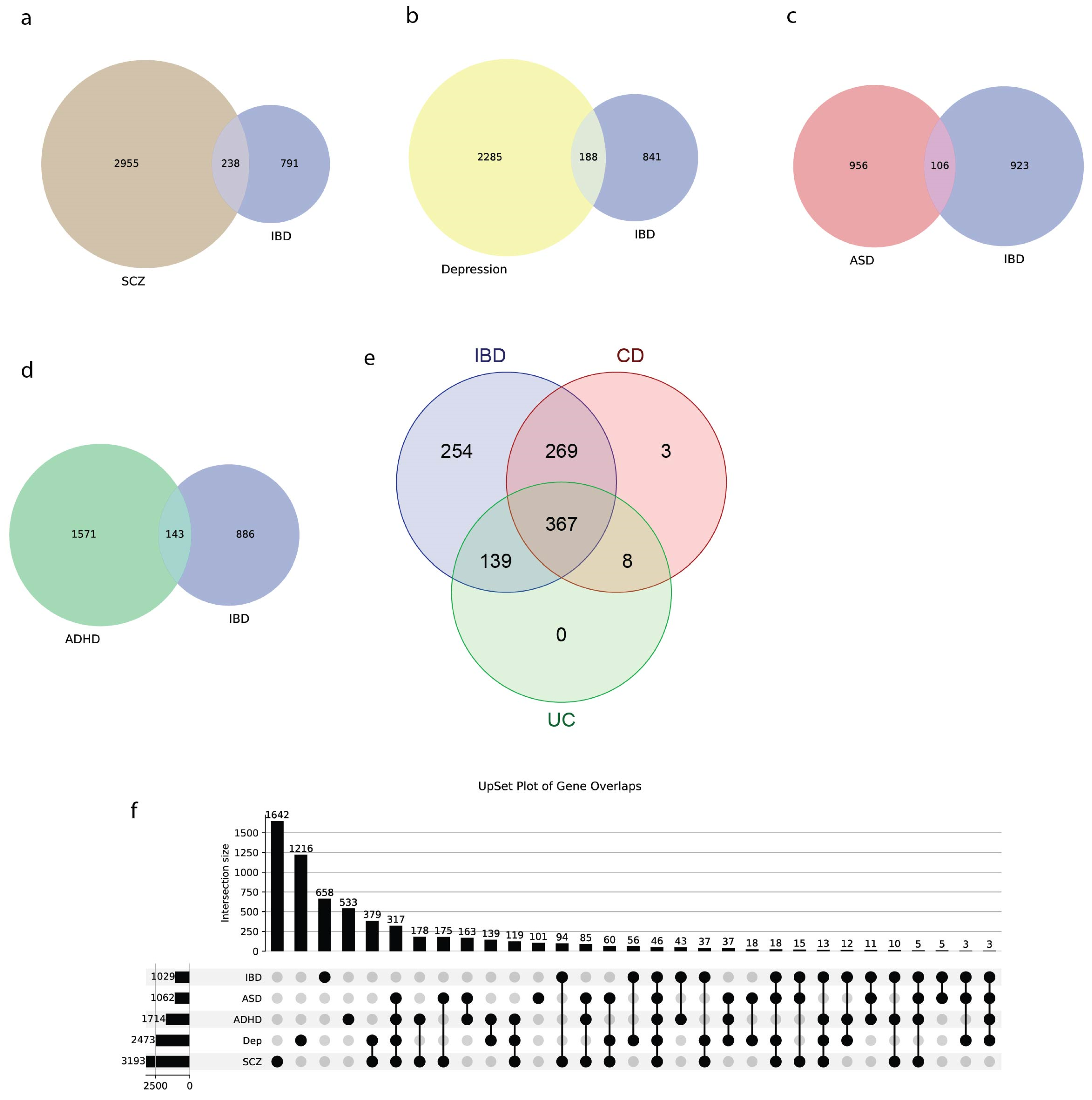
3.6. IBD Shows Significant Genetic Overlap with Neuropsychiatric Disorders
4. Discussion
4.1. Genetic Insights and Pathway Enrichment
4.2. Genetic Architecture and SNP Distribution in IBD
4.3. Expression Patterns Along the Gut–Brain Axis
4.4. Genetic Overlap with Brain Disorders
4.5. Therapeutic and Repurposing Implications Across the Gut-Brain Axis
4.6. Strengths and Limitations
4.7. Future Directions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Franke, A.; McGovern, D.P.B.; Barrett, J.C.; Wang, K.; Radford-Smith, G.L.; Ahmad, T.; Lees, C.W.; Balschun, T.; Lee, J.; Roberts, R.; et al. Genome-Wide Meta-Analysis Increases to 71 the Number of Confirmed Crohn’s Disease Susceptibility Loci. Nat. Genet. 2010, 42, 1118–1125. [Google Scholar] [CrossRef] [PubMed]
- The International IBD Genetics Consortium (IIBDGC); Jostins, L.; Ripke, S.; Weersma, R.K.; Duerr, R.H.; McGovern, D.P.; Hui, K.Y.; Lee, J.C.; Philip Schumm, L.; Sharma, Y.; et al. Host–Microbe Interactions Have Shaped the Genetic Architecture of Inflammatory Bowel Disease. Nature 2012, 491, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Xavier, R.J.; Podolsky, D.K. Unravelling the Pathogenesis of Inflammatory Bowel Disease. Nature 2007, 448, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.Z.; Van Sommeren, S.; Huang, H.; Ng, S.C.; Alberts, R.; Takahashi, A.; Ripke, S.; Lee, J.C.; Jostins, L.; Shah, T.; et al. Association Analyses Identify 38 Susceptibility Loci for Inflammatory Bowel Disease and Highlight Shared Genetic Risk Across Populations. Nat. Genet. 2015, 47, 979–986. [Google Scholar] [CrossRef]
- de Lange, K.M.; Moutsianas, L.; Lee, J.C.; Lamb, C.A.; Luo, Y.; Kennedy, N.A.; Jostins, L.; Rice, D.L.; Gutierrez-Achury, J.; Ji, S.-G.; et al. Genome-Wide Association Study Implicates Immune Activation of Multiple Integrin Genes in Inflammatory Bowel Disease. Nat. Genet. 2017, 49, 256–261. [Google Scholar] [CrossRef]
- Huang, H.; Fang, M.; Jostins, L.; Umićević Mirkov, M.; Boucher, G.; Anderson, C.A.; Andersen, V.; Cleynen, I.; Cortes, A.; Crins, F.; et al. Fine-Mapping Inflammatory Bowel Disease Loci to Single-Variant Resolution. Nature 2017, 547, 173–178. [Google Scholar] [CrossRef]
- Liu, H.; Guan, L.; Su, X.; Zhao, L.; Shu, Q.; Zhang, J. A Broken Network of Susceptibility Genes in the Monocytes of Crohn’s Disease Patients. Life Sci. Alliance 2024, 7, e202302394. [Google Scholar] [CrossRef]
- Zhang, M.; Zhao, J.; Ji, H.; Tan, Y.; Zhou, S.; Sun, J.; Ding, Y.; Li, X. Multi-Omics Insight into the Molecular Networks of Mental Disorder Related Genetic Pathways in the Pathogenesis of Inflammatory Bowel Disease. Transl. Psychiatry 2025, 15, 91. [Google Scholar] [CrossRef]
- Carithers, L.J.; Moore, H.M. The Genotype-Tissue Expression (GTEx) Project. Biopreservation Biobanking 2015, 13, 307–308. [Google Scholar] [CrossRef]
- Danese, S.; Fiocchi, C. Ulcerative Colitis. N. Engl. J. Med. 2011, 365, 1713–1725. [Google Scholar] [CrossRef]
- Baumgart, D.C.; Carding, S.R. Inflammatory Bowel Disease: Cause and Immunobiology. Lancet 2007, 369, 1627–1640. [Google Scholar] [CrossRef]
- Lonsdale, J.; Thomas, J.; Salvatore, M.; Phillips, R.; Lo, E.; Shad, S.; Hasz, R.; Walters, G.; Garcia, F.; Young, N.; et al. The Genotype-Tissue Expression (GTEx) Project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef]
- Bycroft, C.; Freeman, C.; Petkova, D.; Band, G.; Elliott, L.T.; Sharp, K.; Motyer, A.; Vukcevic, D.; Delaneau, O.; O’Connell, J.; et al. The UK Biobank Resource with Deep Phenotyping and Genomic Data. Nature 2018, 562, 203–209. [Google Scholar] [CrossRef]
- Gong, W.; Guo, P.; Li, Y.; Liu, L.; Yan, R.; Liu, S.; Wang, S.; Xue, F.; Zhou, X.; Yuan, Z. Role of the Gut-Brain Axis in the Shared Genetic Etiology Between Gastrointestinal Tract Diseases and Psychiatric Disorders: A Genome-Wide Pleiotropic Analysis. JAMA Psychiatry 2023, 80, 360–370. [Google Scholar] [CrossRef]
- Watanabe, K.; Taskesen, E.; van Bochoven, A.; Posthuma, D. Functional Mapping and Annotation of Genetic Associations with FUMA. Nat. Commun. 2017, 8, 1826. [Google Scholar] [CrossRef]
- MAGMA: Generalized Gene-Set Analysis of GWAS Data|PLOS Computational Biology. Available online: https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1004219 (accessed on 18 September 2025).
- The GTEx Consortium. The GTEx Consortium Atlas of Genetic Regulatory Effects Across Human Tissues. Science 2020, 369, 1318–1330. [Google Scholar] [CrossRef] [PubMed]
- Cryan, J.F.; Dinan, T.G. Mind-Altering Microorganisms: The Impact of the Gut Microbiota on Brain and Behaviour. Nat. Rev. Neurosci. 2012, 13, 701–712. [Google Scholar] [CrossRef] [PubMed]
- Wachsmuth, H.R.; Weninger, S.N.; Duca, F.A. Role of the Gut–Brain Axis in Energy and Glucose Metabolism. Exp. Mol. Med. 2022, 54, 377–392. [Google Scholar] [CrossRef] [PubMed]
- La Torre, D.; Van Oudenhove, L.; Vanuytsel, T.; Verbeke, K. Psychosocial Stress-Induced Intestinal Permeability in Healthy Humans: What Is the Evidence? Neurobiol. Stress 2023, 27, 100579. [Google Scholar] [CrossRef]
- Benros, M.E.; Waltoft, B.L.; Nordentoft, M.; Østergaard, S.D.; Eaton, W.W.; Krogh, J.; Mortensen, P.B. Autoimmune Diseases and Severe Infections as Risk Factors for Mood Disorders: A Nationwide Study. JAMA Psychiatry 2013, 70, 812. [Google Scholar] [CrossRef]
- Mikocka-Walus, A.A.; Turnbull, D.A.; Moulding, N.T.; Wilson, I.G.; Andrews, J.M.; Holtmann, G.J. Controversies Surrounding the Comorbidity of Depression and Anxiety in Inflammatory Bowel Disease Patients: A Literature Review. Inflamm. Bowel Dis. 2007, 13, 225–234. [Google Scholar] [CrossRef]
- Bhamre, R.; Sawrav, S.; Adarkar, S.; Sakaria, R.; Bhatia, S.J. Psychiatric Comorbidities in Patients with Inflammatory Bowel Disease. Indian J. Gastroenterol. 2018, 37, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, C.N.; Hitchon, C.A.; Walld, R.; Bolton, J.M.; Sareen, J.; Walker, J.R.; Graff, L.A.; Patten, S.B.; Singer, A.; Lix, L.M.; et al. Increased Burden of Psychiatric Disorders in Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2019, 25, 360–368. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Luo, G.-Y.; Tian, T.; Zhao, Y.-Q.; Meng, S.-Y.; Wu, J.-H.; Han, W.-X.; Deng, B.; Ni, J. Shared Genetic Basis and Causality between Schizophrenia and Inflammatory Bowel Disease: Evidence from a Comprehensive Genetic Analysis. Psychol. Med. 2024, 54, 2658–2668. [Google Scholar] [CrossRef] [PubMed]
- Ge, L.; Liu, S.; Li, S.; Yang, J.; Hu, G.; Xu, C.; Song, W. Psychological Stress in Inflammatory Bowel Disease: Psychoneuroimmunological Insights into Bidirectional Gut–Brain Communications. Front. Immunol. 2022, 13, 1016578. [Google Scholar] [CrossRef] [PubMed]
- Sung, K.; Zhang, B.; Wang, H.E.; Bai, Y.; Tsai, S.; Su, T.; Chen, T.; Hou, M.; Lu, C.; Wang, Y.; et al. Schizophrenia and Risk of New-onset Inflammatory Bowel Disease: A Nationwide Longitudinal Study. Aliment. Pharmacol. Ther. 2022, 55, 1192–1201. [Google Scholar] [CrossRef]
- Choudhary, A.; Nayak, R.; Peles, D.; Mizrahi, L.; Stern, S. Current Progress in Understanding Schizophrenia Using Genomics and Pluripotent Stem Cells: A Meta-Analytical Overview. Schizophr. Res. 2022, 273, 24–38. [Google Scholar] [CrossRef]
- Romanovsky, E.; Choudhary, A.; Peles, D.; Akel, A.A.; Stern, S. Uncovering Convergence and Divergence between Autism and Schizophrenia Using Genomic Tools and Patients’ Neurons. Mol. Psychiatry 2023, 30, 1019–1028. [Google Scholar] [CrossRef]
- Fakhfouri, G.; Mijailović, N.R.; Rahimian, R. Psychiatric Comorbidities of Inflammatory Bowel Disease: It Is a Matter of Microglia’s Gut Feeling. Cells 2024, 13, 177. [Google Scholar] [CrossRef]
- Waterman, M.; Xu, W.; Stempak, J.M.; Milgrom, R.; Bernstein, C.N.; Griffiths, A.M.; Greenberg, G.R.; Steinhart, H.A.; Silverberg, M.S. Distinct and Overlapping Genetic Loci in Crohn’s Disease and Ulcerative Colitis: Correlations with Pathogenesis. Inflamm. Bowel Dis. 2011, 17, 1936–1942. [Google Scholar] [CrossRef]
- Atreya, I.; Atreya, R.; Neurath, M.F. NF-κB in Inflammatory Bowel Disease. J. Intern. Med. 2008, 263, 591–596. [Google Scholar] [CrossRef]
- Kerber, E.L.; Padberg, C.; Koll, N.; Schuetzhold, V.; Fandrey, J.; Winning, S. The Importance of Hypoxia-Inducible Factors (HIF-1 and HIF-2) for the Pathophysiology of Inflammatory Bowel Disease. Int. J. Mol. Sci. 2020, 21, 8551. [Google Scholar] [CrossRef] [PubMed]
- Coskun, M.; Salem, M.; Pedersen, J.; Nielsen, O.H. Involvement of JAK/STAT Signaling in the Pathogenesis of Inflammatory Bowel Disease. Pharmacol. Res. 2013, 76, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Schindler, C.; Levy, D.E.; Decker, T. JAK-STAT Signaling: From Interferons to Cytokines. J. Biol. Chem. 2007, 282, 20059–20063. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Okamoto, S.; Hisamatsu, T.; Kamada, N.; Chinen, H.; Saito, R.; Kitazume, M.T.; Nakazawa, A.; Sugita, A.; Koganei, K.; et al. IL23 Differentially Regulates the Th1/Th17 Balance in Ulcerative Colitis and Crohn’s Disease. Gut 2008, 57, 1682–1689. [Google Scholar] [CrossRef]
- Song, L.; Zhou, R.; Huang, S.; Zhou, F.; Xu, S.; Wang, W.; Yi, F.; Wang, X.; Xia, B. High Intestinal and Systemic Levels of Interleukin-23/T-Helper 17 Pathway in Chinese Patients with Inflammatory Bowel Disease. Mediat. Inflamm. 2013, 2013, 425915. [Google Scholar] [CrossRef]
- Schmitt, H.; Billmeier, U.; Dieterich, W.; Rath, T.; Sonnewald, S.; Reid, S.; Hirschmann, S.; Hildner, K.; Waldner, M.J.; Mudter, J.; et al. Expansion of IL-23 Receptor Bearing TNFR2+ T Cells Is Associated with Molecular Resistance to Anti-TNF Therapy in Crohn’s Disease. Gut 2019, 68, 814–828. [Google Scholar] [CrossRef]
- Gandal, M.J.; Haney, J.R.; Parikshak, N.N.; Leppa, V.; Ramaswami, G.; Hartl, C.; Schork, A.J.; Appadurai, V.; Buil, A.; Werge, T.M.; et al. Shared Molecular Neuropathology across Major Psychiatric Disorders Parallels Polygenic Overlap. Science 2018, 359, 693–697. [Google Scholar] [CrossRef]
- Lanz, T.A.; Reinhart, V.; Sheehan, M.J.; Rizzo, S.J.S.; Bove, S.E.; James, L.C.; Volfson, D.; Lewis, D.A.; Kleiman, R.J. Postmortem Transcriptional Profiling Reveals Widespread Increase in Inflammation in Schizophrenia: A Comparison of Prefrontal Cortex, Striatum, and Hippocampus among Matched Tetrads of Controls with Subjects Diagnosed with Schizophrenia, Bipolar or Major Depressive Disorder. Transl. Psychiatry 2019, 9, 151. [Google Scholar] [CrossRef]
- Voineagu, I.; Wang, X.; Johnston, P.; Lowe, J.K.; Tian, Y.; Horvath, S.; Mill, J.; Cantor, R.M.; Blencowe, B.J.; Geschwind, D.H. Transcriptomic Analysis of Autistic Brain Reveals Convergent Molecular Pathology. Nature 2011, 474, 380–384. [Google Scholar] [CrossRef]
- Lindholm Carlström, E.; Niazi, A.; Etemadikhah, M.; Halvardson, J.; Enroth, S.; Stockmeier, C.A.; Rajkowska, G.; Nilsson, B.; Feuk, L. Transcriptome Analysis of Post-Mortem Brain Tissue Reveals Up-Regulation of the Complement Cascade in a Subgroup of Schizophrenia Patients. Genes 2021, 12, 1242. [Google Scholar] [CrossRef] [PubMed]
- Huang, E.J.; Reichardt, L.F. Neurotrophins: Roles in Neuronal Development and Function. Annu. Rev. Neurosci. 2001, 24, 677–736. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Song, D.; Yan, Y.; Quan, Z.; Qing, H. The Role of Oxytocin in Early-Life-Stress-Related Neuropsychiatric Disorders. Int. J. Mol. Sci. 2023, 24, 10430. [Google Scholar] [CrossRef] [PubMed]
- Zois, C.D.; Katsanos, K.H.; Kosmidou, M.; Tsianos, E.V. Neurologic Manifestations in Inflammatory Bowel Diseases: Current Knowledge and Novel Insights. J. Crohn’s Colitis 2010, 4, 115–124. [Google Scholar] [CrossRef]
- Pulli, I.; Asghar, M.Y.; Kemppainen, K.; Törnquist, K. Sphingolipid-Mediated Calcium Signaling and Its Pathological Effects. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2018, 1865, 1668–1677. [Google Scholar] [CrossRef]
- Lee, J.Y.; Jin, H.K.; Bae, J. Sphingolipids in Neuroinflammation: A Potential Target for Diagnosis and Therapy. BMB Rep. 2020, 53, 28–34. [Google Scholar] [CrossRef]
- Fre, S.; Bardin, A.; Robine, S.; Louvard, D. Notch Signaling in Intestinal Homeostasis across Species: The Cases of Drosophila, Zebrafish and the Mouse. Exp. Cell Res. 2011, 317, 2740–2747. [Google Scholar] [CrossRef]
- Lasky, J.L.; Wu, H. Notch Signaling, Brain Development, and Human Disease. Pediatr. Res. 2005, 57, 104R–109R. [Google Scholar] [CrossRef]
- Inestrosa, N.C.; Varela-Nallar, L. Wnt Signaling in the Nervous System and in Alzheimer’s Disease. J. Mol. Cell Biol. 2014, 6, 64–74. [Google Scholar] [CrossRef]
- Patapoutian, A.; Reichardt, L.F. Roles of Wnt Proteins in Neural Development and Maintenance. Curr. Opin. Neurobiol. 2000, 10, 392–399. [Google Scholar] [CrossRef]
- Benros, M.E.; Nielsen, P.R.; Nordentoft, M.; Eaton, W.W.; Dalton, S.O.; Mortensen, P.B. Autoimmune Diseases and Severe Infections as Risk Factors for Schizophrenia: A 30-Year Population-Based Register Study. Am. J. Psychiatry 2011, 168, 1303–1310. [Google Scholar] [CrossRef]
- Mikocka-Walus, A.; Pittet, V.; Rossel, J.-B.; Von Känel, R.; Anderegg, C.; Bauerfeind, P.; Beglinger, C.; Begré, S.; Belli, D.; Bengoa, J.M.; et al. Symptoms of Depression and Anxiety Are Independently Associated with Clinical Recurrence of Inflammatory Bowel Disease. Clin. Gastroenterol. Hepatol. 2016, 14, 829–835.e1. [Google Scholar] [CrossRef]
- Masi, A.; Quintana, D.S.; Glozier, N.; Lloyd, A.R.; Hickie, I.B.; Guastella, A.J. Cytokine Aberrations in Autism Spectrum Disorder: A Systematic Review and Meta-Analysis. Mol. Psychiatry 2015, 20, 440–446. [Google Scholar] [CrossRef] [PubMed]
- Estes, M.L.; McAllister, A.K. Immune Mediators in the Brain and Peripheral Tissues in Autism Spectrum Disorder. Nat. Rev. Neurosci. 2015, 16, 469–486. [Google Scholar] [CrossRef] [PubMed]
- Zawadzka, A.; Cieślik, M.; Adamczyk, A. The Role of Maternal Immune Activation in the Pathogenesis of Autism: A Review of the Evidence, Proposed Mechanisms and Implications for Treatment. Int. J. Mol. Sci. 2021, 22, 11516. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Choi, M.J.; Ha, S.; Hwang, J.; Koyanagi, A.; Dragioti, E.; Radua, J.; Smith, L.; Jacob, L.; Salazar de Pablo, G.; et al. Association between Autism Spectrum Disorder and Inflammatory Bowel Disease: A Systematic Review and Meta-Analysis. Autism. Res. 2022, 15, 340–352. [Google Scholar] [CrossRef]
- Sewell, G.W.; Kaser, A. Interleukin-23 in the Pathogenesis of Inflammatory Bowel Disease and Implications for Therapeutic Intervention. J. Crohn’s Colitis 2022, 16, ii3–ii19. [Google Scholar] [CrossRef]
- Louis, E.; Schreiber, S.; Panaccione, R.; Bossuyt, P.; Biedermann, L.; Colombel, J.-F.; Parkes, G.; Peyrin-Biroulet, L.; D’Haens, G.; Hisamatsu, T.; et al. Risankizumab for Ulcerative Colitis: Two Randomized Clinical Trials. JAMA 2024, 332, 881–897. [Google Scholar] [CrossRef]
- Sandborn, W.J.; Su, C.; Sands, B.E.; D’Haens, G.R.; Vermeire, S.; Schreiber, S.; Danese, S.; Feagan, B.G.; Reinisch, W.; Niezychowski, W.; et al. Tofacitinib as Induction and Maintenance Therapy for Ulcerative Colitis. N. Engl. J. Med. 2017, 376, 1723–1736. [Google Scholar] [CrossRef]
- RINVOQ® (Upadacitinib) Receives FDA Approval for the Treatment of Adults with Moderately to Severely Active Ulcerative Colitis. Available online: https://news.abbvie.com/2022-03-16-RINVOQ-R-upadacitinib-Receives-FDA-Approval-for-the-Treatment-of-Adults-with-Moderately-to-Severely-Active-Ulcerative-Colitis (accessed on 22 September 2025).
- Naser, S.A. Role of ATG16L, NOD2 and IL23R in Crohn’s Disease Pathogenesis. World J. Gastroenterol. 2012, 18, 412. [Google Scholar] [CrossRef]
- Kappelmann, N.; Lewis, G.; Dantzer, R.; Jones, P.B.; Khandaker, G.M. Antidepressant Activity of Anti-Cytokine Treatment: A Systematic Review and Meta-Analysis of Clinical Trials of Chronic Inflammatory Conditions. Mol. Psychiatry 2018, 23, 335–343. [Google Scholar] [CrossRef]
- Raison, C.L.; Rutherford, R.E.; Woolwine, B.J.; Shuo, C.; Schettler, P.; Drake, D.F.; Haroon, E.; Miller, A.H. A Randomized Controlled Trial of the Tumor Necrosis Factor Antagonist Infliximab for Treatment-Resistant Depression: The Role of Baseline Inflammatory Biomarkers. JAMA Psychiatry 2013, 70, 31–41. [Google Scholar] [CrossRef]
- Benveniste, E.N.; Liu, Y.; McFarland, B.C.; Qin, H. Involvement of the Janus Kinase/Signal Transducer and Activator of Transcription Signaling Pathway in Multiple Sclerosis and the Animal Model of Experimental Autoimmune Encephalomyelitis. J. Interferon. Cytokine Res. 2014, 34, 577–588. [Google Scholar] [CrossRef] [PubMed]
- Faquetti, M.L.; Slappendel, L.; Bigonne, H.; Grisoni, F.; Schneider, P.; Aichinger, G.; Schneider, G.; Sturla, S.J.; Burden, A.M. Baricitinib and Tofacitinib Off-target Profile, with a Focus on Alzheimer’s Disease. Alzheimer’s Dement. N. Y. 2024, 10, e12445. [Google Scholar] [CrossRef] [PubMed]
- Ghasemi Noghabi, P.; Shahini, N.; Salimi, Z.; Ghorbani, S.; Bagheri, Y.; Derakhshanpour, F. Elevated Serum IL-17 A and CCL20 Levels as Potential Biomarkers in Major Psychotic Disorders: A Case-Control Study. BMC Psychiatry 2024, 24, 677. [Google Scholar] [CrossRef]
- Moulton, C.D.; Malys, M.; Hopkins, C.W.P.; Rokakis, A.S.; Young, A.H.; Powell, N. Activation of the Interleukin-23/Th17 Axis in Major Depression: A Systematic Review and Meta-Analysis. Eur. Arch. Psychiatry Clin. Neurosci. 2024, 275, 1653–1673. [Google Scholar] [CrossRef]
- Al-Hakeim, H.K.; Al-Musawi, A.F.; Al-Mulla, A.; Al-Dujaili, A.H.; Debnath, M.; Maes, M. The Interleukin-6/Interleukin-23/T Helper 17-Axis as a Driver of Neuro-Immune Toxicity in the Major Neurocognitive Psychosis or Deficit Schizophrenia: A Precision Nomothetic Psychiatry Analysis. PLoS ONE 2022, 17, e0275839. [Google Scholar] [CrossRef]
- Hughes, H.K.; Moreno, R.J.; Ashwood, P. Innate Immune Dysfunction and Neuroinflammation in Autism Spectrum Disorder (ASD). Brain Behav. Immun. 2023, 108, 245–254. [Google Scholar] [CrossRef]
- Raup-Konsavage, W.M.; Cooper, T.K.; Yochum, G.S. A Role for MYC in Lithium-Stimulated Repair of the Colonic Epithelium After DSS-Induced Damage in Mice. Dig. Dis. Sci. 2016, 61, 410–422. [Google Scholar] [CrossRef]
- Huang, S.; Hu, S.; Liu, S.; Tang, B.; Liu, Y.; Tang, L.; Lei, Y.; Zhong, L.; Yang, S.; He, S. Lithium Carbonate Alleviates Colon Inflammation through Modulating Gut Microbiota and Treg Cells in a GPR43-Dependent Manner. Pharmacol. Res. 2022, 175, 105992. [Google Scholar] [CrossRef]
- van der Logt, E.M.J.; Blokzijl, T.; Diepstra, A.; Peppelenbosch, M.P.; Huls, G.; Faber, K.N.; Dijkstra, G. Lithium Induces Intestinothrophic Effects in the Healthy Colon, but Does Not Ameliorate Dextran Sulfate Sodium-Induced Colitis in Mice. E-SPEN J. 2012, 7, e16–e22. [Google Scholar] [CrossRef]
- Cadwell, K.; Liu, J.Y.; Brown, S.L.; Miyoshi, H.; Loh, J.; Lennerz, J.K.; Kishi, C.; Kc, W.; Carrero, J.A.; Hunt, S.; et al. A Key Role for Autophagy and the Autophagy Gene ATG16L1 in Mouse and Human Intestinal Paneth Cells. Nature 2008, 456, 259–263. [Google Scholar] [CrossRef]
- Puglisi-Allegra, S.; Lazzeri, G.; Busceti, C.L.; Giorgi, F.S.; Biagioni, F.; Fornai, F. Lithium Engages Autophagy for Neuroprotection and Neuroplasticity: Translational Evidence for Therapy. Neurosci. Biobehav. Rev. 2023, 148, 105148. [Google Scholar] [CrossRef] [PubMed]
- Motoi, Y.; Shimada, K.; Ishiguro, K.; Hattori, N. Lithium and Autophagy. ACS Chem. Neurosci. 2014, 5, 434–442. [Google Scholar] [CrossRef]
- Elbaz, E.M.; Essam, R.M.; Ahmed, K.A.; Safwat, M.H. Donepezil Halts Acetic Acid-Induced Experimental Colitis in Rats and Its Associated Cognitive Impairment through Regulating Inflammatory/Oxidative/Apoptotic Cascades: An Add-on to Its Anti-Dementia Activity. Int. Immunopharmacol. 2023, 116, 109841. [Google Scholar] [CrossRef] [PubMed]
- Suparan, K.; Sriwichaiin, S.; Thonusin, C.; Sripetchwandee, J.; Khuanjing, T.; Maneechote, C.; Nawara, W.; Arunsak, B.; Chattipakorn, N.; Chattipakorn, S.C. Donepezil Ameliorates Gut Barrier Disruption in Doxorubicin-Treated Rats. Food Chem. Toxicol. 2024, 189, 114741. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.-B.; Zhao, D.-Y.; Qi, Q.-Q.; Long, X.; Li, X.; Chen, F.-X.; Zuo, X.-L. BDNF Modulates Intestinal Barrier Integrity through Regulating the Expression of Tight Junction Proteins. Neurogastroenterol. Motil. 2017, 29, e12967. [Google Scholar] [CrossRef]
- Zhao, D.Y.; Zhang, W.X.; Qi, Q.Q.; Long, X.; Li, X.; Yu, Y.B.; Zuo, X.L. Brain-Derived Neurotrophic Factor Modulates Intestinal Barrier by Inhibiting Intestinal Epithelial Cells Apoptosis in Mice. Physiol. Res. 2018, 67, 475–485. [Google Scholar] [CrossRef]
- Molska, M.; Mruczyk, K.; Cisek-Woźniak, A.; Prokopowicz, W.; Szydełko, P.; Jakuszewska, Z.; Marzec, K.; Trocholepsza, M. The Influence of Intestinal Microbiota on BDNF Levels. Nutrients 2024, 16, 2891. [Google Scholar] [CrossRef]
- GEO Accession Viewer. Available online: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE16879 (accessed on 8 October 2025).
- Rike, W.; Stern, S. Proteins and Transcriptional Dysregulation of the Brain Extracellular Matrix in Parkinson’s Disease: A Systematic Review. medRxiv 2023. [Google Scholar] [CrossRef]
- Tripathi, M.K.; Ojha, S.K.; Kartawy, M.; Hamoudi, W.; Choudhary, A.; Stern, S.; Aran, A.; Amal, H. The NO Answer for Autism Spectrum Disorder. Adv. Sci. 2023, 10, 2205783. [Google Scholar] [CrossRef]
- Hussein, Y.; Tripathi, U.; Choudhary, A.; Nayak, R.; Peles, D.; Rosh, I.; Djamus, J.; Spiegel, R.; Garin-Shkolnik, T.; Stern, S. Early Maturation and Hyperexcitability Is a Shared Phenotype of Cortical Neurons Derived from Different ASD-Causing Mutations. Transl. Psychiatry 2022, 13, 246. [Google Scholar] [CrossRef] [PubMed]
- Stern, S.; Zhang, L.; Wang, M.; Wright, R.; Rosh, I.; Hussein, Y.; Stern, T.; Choudhary, A.; Tripathi, U.; Reed, P.; et al. Monozygotic Twins Discordant for Schizophrenia Differ in Maturation and Synaptic Transmission. Mol. Psychiatry 2024, 29, 3208–3222. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, T.; Mendes, A.P.D.; Moreira, D.P.; Goulart, E.; Oliveira, D.; Kobayashi, G.S.; Stern, S.; Kok, F.; Marchetto, M.C.; Santos, R.; et al. Inositol Monophosphatase 1 (IMPA1) Mutation in Intellectual Disability Patients Impairs Neurogenesis but Not Gliogenesis. Mol. Psychiatry 2021, 26, 3558–3571. [Google Scholar] [CrossRef] [PubMed]
- Rosh, I.; Tripathi, U.K.; Hussein, Y.; Rike, W.A.; Djamus, J.; Shklyar, B.; Manole, A.; Houlden, H.; Winkler, J.; Gage, F.H.; et al. Synaptic Dysfunction and Extracellular Matrix Dysregulation in Dopaminergic Neurons from Sporadic and E326K-GBA1 Parkinson’s Disease Patients. npj Park. Dis. 2024, 10, 38. [Google Scholar] [CrossRef]
- Manole, A.; Wong, T.; Rhee, A.; Novak, S.; Chin, S.-M.; Tsimring, K.; Paucar, A.; Williams, A.; Newmeyer, T.F.; Schafer, S.T.; et al. NGLY1 Mutations Cause Protein Aggregation in Human Neurons. Cell Rep. 2023, 42, 113466. [Google Scholar] [CrossRef]
- Tripathi, U.; Rosh, I.; Ezer, R.B.; Nayak, R.; Choudhary, A.; Djamus, J.; Manole, A.; Haulden, H.; Gage, F.H.; Stern, S. Upregulated Extracellular Matrix-Related Genes and Impaired Synaptic Activity in Dopaminergic and Hippocampal Neurons Derived from Parkinson’s Disease Patients with PINK1 and PARK2 Mutations. bioRxiv 2023. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tripathi, U.; Stern, Y.; Dagan, I.; Nayak, R.; Romanovsky, E.; Zittan, E.; Stern, S. From Genome-Wide SNPs to Neuroimmune Crosstalk: Mapping the Genetic Landscape of IBD and Its Brain Overlap. Biology 2025, 14, 1433. https://doi.org/10.3390/biology14101433
Tripathi U, Stern Y, Dagan I, Nayak R, Romanovsky E, Zittan E, Stern S. From Genome-Wide SNPs to Neuroimmune Crosstalk: Mapping the Genetic Landscape of IBD and Its Brain Overlap. Biology. 2025; 14(10):1433. https://doi.org/10.3390/biology14101433
Chicago/Turabian StyleTripathi, Utkarsh, Yam Stern, Inbal Dagan, Ritu Nayak, Eva Romanovsky, Eran Zittan, and Shani Stern. 2025. "From Genome-Wide SNPs to Neuroimmune Crosstalk: Mapping the Genetic Landscape of IBD and Its Brain Overlap" Biology 14, no. 10: 1433. https://doi.org/10.3390/biology14101433
APA StyleTripathi, U., Stern, Y., Dagan, I., Nayak, R., Romanovsky, E., Zittan, E., & Stern, S. (2025). From Genome-Wide SNPs to Neuroimmune Crosstalk: Mapping the Genetic Landscape of IBD and Its Brain Overlap. Biology, 14(10), 1433. https://doi.org/10.3390/biology14101433





