Development and Validation of a 7-eRNA Prognostic Signature for Lung Adenocarcinoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection and Preprocessing
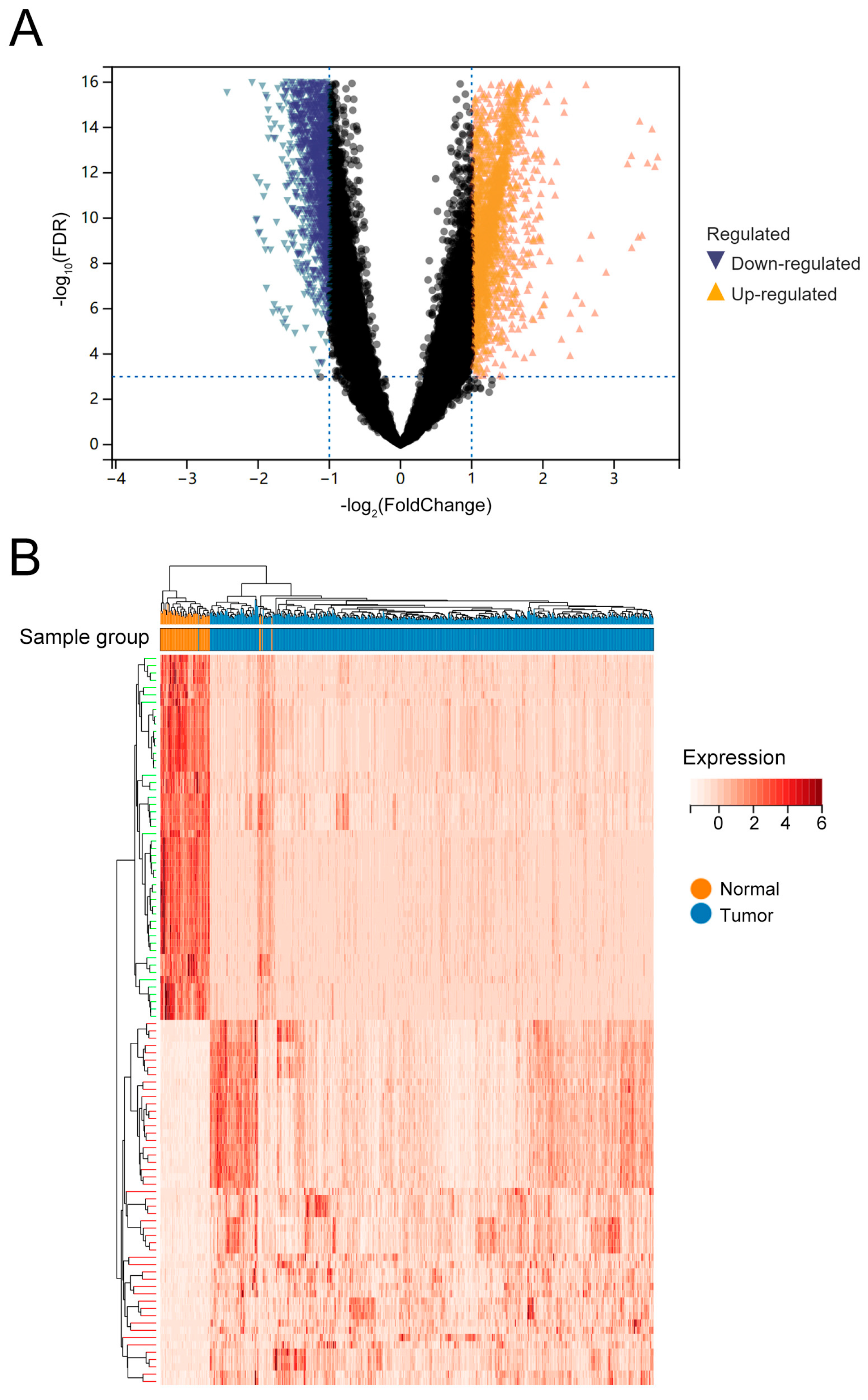
2.2. Differential Expression Analysis of eRNAs
2.3. Establishing and Evaluating Prognostic 7-eRNA Model
2.4. Gene Set Enrichment Analysis (GSEA)
2.5. Drug Sensitivity Analysis
2.6. Tumor Immune Microenvironment and Somatic Mutation Analysis
2.7. eRNA-Gene Interaction Analysis
3. Results
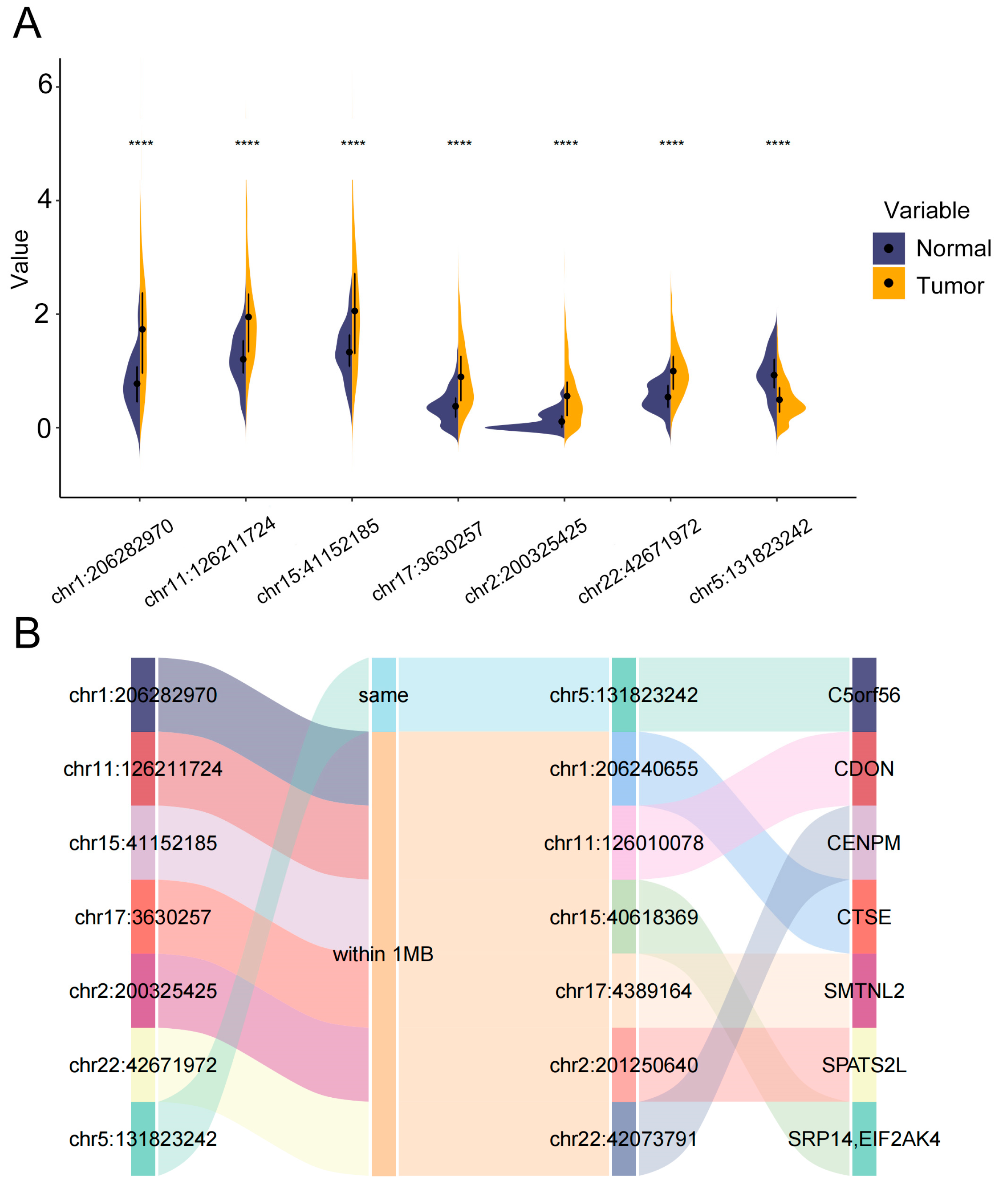
3.1. Differentially Expressed of 6280 eRNAs in LUAD
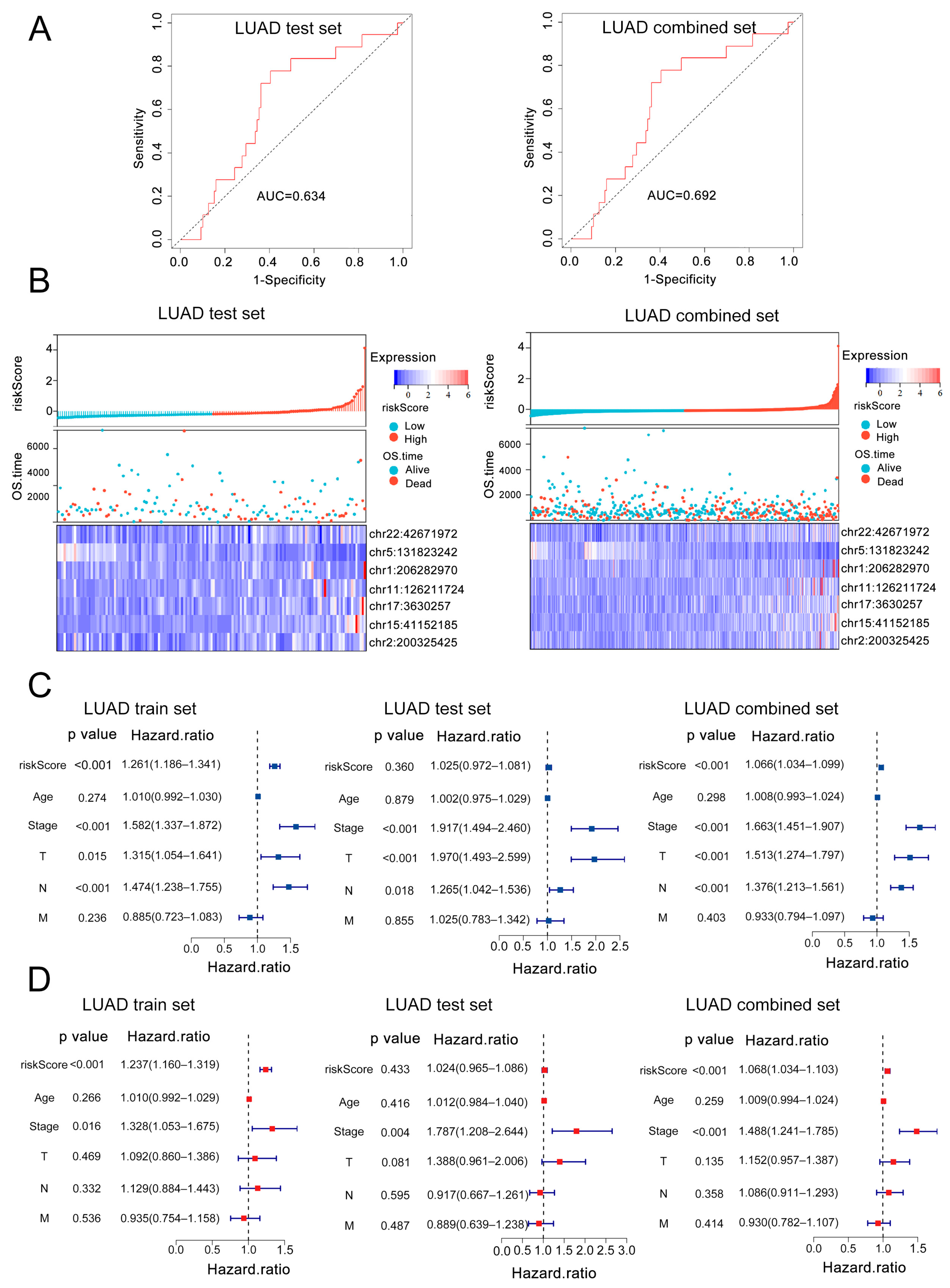
3.2. Construction and Validation of a 7-eRNA Prognostic Model for LUAD
3.3. External Validation and Independent Prognostic Value of the 7-eRNA Model in LUAD
3.4. Clinical Nomogram Integrating 7-eRNA Risk Score and Clinical Factors Enhances LUAD Prognosis Prediction
3.5. Pathway Enrichment in High-Risk LUAD Patients
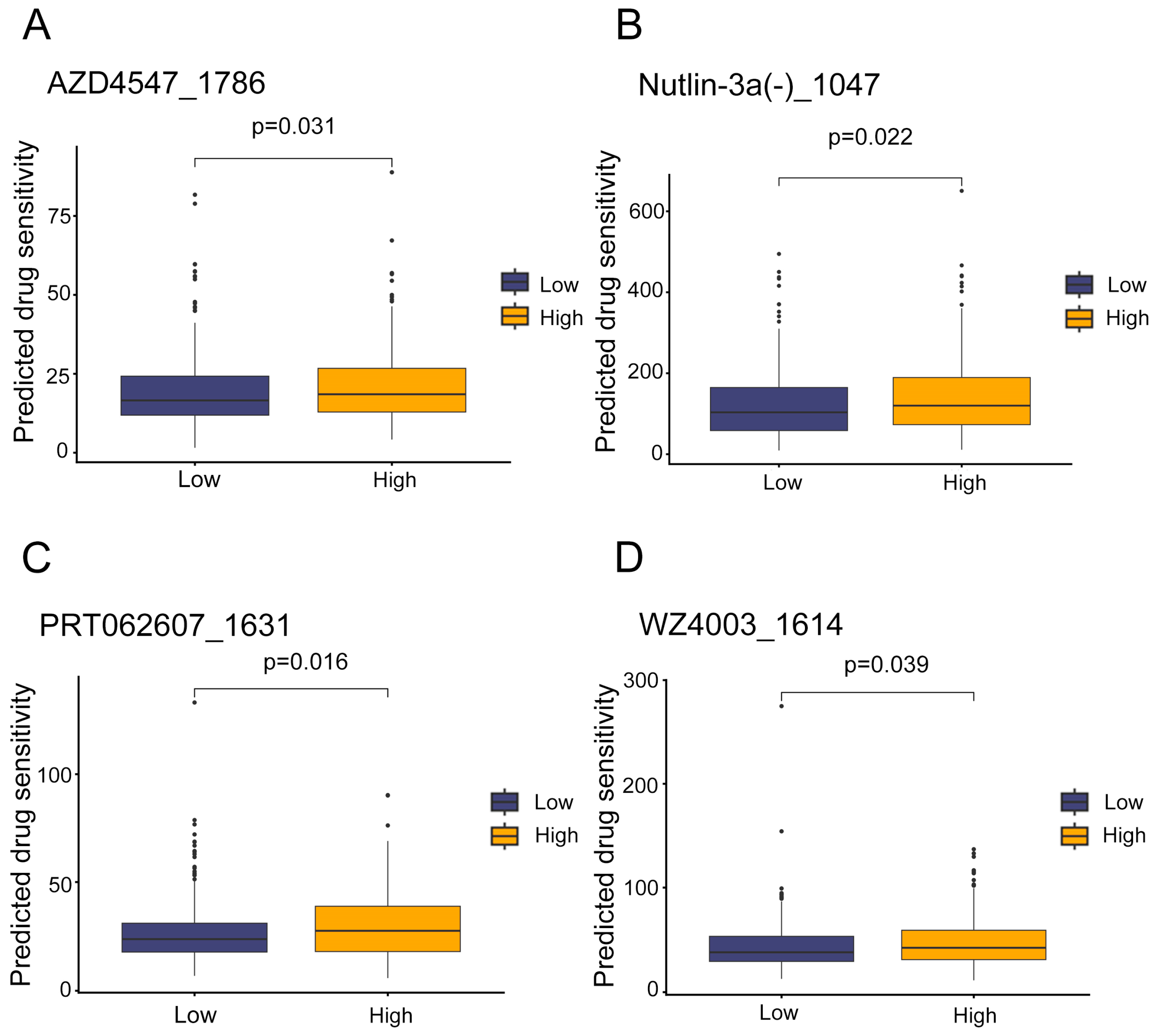
3.6. Potential Therapeutic Agents for High-Risk LUAD Patients
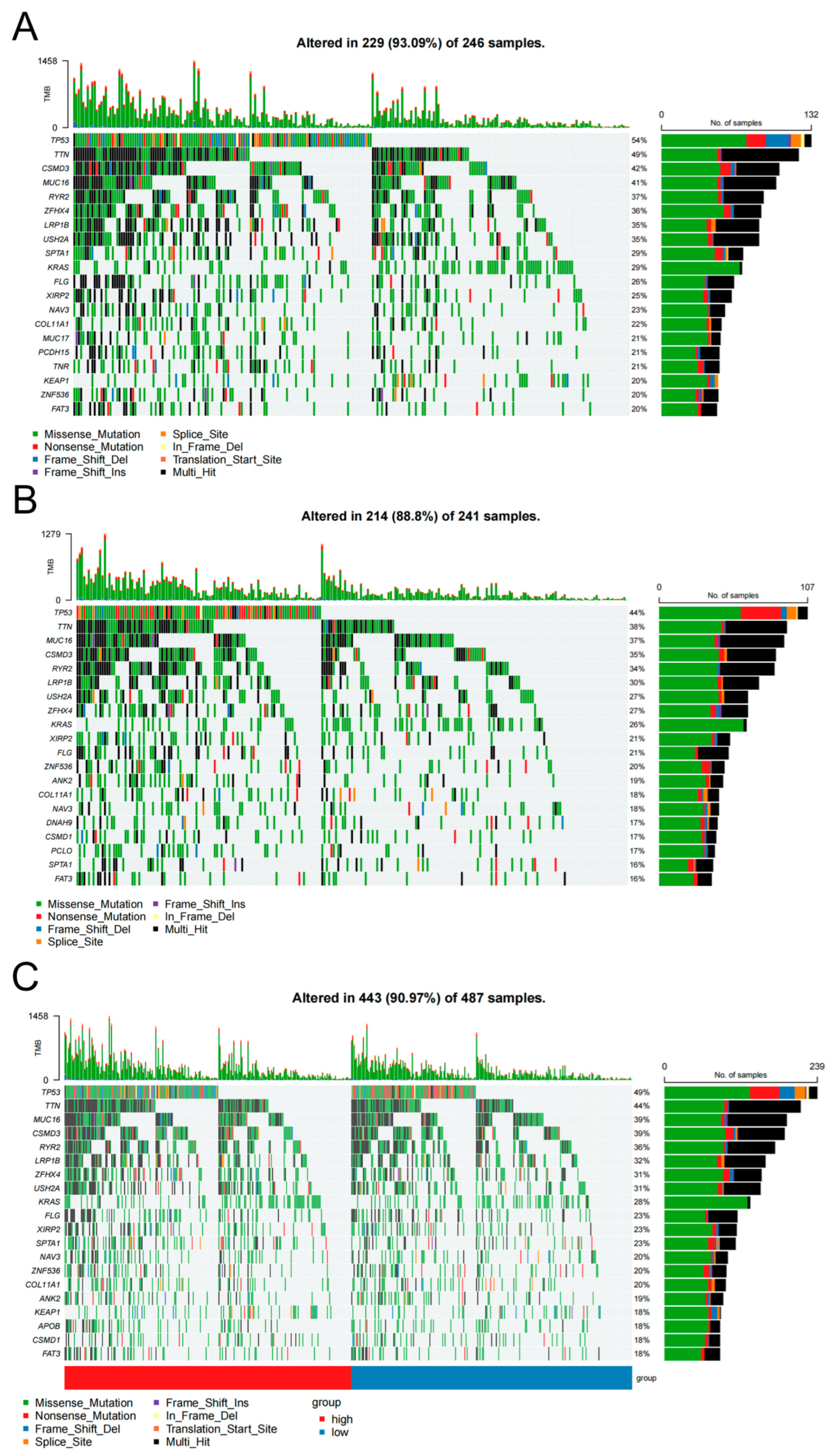
3.7. Somatic Mutation Differences Distinguish High- and Low-Risk LUAD Groups
3.8. Differential Expression and Functional Annotations of eRNAs in the 7-eRNA Model for LUAD
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| LUAD | Lung adenocarcinoma |
| eRNA | Enhancer RNA |
| TCGA | The Cancer Genome Atlas |
| NSCLC | Non-small cell lung cancer |
| KM | Kaplan–Meier |
| ROC | Receiver operating characteristic |
| OS | Overall survival |
| GDSC | Genomics of Drug Sensitivity in Cancer |
| ssGSEA | Single-sample gene set enrichment analysis |
| TNM | tumor node metastasis |
| AUC | Area under the curve |
| IC50 | Half-maximal inhibitory concentration |
| DEG | Differentially expressed genes |
| TIME | Tumor immune microenvironment |
| IBD | Inflammatory bowel disease |
| PVOD | Pulmonary veno-occlusive disease |
| CEA | Carcinoembryonic antigen |
| NSE | Neuron-specific enolase |
| ctDNA | Circulating tumor DNA |
| CTCs | Circulating tumor cells |
| Hi-C | High-throughput chromosomal conformation capture |
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Duma, N.; Santana-Davila, R.; Molina, J.R. Non–small cell lung cancer: Epidemiology, screening, diagnosis, and treatment. Mayo Clin. Proc. 2019, 94, 1623–1640. [Google Scholar] [CrossRef] [PubMed]
- Strand, T.-E.; Rostad, H.; Møller, B.; Norstein, J. Survival after resection for primary lung cancer: A population based study of 3211 resected patients. Thorax 2006, 61, 710–715. [Google Scholar] [CrossRef]
- Goldstraw, P.; Chansky, K.; Crowley, J.; Rami-Porta, R.; Asamura, H.; Eberhardt, W.E.; Nicholson, A.G.; Groome, P.; Mitchell, A.; Bolejack, V. The IASLC lung cancer staging project: Proposals for revision of the TNM stage groupings in the forthcoming (eighth) edition of the TNM classification for lung cancer. J. Thorac. Oncol. 2016, 11, 39–51. [Google Scholar] [CrossRef]
- Zhou, Y.; Tao, L.; Qiu, J.; Xu, J.; Yang, X.; Zhang, Y.; Tian, X.; Guan, X.; Cen, X.; Zhao, Y.; et al. Tumor biomarkers for diagnosis, prognosis and targeted therapy. Signal Transduct. 2024, 9, 132. [Google Scholar] [CrossRef]
- Seijo, L.M.; Peled, N.; Ajona, D.; Boeri, M.; Field, J.K.; Sozzi, G.; Pio, R.; Zulueta, J.J.; Spira, A.; Massion, P.; et al. Biomarkers in lung cancer screening: Achievements, promises, and challenges. J. Thorac. Oncol. 2019, 14, 343–357. [Google Scholar] [CrossRef]
- Oliver, J.; Garcia-Aranda, M.; Chaves, P.; Alba, E.; Cobo-Dols, M.; Onieva, J.L.; Barragan, I. Emerging noninvasive methylation biomarkers of cancer prognosis and drug response prediction. Semin. Cancer Biol. 2022, 83, 584–595. [Google Scholar] [CrossRef] [PubMed]
- Ferraro, S.; Biganzoli, E.M.; Castaldi, S.; Plebani, M. Health technology assessment to assess value of biomarkers in the decision-making process. Clin. Chem. Lab. Med. 2022, 60, 647–654. [Google Scholar] [CrossRef]
- Han, Z.; Li, W.J.C.P. Enhancer RNA: What we know and what we can achieve. Cell Prolif. 2022, 55, e13202. [Google Scholar] [CrossRef]
- Sartorelli, V.; Lauberth, S.M. Enhancer RNAs are an important regulatory layer of the epigenome. Nat. Struct. Mol. Biol. 2020, 27, 521–528. [Google Scholar] [CrossRef]
- Christophorou, M.; Ringshausen, I.; Finch, A.; Swigart, L.B.; Evan, G.J. The pathological response to DNA damage does not contribute to p53-mediated tumour suppression. Nature 2006, 443, 214–217. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Notani, D.; Rosenfeld, M.G. Enhancers as non-coding RNA transcription units: Recent insights and future perspectives. Nat. Rev. Genet. 2016, 17, 207–223. [Google Scholar] [CrossRef]
- Mousavi, K.; Zare, H.; Dell’Orso, S.; Grontved, L.; Gutierrez-Cruz, G.; Derfoul, A.; Hager, G.L.; Sartorelli, V. eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol. Cell 2013, 51, 606–617. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, A.; Mimura, J.; Itoh, K. Non-coding RNA derived from the region adjacent to the human HO-1 E2 enhancer selectively regulates HO-1 gene induction by modulating Pol II binding. Nucleic Acids Res. 2014, 42, 13599–13614. [Google Scholar] [CrossRef]
- Li, W.; Notani, D.; Ma, Q.; Tanasa, B.; Nunez, E.; Chen, A.Y.; Merkurjev, D.; Zhang, J.; Ohgi, K.; Song, X. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature 2013, 498, 516–520. [Google Scholar] [CrossRef]
- Li, W.; Hu, Y.; Oh, S.; Ma, Q.; Merkurjev, D.; Song, X.; Zhou, X.; Liu, Z.; Tanasa, B.; He, X. Condensin I and II complexes license full estrogen receptor α-dependent enhancer activation. Mol. Cell 2015, 59, 188–202. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Shen, W.; Song, Z.; Zhong, X.; Huang, M.; Shen, D.; Gao, P.; Qian, X.; Wang, M.; He, X.; Wang, T. Sangerbox: A comprehensive, interaction-friendly clinical bioinformatics analysis platform. Imeta 2022, 1, e36. [Google Scholar] [CrossRef]
- Ranstam, J.; Cook, J.A. LASSO regression. J. Br. Surg. 2018, 105, 1348. [Google Scholar] [CrossRef]
- Rich, J.T.; Neely, J.G.; Paniello, R.C.; Voelker, C.C.; Nussenbaum, B.; Wang, E.W. A practical guide to understanding Kaplan-Meier curves. Otolaryngol. Head Neck Surg. 2010, 143, 331–336. [Google Scholar] [CrossRef] [PubMed]
- Maeser, D.; Gruener, R.F.; Huang, R.S. oncoPredict: An R package for predicting in vivo or cancer patient drug response and biomarkers from cell line screening data. Brief. Bioinform. 2021, 22, bbab260. [Google Scholar] [CrossRef]
- Wang, Z.-W.; Zou, F.-M.; Wang, A.-L.; Yang, J.; Jin, R.; Wang, B.-L.; Shen, L.-J.; Qi, S.; Liu, J.; Liu, J. Repurposing of the FGFR inhibitor AZD4547 as a potent inhibitor of necroptosis by selectively targeting RIPK1. Acta Pharmacol. Sin. 2023, 44, 801–810. [Google Scholar] [CrossRef]
- Wiechens, E.; Vigliotti, F.; Siniuk, K.; Schwarz, R.; Schwab, K.; Riege, K.; van Bömmel, A.; Görlich, I.; Bens, M.; Sahm, A.; et al. Gene regulation by convergent promoters. Nat. Genet. 2025, 57, 206–217. [Google Scholar] [CrossRef]
- Coffey, G.; DeGuzman, F.; Inagaki, M.; Pak, Y.; Delaney, S.M.; Ives, D.; Betz, A.; Jia, Z.J.; Pandey, A.; Baker, D. Specific inhibition of spleen tyrosine kinase suppresses leukocyte immune function and inflammation in animal models of rheumatoid arthritis. J. Pharmacol. Exp. Ther. 2012, 340, 350–359. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Buhrlage, S.J.; Huang, H.-T.; Deng, X.; Zhou, W.; Wang, J.; Traynor, R.; Prescott, A.R.; Alessi, D.R.; Gray, N.S. Characterization of WZ4003 and HTH-01-015 as selective inhibitors of the LKB1-tumour-suppressor-activated NUAK kinases. Biochem. J. 2014, 457, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Zubair, T.; Bandyopadhyay, D. Small molecule EGFR inhibitors as anti-cancer agents: Discovery, mechanisms of action, and opportunities. Int. J. Mol. Sci. 2023, 24, 2651. [Google Scholar] [CrossRef]
- Cao, Q.; Wang, N.; Ren, L.; Tian, J.; Yang, S.; Cheng, H. miR-125a-5p post-transcriptionally suppresses GALNT7 to inhibit proliferation and invasion in cervical cancer cells via the EGFR/PI3K/AKT pathway. Cancer Cell Int. 2020, 20, 117. [Google Scholar] [CrossRef]
- Burotto, M.; Chiou, V.L.; Lee, J.M.; Kohn, E.C. The MAPK pathway across different malignancies: A new perspective. Cancer 2014, 120, 3446–3456. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Liu, W.-Z.; Liu, T.; Feng, X.; Yang, N.; Zhou, H.-F. Signaling pathway of MAPK/ERK in cell proliferation, differentiation, migration, senescence and apoptosis. J. Recept. Signal Transduct. 2015, 35, 600–604. [Google Scholar] [CrossRef]
- Chang, F.; Lee, J.; Navolanic, P.; Steelman, L.; Shelton, J.; Blalock, W.; Franklin, R.; McCubrey, J.A. Involvement of PI3K/Akt pathway in cell cycle progression, apoptosis, and neoplastic transformation: A target for cancer chemotherapy. Leukemia 2003, 17, 590–603. [Google Scholar] [CrossRef]
- Yoshihara, K.; Shahmoradgoli, M.; Martínez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Treviño, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013, 4, 2612. [Google Scholar] [CrossRef] [PubMed]
- Mogi, A.; Kuwano, H. TP53 mutations in nonsmall cell lung cancer. BioMed Res. Int. 2011, 2011, 583929. [Google Scholar] [CrossRef]
- Xue, D.; Lin, H.; Lin, L.; Wei, Q.; Yang, S.; Chen, X. TTN/TP53 mutation might act as the predictor for chemotherapy response in lung adenocarcinoma and lung squamous carcinoma patients. Transl. Cancer Res. 2021, 10, 1284. [Google Scholar] [CrossRef] [PubMed]
- Pontious, C.; Kaul, S.; Hong, M.; Hart, P.A.; Krishna, S.G.; Lara, L.F.; Conwell, D.L.; Cruz-Monserrate, Z. Cathepsin E expression and activity: Role in the detection and treatment of pancreatic cancer. Pancreatology 2019, 19, 951–956. [Google Scholar] [CrossRef] [PubMed]
- Kawakubo, T.; Okamoto, K.; Iwata, J.-I.; Shin, M.; Okamoto, Y.; Yasukochi, A.; Nakayama, K.I.; Kadowaki, T.; Tsukuba, T.; Yamamoto, K. Cathepsin E prevents tumor growth and metastasis by catalyzing the proteolytic release of soluble TRAIL from tumor cell surface. Cancer Res. 2007, 67, 10869–10878. [Google Scholar] [CrossRef] [PubMed]
- Yasukochi, A.; Kawakubo, T.; Nakamura, S.; Yamamoto, K. Cathepsin E enhances anticancer activity of doxorubicin on human prostate cancer cells showing resistance to TRAIL-mediated apoptosis. Biol. Chem. 2010, 391, 947–958. [Google Scholar] [CrossRef]
- Ullmann, R.; Morbini, P.; Halbwedl, I.; Bongiovanni, M.; Gogg-Kammerer, M.; Papotti, M.; Gabor, S.; Renner, H.; Popper, H.H. Protein expression profiles in adenocarcinomas and squamous cell carcinomas of the lung generated using tissue microarrays. J. Pathol. J. Pathol. Soc. Great Br. Irel. 2004, 203, 798–807. [Google Scholar] [CrossRef]
- Abd-Elgaliel, W.R.; Cruz-Monserrate, Z.; Wang, H.; Logsdon, C.D.; Tung, C.-H. Pancreatic cancer-associated Cathepsin E as a drug activator. J. Control. Release 2013, 167, 221–227. [Google Scholar] [CrossRef]
- John, C.; Guyatt, A.L.; Shrine, N.; Packer, R.; Olafsdottir, T.A.; Liu, J.; Hayden, L.P.; Chu, S.H.; Koskela, J.T.; Luan, J.; et al. Genetic associations and architecture of asthma-COPD overlap. Chest 2022, 161, 1155–1166. [Google Scholar] [CrossRef]
- Ma, H.; Hu, T.; Tao, W.; Tong, J.; Han, Z.; Herndler-Brandstetter, D.; Wei, Z.; Liu, R.; Zhou, T.; Liu, Q.; et al. A lncRNA from an inflammatory bowel disease risk locus maintains intestinal host-commensal homeostasis. Cell Res. 2023, 33, 372–388. [Google Scholar] [CrossRef]
- Faoro, C.; Ataide, S.F. Noncanonical functions and cellular dynamics of the mammalian signal recognition particle components. Front. Mol. Biosci. 2021, 8, 679584. [Google Scholar] [CrossRef]
- Gussakovsky, D.; Black, N.A.; Booy, E.P.; McKenna, S.A. The role of SRP9/SRP14 in regulating Alu RNA. RNA Biol. 2024, 21, 1239–1250. [Google Scholar] [CrossRef]
- Eyries, M.; Montani, D.; Girerd, B.; Perret, C.; Leroy, A.; Lonjou, C.; Chelghoum, N.; Coulet, F.; Bonnet, D.; Dorfmüller, P.; et al. EIF2AK4 mutations cause pulmonary veno-occlusive disease, a recessive form of pulmonary hypertension. Nat. Genet. 2014, 46, 65–69. [Google Scholar] [CrossRef] [PubMed]
- Grunnet, M.; Sorensen, J. Carcinoembryonic antigen (CEA) as tumor marker in lung cancer. Lung Cancer 2012, 76, 138–143. [Google Scholar] [CrossRef] [PubMed]
- Pavićević, R.; Miličić, J.; Bubanović, G.; Šupe, S. Serum Tumor Marker CYFRA 21-1 in the Diagnostics of NSCLC Lung. Coll. Antropol. 1998, 22, 629–635. [Google Scholar]
- Isgrò, M.A.; Bottoni, P.; Scatena, R. Neuron-specific enolase as a biomarker: Biochemical and clinical aspects. Adv. Cancer Biomark. Biochem. Clin. A Crit. Revis. 2015, 867, 125–143. [Google Scholar]
- Araki, T.; Kanda, S.; Horinouchi, H.; Ohe, Y. Current treatment strategies for EGFR-mutated non-small cell lung cancer: From first line to beyond osimertinib resistance. Jpn. J. Clin. Oncol. 2023, 53, 547–561. [Google Scholar] [CrossRef]
- Sasaki, T.; Kuno, H.; Hiyama, T.; Oda, S.; Masuoka, S.; Miyasaka, Y.; Taki, T.; Nagasaki, Y.; Ohtani-Kim, S.J.-Y.; Ishii, G.; et al. 2021 WHO classification of lung cancer: Molecular biology research and radiologic-pathologic correlation. Radiographics 2024, 44, e230136. [Google Scholar] [CrossRef]
- Li, W.; Chang, F.; Zhang, H.; Meng, F.; Ke, Z.; Zhang, Y. Clinical Pathological Characteristics and Prognosis of Multigene Co-Mutations in Elderly Patients With Non-Small Cell Lung Cancer: A Retrospective Analysis. Clin. Med. Insights Oncol. 2023, 17, 11795549231211505. [Google Scholar] [CrossRef]
- Vallejo, J.; Singh, H.; Larkins, E.; Drezner, N.; Ricciuti, B.; Mishra-Kalyani, P.; Tang, S.; Beaver, J.A.; Awad, M. Impact of increasing PD-L1 levels on outcomes to PD-1/PD-L1 inhibition in patients with NSCLC: A pooled analysis of 11 prospective clinical trials. Oncologist 2024, 29, 422–430. [Google Scholar] [CrossRef]
- Zhang, N.; Yang, X.; Piao, M.; Xun, Z.; Wang, Y.; Ning, C.; Zhang, X.; Zhang, L.; Wang, Y.; Wang, S.; et al. Biomarkers and prognostic factors of PD-1/PD-L1 inhibitor-based therapy in patients with advanced hepatocellular carcinoma. Biomark. Res. 2024, 12, 26. [Google Scholar] [CrossRef] [PubMed]
- Klug, M.; Kirshenboim, Z.; Truong, M.T.; Sorin, V.; Ofek, E.; Agrawal, R.; Marom, E.M. Proposed Ninth Edition TNM Staging System for Lung Cancer: Guide for Radiologists. RadioGraphics 2024, 44, e240057. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Luo, J.; Wu, S.; Si, H.; Gao, C.; Xu, W.; Abdullah, S.E.; Higgs, B.W.; Dennis, P.A.; van der Heijden, M.S.; et al. Prognostic and predictive impact of circulating tumor DNA in patients with advanced cancers treated with immune checkpoint blockade. Cancer Discov. 2020, 10, 1842–1853. [Google Scholar] [CrossRef]
- Capuozzo, M.; Ferrara, F.; Santorsola, M.; Zovi, A.; Ottaiano, A. Circulating tumor cells as predictive and prognostic biomarkers in solid tumors. Cells 2023, 12, 2590. [Google Scholar] [CrossRef]
- Fang, W.; Jin, H.; Zhou, H.; Hong, S.; Ma, Y.; Zhang, Y.; Su, X.; Chen, L.; Yang, Y.; Xu, S. Intratumoral heterogeneity as a predictive biomarker in anti-PD-(L) 1 therapies for non-small cell lung cancer. Mol. Cancer 2021, 20, 37. [Google Scholar] [CrossRef]
- Zhang, Z.; Hong, W.; Ruan, H.; Jing, Y.; Li, S.; Liu, Y.; Wang, J.; Li, W.; Diao, L.; Han, L. HeRA: An atlas of enhancer RNAs across human tissues. Nucleic Acids Res. 2021, 49, D932–D938. [Google Scholar] [CrossRef]
- Zhang, Z.; Lee, J.-H.; Ruan, H.; Ye, Y.; Krakowiak, J.; Hu, Q.; Xiang, Y.; Gong, J.; Zhou, B.; Wang, L.; et al. Transcriptional landscape and clinical utility of enhancer RNAs for eRNA-targeted therapy in cancer. Nat. Commun. 2019, 10, 4562. [Google Scholar] [CrossRef] [PubMed]
- Feng, G.; Wang, T.; Xue, F.; Qi, Y.; Wang, R.; Yuan, H. Identification of enhancer RNAs for the prognosis of head and neck squamous cell carcinoma. Head Neck 2021, 43, 3820–3831. [Google Scholar] [CrossRef]
- Cai, S.; Hu, X.; Chen, R.; Zhang, Y. Identification and validation of an immune-related eRNA prognostic signature for hepatocellular carcinoma. Front. Genet. 2021, 12, 657051. [Google Scholar] [CrossRef]
- Wu, P.; Shi, J.; Wang, Z.; Sun, W.; Zhang, H. Evaluate the immune-related eRNA models and signature score to predict the response to immunotherapy in thyroid carcinoma. Cancer Cell Int. 2022, 22, 307. [Google Scholar] [CrossRef]
- Monahan, K.; Horta, A.; Lomvardas, S. LHX2-and LDB1-mediated trans interactions regulate olfactory receptor choice. Nature 2019, 565, 448–453. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.-S.; Zheng, Y.-Z.; Qin, X.-Y.; Yang, X.-P.; Shen, P.; Cai, W.-J.; Li, X.-Q.; Liao, H.-Y. Establishing a TNM-like risk classification for metachronous second pulmonary adenocarcinoma in patients with previously resected pulmonary adenocarcinoma. J. Thorac. Dis. 2022, 14, 1306. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Chen, Z.; Li, T.; Feng, D.; Liu, X.; Wang, X.; Han, H.; Yu, L.; Li, X.; Li, B.; et al. Enhancer RNA in cancer: Identification, expression, resources, relationship with immunity, drugs, and prognosis. Brief. Funct. Genom. 2025, 24, elaf007. [Google Scholar] [CrossRef] [PubMed]



| Clinical Information | Classification | Numerical Code |
|---|---|---|
| Survival status | Alive | 0 |
| Dead | 1 | |
| Stage | Stage Ⅰ | 1 |
| Stage Ⅱ | 2 | |
| Stage Ⅲ | 3 | |
| Stage Ⅳ | 4 | |
| T Classification | T1 | 1 |
| T2 | 2 | |
| T3 | 3 | |
| T4 | 4 | |
| TX | 5 | |
| N Classification | N0 | 0 |
| N1 | 1 | |
| N2 | 2 | |
| N3 | 3 | |
| M Classification | M0 | 0 |
| M1 | 1 | |
| MX | 2 |
| ID | Coefficient | HR | 95% CI | p Value |
|---|---|---|---|---|
| chr1:206282970 | 0.04578 | 1.04684 | 1.02279–1.07146 | 0.00011 |
| chr2:200325425 | 0.20554 | 1.22819 | 0.98391–1.53313 | 0.06928 |
| chr5:131823242 | −1.11341 | 0.32844 | 0.17302–0.62344 | 0.00066 |
| chr11:126211724 | 0.02812 | 1.02852 | 0.99748–1.06053 | 0.07210 |
| chr15:41152185 | 0.05767 | 1.05936 | 1.02252–1.09753 | 0.00141 |
| chr17:3630257 | 0.22138 | 1.24780 | 1.02789–1.51475 | 0.02521 |
| chr22:42671972 | 0.24401 | 1.27636 | 1.02369–1.59140 | 0.03016 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Y.; Chen, K.; Zhang, J.; Hu, Z.; Xiong, M.; Fang, Z.; Chen, G.; Meng, X.; Liao, B.; Xiong, Y.; et al. Development and Validation of a 7-eRNA Prognostic Signature for Lung Adenocarcinoma. Biology 2025, 14, 1431. https://doi.org/10.3390/biology14101431
Sun Y, Chen K, Zhang J, Hu Z, Xiong M, Fang Z, Chen G, Meng X, Liao B, Xiong Y, et al. Development and Validation of a 7-eRNA Prognostic Signature for Lung Adenocarcinoma. Biology. 2025; 14(10):1431. https://doi.org/10.3390/biology14101431
Chicago/Turabian StyleSun, Yiwen, Keng Chen, Jingkai Zhang, Zhijie Hu, Mingmei Xiong, Zhigang Fang, Guanmei Chen, Xiaomei Meng, Baolin Liao, Yuanyan Xiong, and et al. 2025. "Development and Validation of a 7-eRNA Prognostic Signature for Lung Adenocarcinoma" Biology 14, no. 10: 1431. https://doi.org/10.3390/biology14101431
APA StyleSun, Y., Chen, K., Zhang, J., Hu, Z., Xiong, M., Fang, Z., Chen, G., Meng, X., Liao, B., Xiong, Y., & Lin, L. (2025). Development and Validation of a 7-eRNA Prognostic Signature for Lung Adenocarcinoma. Biology, 14(10), 1431. https://doi.org/10.3390/biology14101431







