Biparental Inheritance and Instability of kDNA in Experimental Hybrids of Trypanosoma cruzi: A Proposal for a Mechanism
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Source
2.2. Sequence Data Processing and mHVR Frequency Estimation
2.3. Variant Calling in Maxicircles
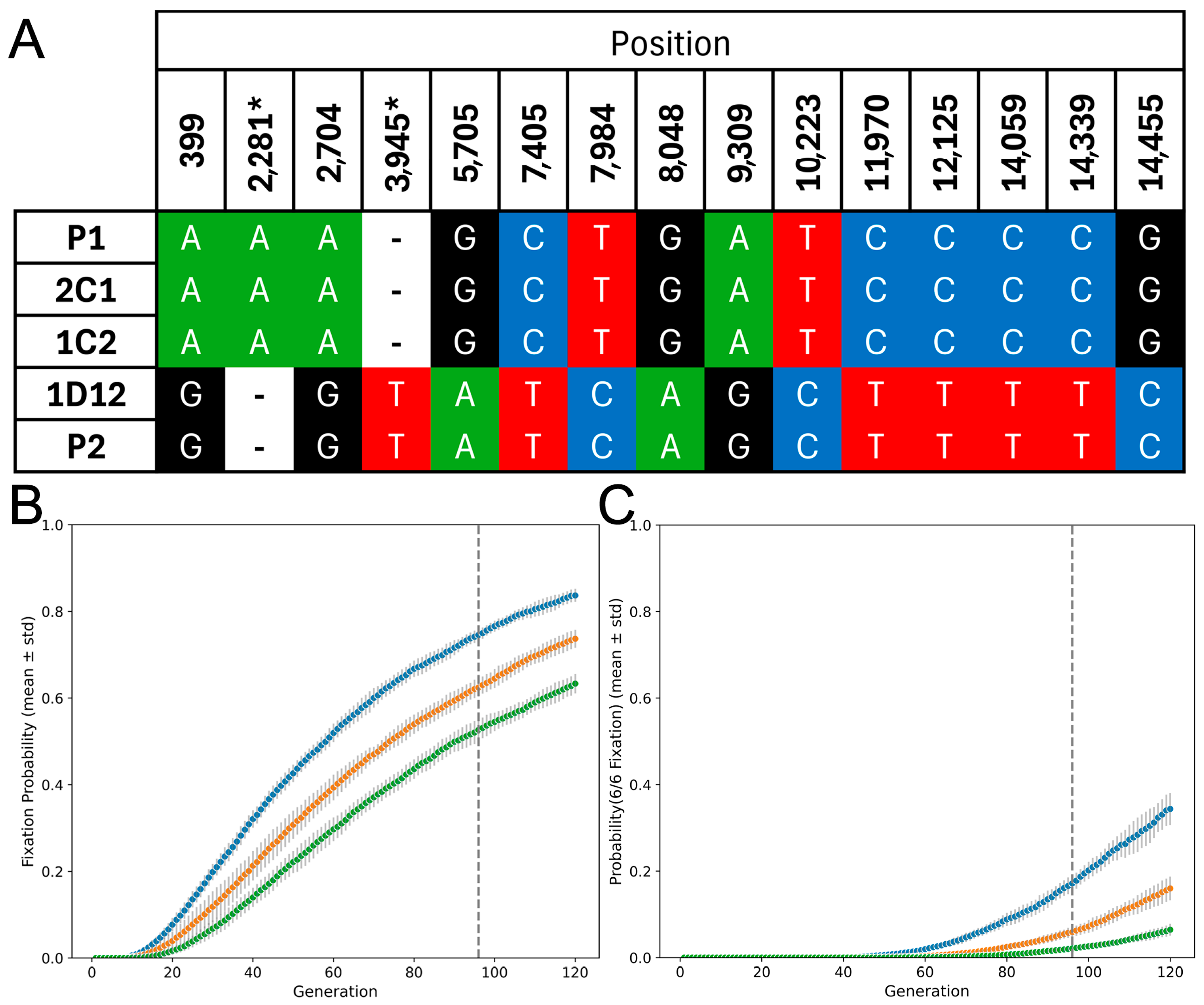
2.4. Simulation Model for Maxicircle Drift
2.5. kDNA Quantification by Read Depth
2.6. kDNA Quantification from PI Flow Cytometry and Nuclear Read Depth
2.7. Estimation of Parental Contributions
3. Results
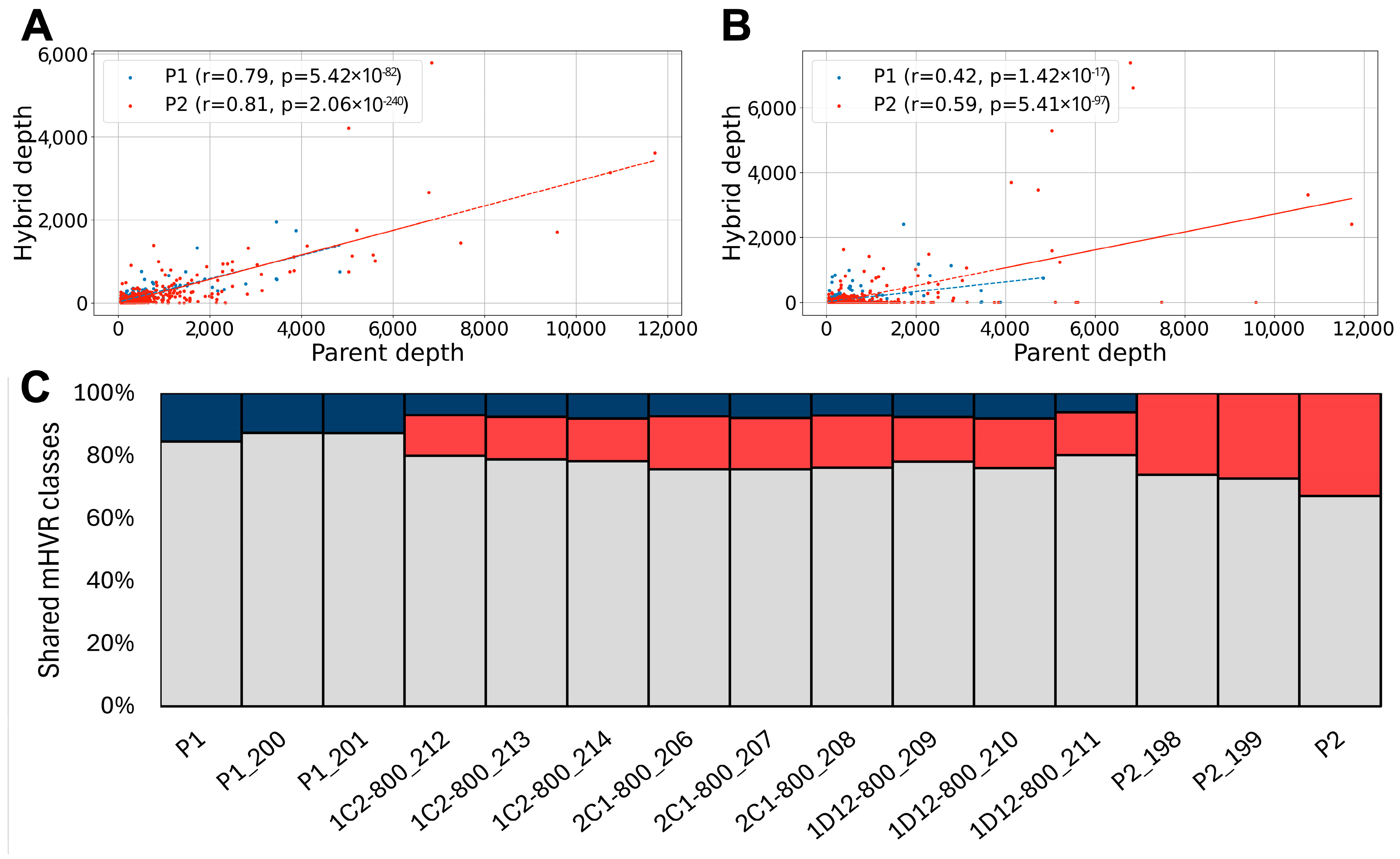
3.1. Experimental Hybrids Exhibit mHVRs from Both Parents
3.2. Minicircles from Both Parents Persist After 800 Generations in the Hybrids
3.3. Hybrids Retain Maxicircles from a Single Parent
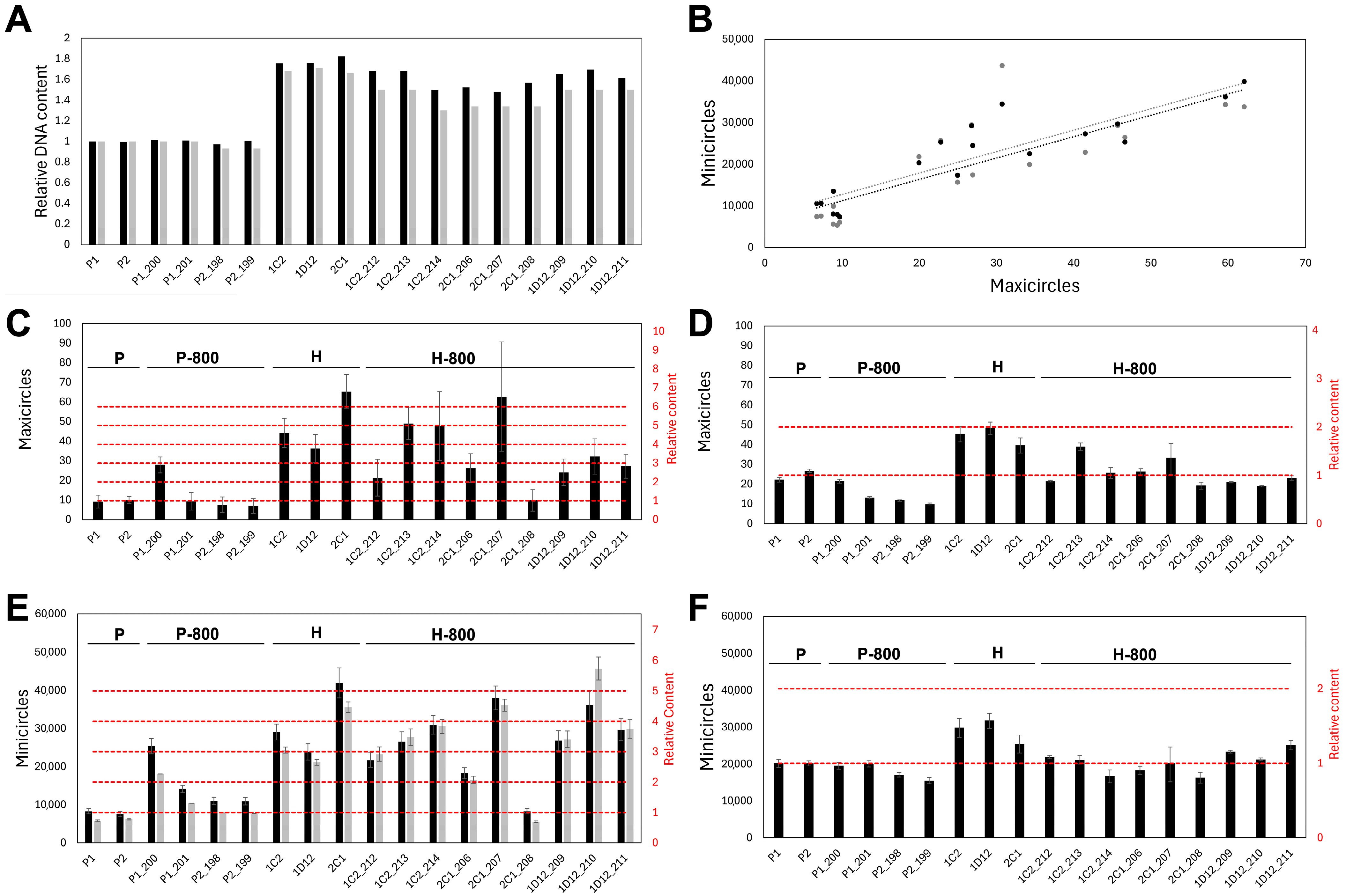
3.4. kDNA Content Is Increased and Unstable in Hybrids
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| mHVR | Minicircle hypervariable region |
| kDNA | Kinetoplastic DNA |
| PI | Propidium Iodide |
| BB | Basal body |
| TAC | Tripartite attachment complex |
| APS | Antipodal sites |
References
- Shapiro, T.A.; Englund, P.T. The structure and replication of kinetoplast DNA. Annu. Rev. Microbiol. 1995, 49, 117–143. [Google Scholar] [CrossRef]
- Simpson, L.; Sbicego, S.; Aphasizhev, R. Uridine insertion/deletion RNA editing in trypanosome mitochondria: A complex business. RNA 2003, 9, 265–276. [Google Scholar] [CrossRef]
- Aphasizheva, I.; Alfonzo, J.; Carnes, J.; Cestari, I.; Cruz-Reyes, J.; Göringer, H.U.; Hajduk, S.; Lukeš, J.; Madison-Antenucci, S.; Maslov, D.A.; et al. Lexis and grammar of mitochondrial RNA processing in trypanosomes. Trends Parasitol. 2020, 36, 337–355. [Google Scholar] [CrossRef]
- Rusman, F.; Floridia-Yapur, N.; Tomasini, N.; Diosque, P. Guide RNA Repertoires in the Main Lineages of Trypanosoma cruzi: High Diversity and Variable Redundancy Among Strains. Front. Cell. Infect. Microbiol. 2021, 11, 663416. [Google Scholar] [CrossRef]
- Zimmer, S.L. Revisiting trypanosome mitochondrial genome mysteries: Broader and deeper. Trends Parasitol. 2019, 35, 102–104. [Google Scholar] [CrossRef]
- Schnaufer, A.; Panigrahi, A.K.; Panicucci, B.; Igo, R.P.; Wirtz, E.; Salavati, R.; Stuart, K. An RNA ligase essential for RNA editing and survival of the bloodstream form of Trypanosoma brucei. Science 2001, 291, 2159–2162. [Google Scholar] [CrossRef] [PubMed]
- Aeschlimann, S.; Stettler, P.; Schneider, A. DNA segregation in mitochondria and beyond: Insights from the trypanosomal tripartite attachment complex. Trends Biochem. Sci. 2023, 48, 1058–1070. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, S.R.; Godinho, J.L.P.; Vinayagam, J.; Zuma, A.A.; Silva, S.T.D.M.; Jaisankar, P.; Rodrigues, J.C.F.; De Souza, W.; Majumder, H.K. Isobenzofuranone derivative JVPH3, an inhibitor of L. donovani topoisomerase II, disrupts mitochondrial architecture in trypanosomatid parasites. Sci. Rep. 2018, 8, 11940. [Google Scholar] [CrossRef]
- Motta, M.C.M. Kinetoplast as a potential chemotherapeutic target of trypanosomatids. Curr. Pharm. Des. 2008, 14, 847–854. [Google Scholar] [CrossRef] [PubMed]
- Amodeo, S.; Bregy, I.; Ochsenreiter, T. Mitochondrial genome maintenance-the kinetoplast story. FEMS Microbiol. Rev. 2023, 47, fuac047. [Google Scholar] [CrossRef]
- Povelones, M.L. Beyond replication: Division and segregation of mitochondrial DNA in kinetoplastids. Mol. Biochem. Parasitol. 2014, 196, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Englund, P.T. The rotational dynamics of kinetoplast DNA replication. Mol. Microbiol. 2007, 64, 676–690. [Google Scholar] [CrossRef] [PubMed]
- Kitchin, P.A.; Klein, V.A.; Englund, P.T. Intermediates in the replication of kinetoplast DNA minicircles. J. Biol. Chem. 1985, 260, 3844–3851. [Google Scholar] [CrossRef]
- Cavalcanti, D.P.; de Souza, W. The kinetoplast of trypanosomatids: From early studies of electron microscopy to recent advances in atomic force microscopy. Scanning 2018, 2018, 9603051. [Google Scholar] [CrossRef] [PubMed]
- Hajduk, S.L.; Klein, V.A.; Englund, P.T. Replication of kinetoplast DNA maxicircles. Cell 1984, 36, 483–492. [Google Scholar] [CrossRef]
- Schneider, A.; Ochsenreiter, T. Failure is not an option—Mitochondrial genome segregation in trypanosomes. J. Cell Sci. 2018, 131, jcs221820. [Google Scholar] [CrossRef]
- Hoffmann, A.; Käser, S.; Jakob, M.; Amodeo, S.; Peitsch, C.; Týč, J.; Vaughan, S.; Zuber, B.; Schneider, A.; Ochsenreiter, T. Molecular model of the mitochondrial genome segregation machinery in Trypanosoma brucei. Proc. Natl. Acad. Sci. USA 2018, 115, E1809–E1818. [Google Scholar] [CrossRef]
- Turner, C.M.; Hide, G.; Buchanan, N.; Tait, A. Trypanosoma brucei: Inheritance of kinetoplast DNA maxicircles in a genetic cross and their segregation during vegetative growth. Exp. Parasitol. 1995, 80, 234–241. [Google Scholar] [CrossRef]
- Gibson, W.; Garside, L. Kinetoplast DNA minicircles are inherited from both parents in genetic hybrids of Trypanosoma brucei. Mol. Biochem. Parasitol. 1990, 42, 45–53. [Google Scholar] [CrossRef]
- Gibson, W.; Crow, M.; Kearns, J. Kinetoplast DNA minicircles are inherited from both parents in genetic crosses of Trypanosoma brucei. Parasitol. Res. 1997, 83, 483–488. [Google Scholar] [CrossRef]
- Tomasini, N.; Diosque, P. Evolution of Trypanosoma cruzi: Clarifying hybridisations, mitochondrial introgressions and phylogenetic relationships between major lineages. Mem. Inst. Oswaldo Cruz 2015, 110, 403–413. [Google Scholar] [CrossRef] [PubMed]
- Machado, C.A.; Ayala, F.J. Nucleotide sequences provide evidence of genetic exchange among distantly related lineages of Trypanosoma cruzi. Proc. Natl. Acad. Sci. USA 2001, 98, 7396–7401. [Google Scholar] [CrossRef] [PubMed]
- Gaunt, M.W.; Yeo, M.; Frame, I.A.; Stothard, J.R.; Carrasco, H.J.; Taylor, M.C.; Mena, S.S.; Veazey, P.; Miles, G.A.J.; Acosta, N.; et al. Mechanism of genetic exchange in American trypanosomes. Nature 2003, 421, 936–939. [Google Scholar] [CrossRef]
- Rusman, F.; Tomasini, N.; Yapur, N.-F.; Puebla, A.F.; Ragone, P.G.; Diosque, P. Elucidating diversity in the class composition of the minicircle hypervariable region of Trypanosoma cruzi: New perspectives on typing and kDNA inheritance. PLoS Negl. Trop. Dis. 2019, 13, e0007536. [Google Scholar] [CrossRef]
- Rusman, F.; Floridia-Yapur, N.; Ragone, P.G.; Diosque, P.; Tomasini, N. Evidence of hybridization, mitochondrial introgression and biparental inheritance of the kDNA minicircles in Trypanosoma cruzi I. PLoS Negl. Trop. Dis. 2020, 14, e0007770. [Google Scholar] [CrossRef]
- Matos, G.M.; Lewis, M.D.; Talavera-López, C.; Yeo, M.; Grisard, E.C.; Messenger, L.A.; Miles, M.A.; Andersson, B. Microevolution of Trypanosoma cruzi reveals hybridization and clonal mechanisms driving rapid genome diversification. eLife 2022, 11, e75237. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Rusman, F.; Díaz, A.G.; Ponce, T.; Floridia-Yapur, N.; Barnabé, C.; Diosque, P.; Tomasini, N. Wide reference databases for typing Trypanosoma cruzi based on amplicon sequencing of the minicircle hypervariable region. PLoS Negl. Trop. Dis. 2023, 17, e0011764. [Google Scholar] [CrossRef]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Souza, R.T.; Lima, F.M.; Barros, R.M.; Cortez, D.R.; Santos, M.F.; Cordero, E.M.; Ruiz, J.C.; Goldenberg, S.; Teixeira, M.M.G.; da Silveira, J.F. Genome size, karyotype polymorphism and chromosomal evolution in Trypanosoma cruzi. PLoS ONE 2011, 6, e23042. [Google Scholar] [CrossRef] [PubMed]
- de Souza, W. A short review on the morphology of Trypanosoma cruzi: From 1909 to 1999. Mem. Inst. Oswaldo Cruz 1999, 94, 17–36. [Google Scholar] [CrossRef]
- Thomas, S.; Martinez, L.L.I.T.; Westenberger, S.J.; Sturm, N.R. A population study of the minicircles in Trypanosoma cruzi: Predicting guide RNAs in the absence of empirical RNA editing. BMC Genom. 2007, 8, 133. [Google Scholar] [CrossRef]
- Berná, L.; Greif, G.; Pita, S.; Faral-Tello, P.; Díaz-Viraqué, F.; Souza, R.D.C.M.D.; Vallejo, G.A.; Alvarez-Valin, F.; Robello, C. Maxicircle architecture and evolutionary insights into Trypanosoma cruzi complex. PLoS Negl. Trop. Dis. 2021, 15, e0009719. [Google Scholar] [CrossRef]
- Westenberger, S.J.; Barnabé, C.; Campbell, D.A.; Sturm, N.R. Two hybridization events define the population structure of Trypanosoma cruzi. Genetics 2005, 171, 527–543. [Google Scholar] [CrossRef]
- Shapiro, T.A. Kinetoplast DNA maxicircles: Networks within networks. Proc. Natl. Acad. Sci. USA 1993, 90, 7809–7813. [Google Scholar] [CrossRef]
- Carpenter, L.R.; Englund, P.T. Kinetoplast maxicircle DNA replication in Crithidia fasciculata and Trypanosoma brucei. Mol. Cell. Biol. 1995, 15, 6794–6803. [Google Scholar] [CrossRef] [PubMed]
- Vieira-da-Rocha, J.P.; Passos-Silva, D.G.; Mendes, I.C.; Rocha, E.A.; Gomes, D.A.; Machado, C.R.; McCulloch, R. The DNA damage response is developmentally regulated in the African trypanosome. DNA Repair. 2019, 73, 78–90. [Google Scholar] [CrossRef] [PubMed]
- Savill, N.J.; Higgs, P.G. A theoretical study of random segregation of minicircles in trypanosomatids. Proc. Biol. Sci. 1999, 266, 611–620. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, K.N.; de Graffenried, C.L. Polo-like kinase is necessary for flagellum inheritance in Trypanosoma brucei. J. Cell Sci. 2012, 125, 3173–3184. [Google Scholar] [CrossRef]
- Ogbadoyi, E.O.; Robinson, D.R.; Gull, K. A high-order trans-membrane structural linkage is responsible for mitochondrial genome positioning and segregation by flagellar basal bodies in trypanosomes. Mol. Biol. Cell 2003, 14, 1769–1779. [Google Scholar] [CrossRef] [PubMed]
- Gluenz, E.; Povelones, M.L.; Englund, P.T.; Gull, K. The kinetoplast duplication cycle in Trypanosoma brucei is orchestrated by cytoskeleton-mediated cell morphogenesis. Mol. Cell. Biol. 2011, 31, 1012–1021. [Google Scholar] [CrossRef] [PubMed]
- Aeschlimann, S.; Kalichava, A.; Schimanski, B.; Berger, B.M.; Jetishi, C.; Stettler, P.; Ochsenreiter, T.; Schneider, A. Single p197 molecules of the mitochondrial genome segregation system of Trypanosoma brucei determine the distance between basal body and outer membrane. Proc. Natl. Acad. Sci. USA 2022, 119, e2204294119. [Google Scholar] [CrossRef]
- Käser, S.; Willemin, M.; Schnarwiler, F.; Schimanski, B.; Poveda-Huertes, D.; Oeljeklaus, S.; Haenni, B.; Zuber, B.; Warscheid, B.; Meisinger, C.; et al. Biogenesis of the mitochondrial DNA inheritance machinery in the mitochondrial outer membrane of Trypanosoma brucei. PLoS Pathog. 2017, 13, e1006808. [Google Scholar] [CrossRef]
- Schimanski, B.; Aeschlimann, S.; Stettler, P.; Käser, S.; Gomez-Fabra Gala, M.; Bender, J.; Warscheid, B.; Vögtle, F.-N.; Schneider, A. p166 links membrane and intramitochondrial modules of the trypanosomal tripartite attachment complex. PLoS Pathog. 2022, 18, e1010207. [Google Scholar] [CrossRef]
- Trikin, R.; Doiron, N.; Hoffmann, A.; Haenni, B.; Jakob, M.; Schnaufer, A.; Schimanski, B.; Zuber, B.; Ochsenreiter, T. TAC102 is a novel component of the mitochondrial genome segregation machinery in trypanosomes. PLoS Pathog. 2016, 12, e1005586. [Google Scholar] [CrossRef]
- Drew, M.E.; Englund, P.T. Intramitochondrial location and dynamics of Crithidia fasciculata kinetoplast minicircle replication intermediates. J. Cell Biol. 2001, 153, 735–744. [Google Scholar] [CrossRef]
- Englund, P.T. Free minicircles of kinetoplast DNA in Crithidia fasciculata. J. Biol. Chem. 1979, 254, 4895–4900. [Google Scholar] [CrossRef]
- Ferguson, M.; Torri, A.F.; Ward, D.C.; Englund, P.T. In situ hybridization to the Crithidia fasciculata kinetoplast reveals two antipodal sites involved in kinetoplast DNA replication. Cell 1992, 70, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Morga, D.L.; Englund, P.T. The attachment of minicircles to kinetoplast DNA networks during replication. Cell 1993, 74, 703–711. [Google Scholar] [CrossRef] [PubMed]
- Hines, J.C.; Ray, D.S. A mitochondrial DNA primase is essential for cell growth and kinetoplast DNA replication in Trypanosoma brucei. Mol. Cell. Biol. 2010, 30, 1319–1328. [Google Scholar] [CrossRef] [PubMed]
- Hines, J.C.; Ray, D.S. A second mitochondrial DNA primase is essential for cell growth and kinetoplast minicircle DNA replication in Trypanosoma brucei. Eukaryotic Cell 2011, 10, 445–454. [Google Scholar] [CrossRef]
- Liu, B.; Molina, H.; Kalume, D.; Pandey, A.; Griffith, J.D.; Englund, P.T. Role of p38 in replication of Trypanosoma brucei kinetoplast DNA. Mol. Cell. Biol. 2006, 26, 5382–5393. [Google Scholar] [CrossRef]
- Liu, B.; Wang, J.; Yildirir, G.; Englund, P.T. TbPIF5 is a Trypanosoma brucei mitochondrial DNA helicase involved in processing of minicircle Okazaki fragments. PLoS Pathog. 2009, 5, e1000589. [Google Scholar] [CrossRef]
- Lewis, M.D.; Llewellyn, M.S.; Gaunt, M.W.; Yeo, M.; Carrasco, H.J.; Miles, M.A. Flow cytometric analysis and microsatellite genotyping reveal extensive DNA content variation in Trypanosoma cruzi populations and expose contrasts between natural and experimental hybrids. Int. J. Parasitol. 2009, 39, 1305–1317. [Google Scholar] [CrossRef]
- Zingales, B.; Miles, M.A.; Campbell, D.A.; Tibayrenc, M.; Macedo, A.M.; Teixeira, M.M.G.; Schijman, A.G.; Llewellyn, M.S.; Lages-Silva, E.; Machado, C.R.; et al. The revised Trypanosoma cruzi subspecific nomenclature: Rationale, epidemiological relevance and research applications. Infect. Genet. Evol. 2012, 12, 240–253. [Google Scholar] [CrossRef]
- Breton, S.; Stewart, D.T. Atypical mitochondrial inheritance patterns in eukaryotes. Genome 2015, 58, 423–431. [Google Scholar] [CrossRef]
- Bergstrom, C.T.; Pritchard, J. Germline bottlenecks and the evolutionary maintenance of mitochondrial genomes. Genetics 1998, 149, 2135–2146. [Google Scholar] [CrossRef]
- Sasaki, T.; Sato, M. Degradation of paternal mitochondria via mitophagy. Biochim. Biophys. Acta Gen. Subj. 2021, 1865, 129886. [Google Scholar] [CrossRef] [PubMed]


| Hybrid Line | Maximum Log-Likelihood | P1 Proportion (Likelihood) 1 | Maximum Pearson’s r | P1 Proportion (Pearson’s r) 2 |
|---|---|---|---|---|
| 1C2 | 9.4 × 106 | 0.46 | 0.80 | 0.41 |
| 2C1 | 1.7 × 107 | 0.44 | 0.69 | 0.42 |
| 1D12 | 6.5 × 106 | 0.44 | 0.69 | 0.44 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tomasini, N.; Ponce, T.; Rusman, F.; Hodi, S.; Floridia-Yapur, N.; Díaz, A.G.; Aguirre, J.J.; Matos, G.M.; Andersson, B.; Lewis, M.D.; et al. Biparental Inheritance and Instability of kDNA in Experimental Hybrids of Trypanosoma cruzi: A Proposal for a Mechanism. Biology 2025, 14, 1394. https://doi.org/10.3390/biology14101394
Tomasini N, Ponce T, Rusman F, Hodi S, Floridia-Yapur N, Díaz AG, Aguirre JJ, Matos GM, Andersson B, Lewis MD, et al. Biparental Inheritance and Instability of kDNA in Experimental Hybrids of Trypanosoma cruzi: A Proposal for a Mechanism. Biology. 2025; 14(10):1394. https://doi.org/10.3390/biology14101394
Chicago/Turabian StyleTomasini, Nicolás, Tatiana Ponce, Fanny Rusman, Soledad Hodi, Noelia Floridia-Yapur, Anahí Guadalupe Díaz, Juan José Aguirre, Gabriel Machado Matos, Björn Andersson, Michael D. Lewis, and et al. 2025. "Biparental Inheritance and Instability of kDNA in Experimental Hybrids of Trypanosoma cruzi: A Proposal for a Mechanism" Biology 14, no. 10: 1394. https://doi.org/10.3390/biology14101394
APA StyleTomasini, N., Ponce, T., Rusman, F., Hodi, S., Floridia-Yapur, N., Díaz, A. G., Aguirre, J. J., Matos, G. M., Andersson, B., Lewis, M. D., & Diosque, P. (2025). Biparental Inheritance and Instability of kDNA in Experimental Hybrids of Trypanosoma cruzi: A Proposal for a Mechanism. Biology, 14(10), 1394. https://doi.org/10.3390/biology14101394






