Hepatocyte Growth Factor Differentially Modulates Oral Microbiota in Early vs. Late Experimental Periodontitis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Ligature-Induced Periodontitis (LIP) Model
2.3. Protein Expression Analysis by ELISA
2.4. 16S rRNA Sequencing, Bioinformatic, and Statistical Analysis
3. Results
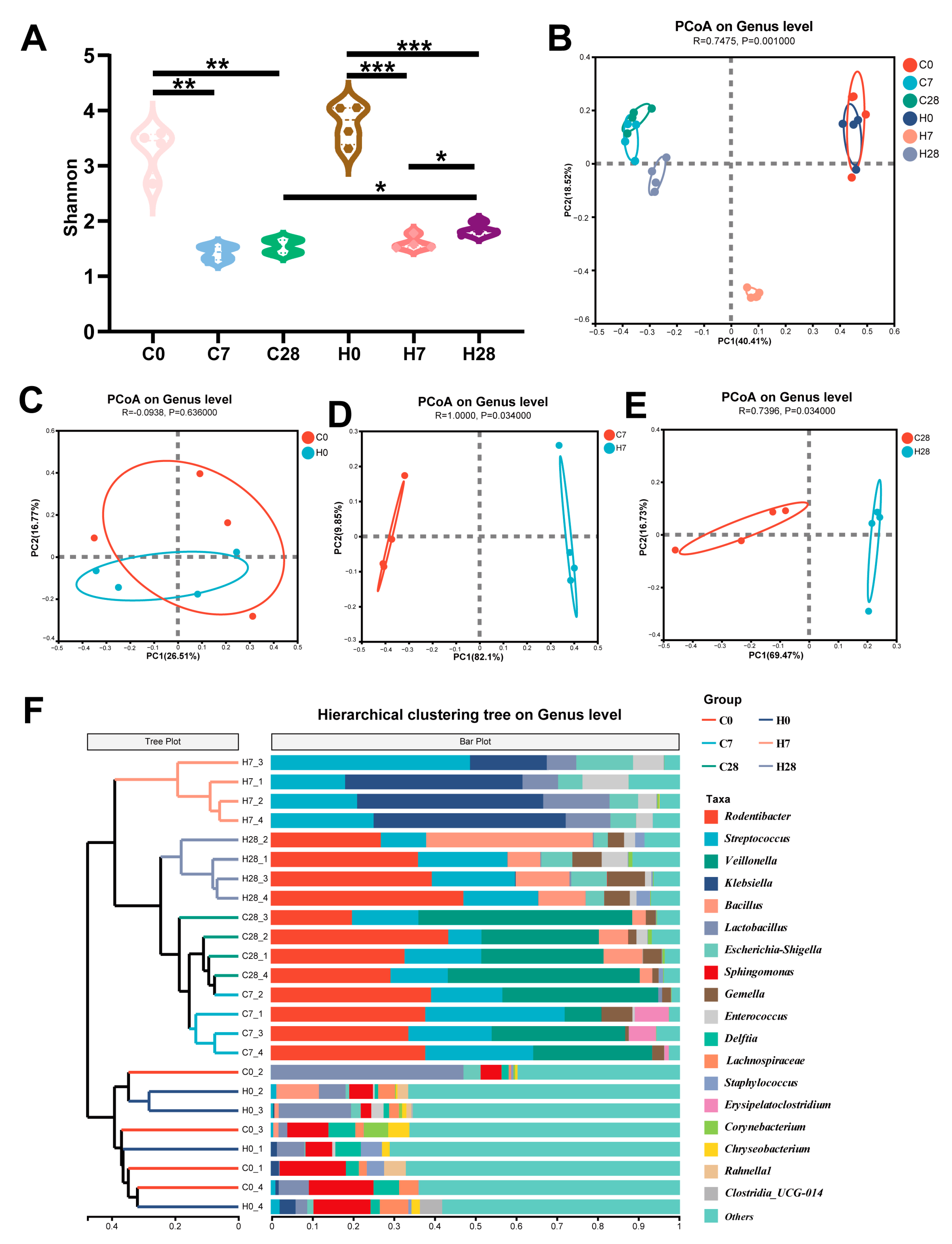
3.1. HGF Altered LIP Microbial Diversity During Periodontitis Development
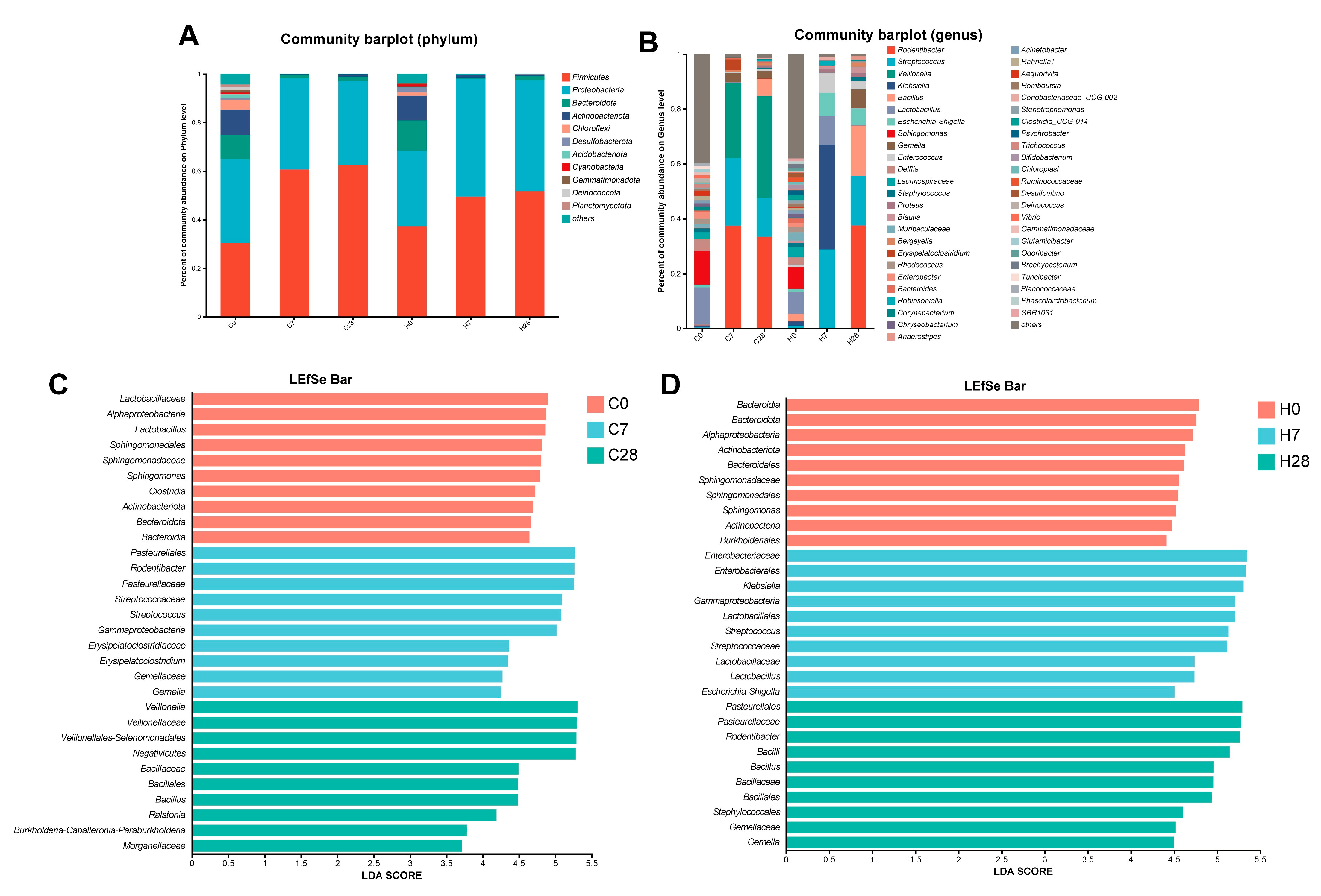
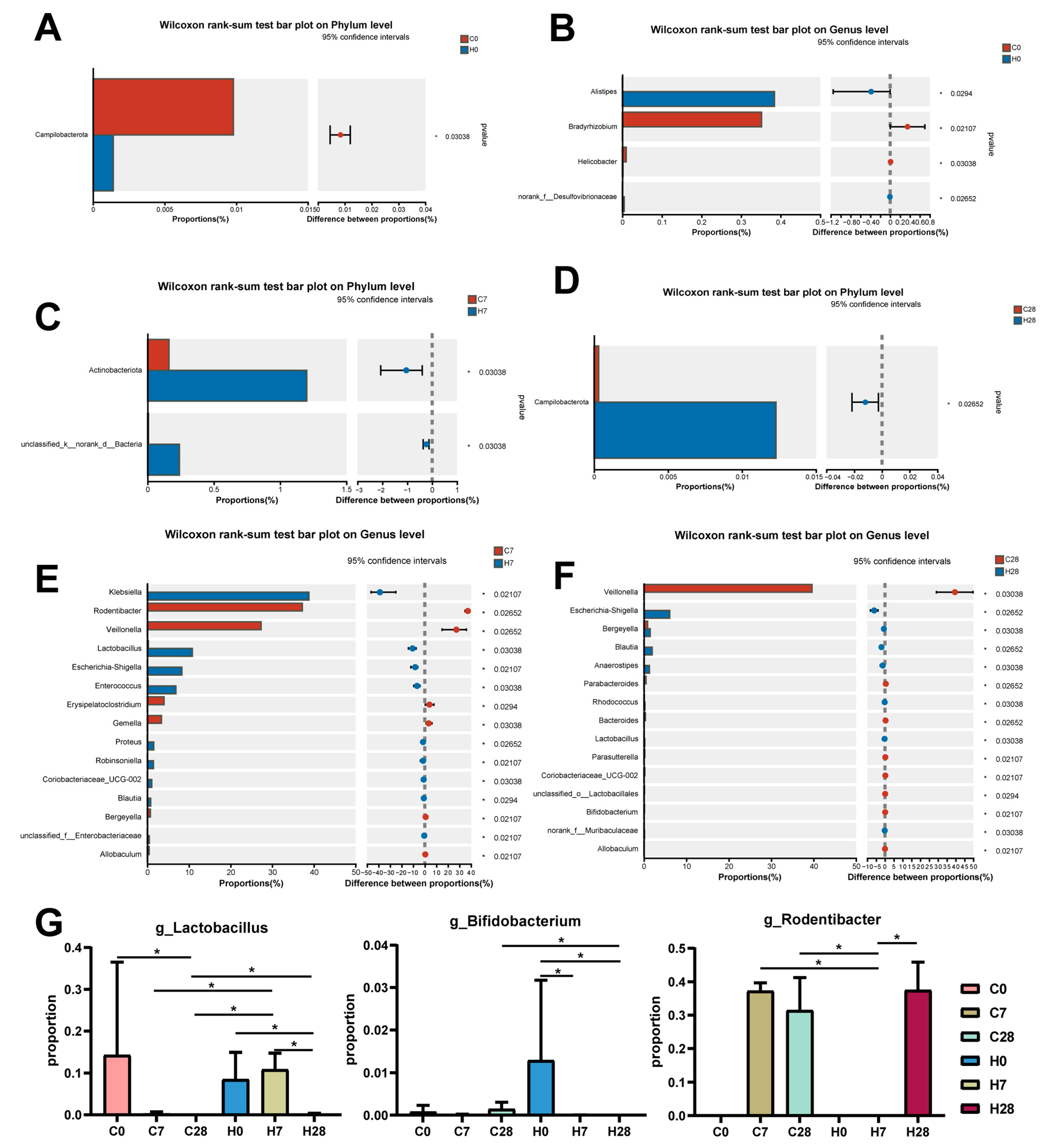
3.2. HGF Contributed to Community Shifts in LIP During Periodontitis Progression
3.3. Identification of LIP Microbiota Correlated with Inflammation and Bone Metabolism
3.4. HGF Shifted Microbial Function During Periodontitis Progression
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| HGF | Hepatocyte Growth Factor |
| WT | Wild Type |
| HGF-Tg | Hepatocyte Growth Factor High-Expression Transgenic |
| TNF-α | Tumor Necrosis Factor-α |
| IL-6 | Interleukin-6 |
| IL-17 | Interleukin-17 |
| LIP | Ligature-induced periodontitis |
| CTXI | C-terminal telopeptide of type I collagen |
| PINP | N-terminal pro-peptide of type I procollagen |
| MAPK | mitogen-activated protein kinase |
| PI3K/Akt | Phosphatidylinositol-3-kinase/protein kinase B |
| FoxO | Forkhead box O |
| TRAF6 | TNF receptor-associated factor 6 |
| RANKL | Receptor Activator of Nuclear Factor-κ B Ligand |
| OPG | Osteoclastogenesis inhibitory factor |
| OTU | Operational Taxonomic Units |
| PCoA | Principal coordinate analysis |
| LDA | Linear Discriminant Analysis |
| LEfSe | Linear Discriminant Analysis Effect Size |
| PICRUSt | Phylogenetic Investigation of Communities by Reconstruction of Unobserved States |
| RDA | Redundancy Analysis |
References
- Kassebaum, N.J.; Bernabé, E.; Dahiya, M.; Bhandari, B.; Murray, C.J.L.; Marcenes, W. Global burden of severe periodontitis in 1990–2010: A systematic review and meta-regression. J. Dent. Res. 2014, 93, 1045–1053. [Google Scholar] [CrossRef] [PubMed]
- Curtis, M.A.; Diaz, P.I.; Van Dyke, T.E. The role of the microbiota in periodontal disease. Periodontology 2000 2020, 83, 14–25. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zhong, Y.; Chen, L.; Liu, W.; Lin, C.; Chen, Y.; Wang, X. HGF Aggravated Periodontitis-Associated Gut Barrier and Microbial Dysfunction: Implications for Oral–Gut Axis Regulation. Biology 2025, 14, 496. [Google Scholar] [CrossRef]
- Blasco-Baque, V.; Garidou, L.; Pomié, C.; Escoula, Q.; Loubieres, P.; Le Gall-David, S.; Lemaitre, M.; Nicolas, S.; Klopp, P.; Waget, A.; et al. Periodontitis induced by Porphyromonas gingivalis drives periodontal microbiota dysbiosis and insulin resistance via an impaired adaptive immune response. Gut 2017, 66, 872–885. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.; Liu, J.; Wang, H.; Song, J.; Tan, L.; Zhao, H. Porphyromonas gingivalis can invade periodontal ligament stem cells. BMC Microbiol. 2017, 17, 38. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, L.; Sun, T.; Shen, S.; Li, Z.; Ma, X.; Gu, X.; Zhang, X.; Peng, A.; Xu, X.; et al. Study of the inflammatory activating process in the early stage of Fusobacterium nucleatum infected PDLSCs. Int. J. Oral Sci. 2023, 15, 8. [Google Scholar] [CrossRef]
- Hajishengallis, G.; Liang, S.; Payne, M.A.; Hashim, A.; Jotwani, R.; Eskan, M.A.; McIntosh, M.L.; Alsam, A.; Kirkwood, K.L.; Lambris, J.D.; et al. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe 2011, 10, 497–506. [Google Scholar] [CrossRef]
- Homayouni Rad, A.; Pourjafar, H.; Mirzakhani, E. A comprehensive review of the application of probiotics and postbiotics in oral health. Front. Cell. Infect. Microbiol. 2023, 13, 1120995. [Google Scholar] [CrossRef]
- van Winkelhoff, A.J.; Rurenga, P.; Wekema-Mulder, G.J.; Singadji, Z.M.; Rams, T.E. Non-oral gram-negative facultative rods in chronic periodontitis microbiota. Microb. Pathog. 2016, 94, 117–122. [Google Scholar] [CrossRef]
- Ribeiro, A.A.; Jiao, Y.; Girnary, M.; Alves, T.; Chen, L.; Farrell, A.; Wu, D.; Teles, F.; Inohara, N.; Swanson, K.V.; et al. Oral biofilm dysbiosis during experimental periodontitis. Mol. Oral Microbiol. 2022, 37, 256–265. [Google Scholar] [CrossRef]
- Dutzan, N.; Kajikawa, T.; Abusleme, L.; Greenwell-Wild, T.; Zuazo, C.E.; Ikeuchi, T.; Brenchley, L.; Abe, T.; Hurabielle, C.; Martin, D.; et al. A dysbiotic microbiome triggers TH17 cells to mediate oral mucosal immunopathology in mice and humans. Sci. Transl. Med. 2018, 10, eaat0797. [Google Scholar] [CrossRef]
- Kittaka, M.; Yoshimoto, T.; Schlosser, C.; Kajiya, M.; Kurihara, H.; Reichenberger, E.J.; Ueki, Y. Microbe-Dependent Exacerbated Alveolar Bone Destruction in Heterozygous Cherubism Mice. JBMR Plus 2020, 4, e10352. [Google Scholar] [CrossRef]
- Lee, C.-T.; Teles, R.; Kantarci, A.; Chen, T.; McCafferty, J.; Starr, J.R.; Brito, L.C.N.; Paster, B.J.; Van Dyke, T.E. Resolvin E1 Reverses Experimental Periodontitis and Dysbiosis. J. Immunol. 2016, 197, 2796–2806. [Google Scholar] [CrossRef]
- Afacan, B.; Keleş Yücel, Z.P.; Paşali, Ç.; Atmaca İlhan, H.; Köse, T.; Emingil, G. Effect of non-surgical periodontal treatment on gingival crevicular fluid hypoxia inducible factor-1 alpha, vascular endothelial growth factor and tumor necrosis factor-alpha levels in generalized aggressive periodontitis patients. J. Periodontol. 2020, 91, 1495–1502. [Google Scholar] [CrossRef]
- Molnarfi, N.; Benkhoucha, M.; Funakoshi, H.; Nakamura, T.; Lalive, P.H. Hepatocyte growth factor: A regulator of inflammation and autoimmunity. Autoimmun. Rev. 2015, 14, 293–303. [Google Scholar] [CrossRef]
- Nagaraja, C.; Pradeep, A.R. Hepatocyte growth factor levels in gingival crevicular fluid in health, disease, and after treatment. J. Periodontol. 2007, 78, 742–747. [Google Scholar] [CrossRef] [PubMed]
- Alamri, T.; Alkhaldy, A.A.; Gauthaman, K.; Pushparaj, P.N.; Moulay, M.; Mirza, A.A.; Azhar, E.I.; Barnawi, S.; Papadopoulou, G.; Karamitros, T.; et al. Growth factors in relation to obesity, food habits, and microbiota among healthy Saudis: Preliminary results. Eur. Rev. Med. Pharmacol. Sci. 2022, 26, 9311–9326. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Liu, W.; Wu, Z.; He, X.; Tang, Y.; He, Q.; Lin, C.; Chen, Y.; Luo, G.; Yu, T.; et al. Hepatocyte growth factor is protective in early stage but bone-destructive in late stage of experimental periodontitis. J. Periodontal Res. 2024, 59, 565–575. [Google Scholar] [CrossRef]
- Wang, X.; Yan, L.; Tang, Y.; He, X.; Zhao, X.; Liu, W.; Wu, Z.; Luo, G. Anti-inflammatory effect of HGF responses to oral traumatic ulcers using an HGF-Tg mouse model. Exp. Anim. 2022, 71, 204–213. [Google Scholar] [CrossRef]
- de Molon, R.S.; Park, C.H.; Jin, Q.; Sugai, J.; Cirelli, J.A. Characterization of ligature-induced experimental periodontitis. Microsc. Res. Tech. 2018, 81, 1412–1421. [Google Scholar] [CrossRef]
- Liu, C.; Zhao, D.; Ma, W.; Guo, Y.; Wang, A.; Wang, Q.; Lee, D.-J. Denitrifying sulfide removal process on high-salinity wastewaters in the presence of Halomonas sp. Appl. Microbiol. Biotechnol. 2016, 100, 1421–1426. [Google Scholar] [CrossRef]
- Ren, Y.; Yu, G.; Shi, C.; Liu, L.; Guo, Q.; Han, C.; Zhang, D.; Zhang, L.; Liu, B.; Gao, H.; et al. Majorbio Cloud: A one-stop, comprehensive bioinformatic platform for multiomics analyses. iMeta 2022, 1, e12. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Oksanen, J.; Kindt, R.; Legendre, P.; O’hara, B.; Stevens, M.; Oksanen, M.; Suggests, M. The Vegan Package: Community Ecology Package; R Foundation: Vienna, Austria, 2007; Volume 1, pp. 1–190. [Google Scholar]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- Parks, D.H.; Tyson, G.W.; Hugenholtz, P.; Beiko, R.G. STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 2014, 30, 3123–3124. [Google Scholar] [CrossRef]
- Eastell, R.; Szulc, P. Use of bone turnover markers in postmenopausal osteoporosis. Lancet Diabetes Endocrinol. 2017, 5, 908–923. [Google Scholar] [CrossRef]
- Petersen, C.; Round, J.L. Defining dysbiosis and its influence on host immunity and disease. Cell. Microbiol. 2014, 16, 1024–1033. [Google Scholar] [CrossRef]
- Arce, M.; Endo, N.; Dutzan, N.; Abusleme, L. A reappraisal of microbiome dysbiosis during experimental periodontitis. Mol. Oral Microbiol. 2022, 37, 180–195. [Google Scholar] [CrossRef]
- Invernici, M.M.; Salvador, S.L.; Silva, P.H.F.; Soares, M.S.M.; Casarin, R.; Palioto, D.B.; Souza, S.L.S.; Taba, M.; Novaes, A.B.; Furlaneto, F.A.C.; et al. Effects of Bifidobacterium probiotic on the treatment of chronic periodontitis: A randomized clinical trial. J. Clin. Periodontol. 2018, 45, 1198–1210. [Google Scholar] [CrossRef]
- İnce, G.; Gürsoy, H.; İpçi, Ş.D.; Cakar, G.; Emekli-Alturfan, E.; Yılmaz, S. Clinical and Biochemical Evaluation of Lozenges Containing Lactobacillus reuteri as an Adjunct to Non-Surgical Periodontal Therapy in Chronic Periodontitis. J. Periodontol. 2015, 86, 746–754. [Google Scholar] [CrossRef]
- Ebersole, J.; Kirakodu, S.; Chen, J.; Nagarajan, R.; Gonzalez, O.A. Oral Microbiome and Gingival Transcriptome Profiles of Ligature-Induced Periodontitis. J. Dent. Res. 2020, 99, 746–757. [Google Scholar] [CrossRef]
- Richard, M.L.; Liguori, G.; Lamas, B.; Brandi, G.; da Costa, G.; Hoffmann, T.W.; Pierluigi Di Simone, M.; Calabrese, C.; Poggioli, G.; Langella, P.; et al. Mucosa-associated microbiota dysbiosis in colitis associated cancer. Gut Microbes 2018, 9, 131–142. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.-T.; Liu, J.-X.; Wang, D.-H.; Sun, H.-J.; Zhang, X.-Y. Melatonin reduced colon inflammation but had no effect on energy metabolism in ageing Mongolian gerbils (Meriones unguiculatus). Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2023, 273, 109731. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Zhang, J.; Ding, F.; Sun, S. Relationships among gut microbes, the interleukin family, and hypertension: A mediation Mendelian randomization study. Front. Nutr. 2023, 10, 1293170. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Luo, Z.; Chen, Y.; Yan, Z.; Fu, J.; Jiang, Y.; Xu, J.; Liu, Y. Butyrate Inhibits Dendritic Cell Activation and Alleviates Periodontitis. J. Dent. Res. 2023, 102, 1326–1336. [Google Scholar] [CrossRef]
- Wexler, H.M. Bacteroides: The good, the bad, and the nitty-gritty. Clin. Microbiol. Rev. 2007, 20, 593–621. [Google Scholar] [CrossRef]
- Kim, M.E.; Kim, D.H.; Lee, J.S. FoxO Transcription Factors: Applicability as a Novel Immune Cell Regulators and Therapeutic Targets in Oxidative Stress-Related Diseases. Int. J. Mol. Sci. 2022, 23, 11877. [Google Scholar] [CrossRef]
- Hedrick, S.M.; Hess Michelini, R.; Doedens, A.L.; Goldrath, A.W.; Stone, E.L. FOXO transcription factors throughout T cell biology. Nat. Rev. Immunol. 2012, 12, 649–661. [Google Scholar] [CrossRef]
- Xiao, E.; Mattos, M.; Vieira, G.H.A.; Chen, S.; Corrêa, J.D.; Wu, Y.; Albiero, M.L.; Bittinger, K.; Graves, D.T. Diabetes Enhances IL-17 Expression and Alters the Oral Microbiome to Increase Its Pathogenicity. Cell Host Microbe 2017, 22, 120–128.e4. [Google Scholar] [CrossRef]
- Chen, Z.; Weng, J.; Du, X.; Ji, R.; Yang, X.; Yang, Y.; Ma, M. Quaternized chitosan/glycyrrhizic acid co-decorated titanium with enhanced antimicrobial, immunomodulatory, and osteogenic properties for dental implant applications. Carbohydr. Polym. 2025, 367, 123984. [Google Scholar] [CrossRef]
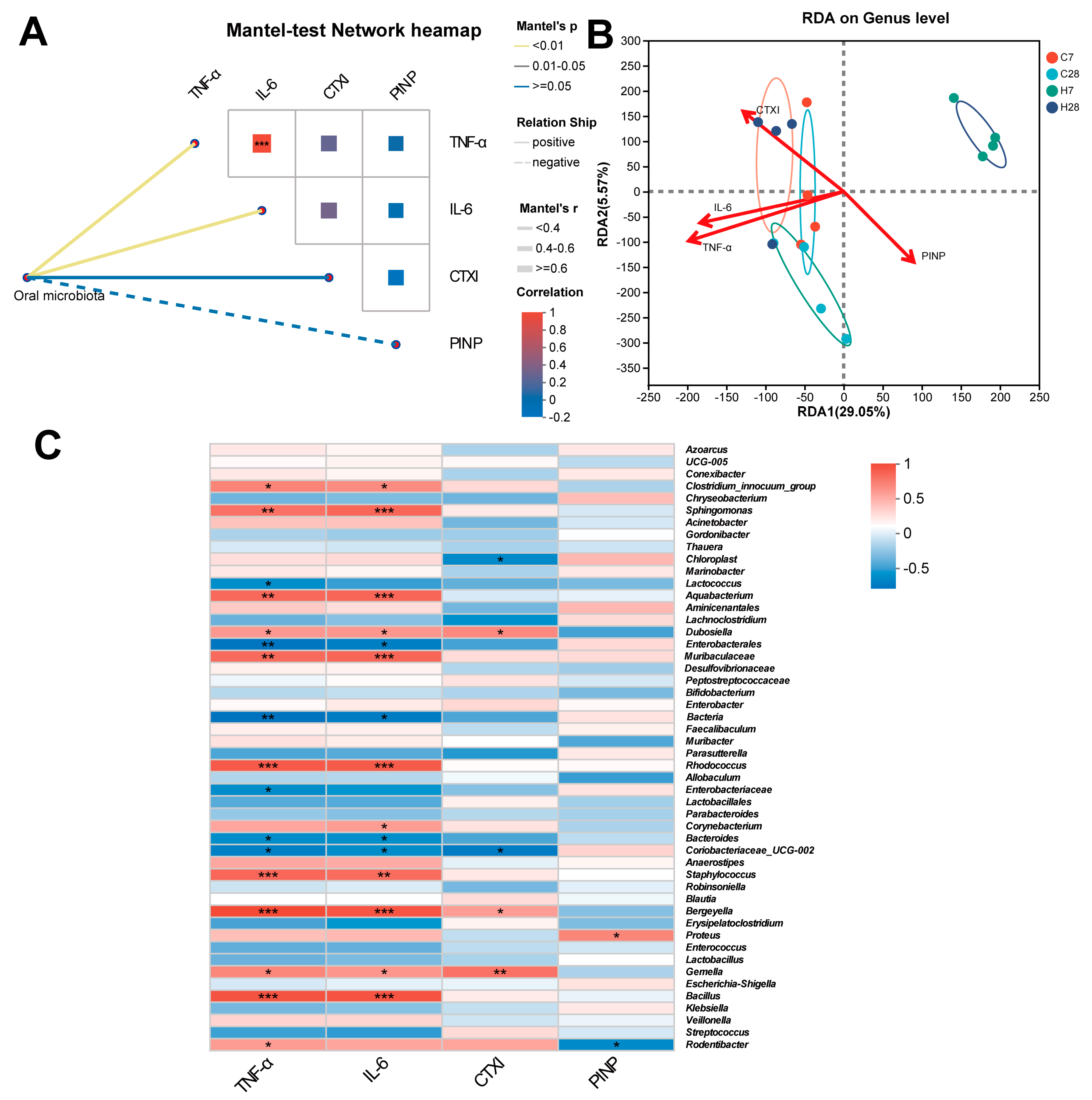

| RDA1 | RDA2 | R2 | p Value | |
|---|---|---|---|---|
| TNF-α | −0.9987 | −0.0515 | 0.4289 | 0.034 |
| IL-6 | −0.9992 | 0.0404 | 0.3657 | 0.045 |
| CTXI | −0.8209 | 0.571 | 0.4613 | 0.013 |
| PINP | 0.7857 | −0.6186 | 0.2793 | 0.104 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ji, R.; Zhao, X.; Chen, Z.; Ge, Y.; Wu, Z.; Wang, X. Hepatocyte Growth Factor Differentially Modulates Oral Microbiota in Early vs. Late Experimental Periodontitis. Biology 2025, 14, 1393. https://doi.org/10.3390/biology14101393
Ji R, Zhao X, Chen Z, Ge Y, Wu Z, Wang X. Hepatocyte Growth Factor Differentially Modulates Oral Microbiota in Early vs. Late Experimental Periodontitis. Biology. 2025; 14(10):1393. https://doi.org/10.3390/biology14101393
Chicago/Turabian StyleJi, Ruotong, Xiaomin Zhao, Zhen Chen, Yifei Ge, Zhicong Wu, and Xinhong Wang. 2025. "Hepatocyte Growth Factor Differentially Modulates Oral Microbiota in Early vs. Late Experimental Periodontitis" Biology 14, no. 10: 1393. https://doi.org/10.3390/biology14101393
APA StyleJi, R., Zhao, X., Chen, Z., Ge, Y., Wu, Z., & Wang, X. (2025). Hepatocyte Growth Factor Differentially Modulates Oral Microbiota in Early vs. Late Experimental Periodontitis. Biology, 14(10), 1393. https://doi.org/10.3390/biology14101393




