Dynamic Metabolome and Transcriptome Profiling Provide Molecular Insights into Floral Bud Differentiation in Michelia ‘Xin’
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Morphological Observation
2.3. Metabolite Profiling of Developing Floral Buds
2.4. Transcriptome Sequencing and Analysis
2.5. RT-qPCR (Real-Time Quantitative Polymerase Chain Reaction)
3. Results
3.1. Morphological Changes During Flower Bud Differentiation in Michelia ‘Xin’
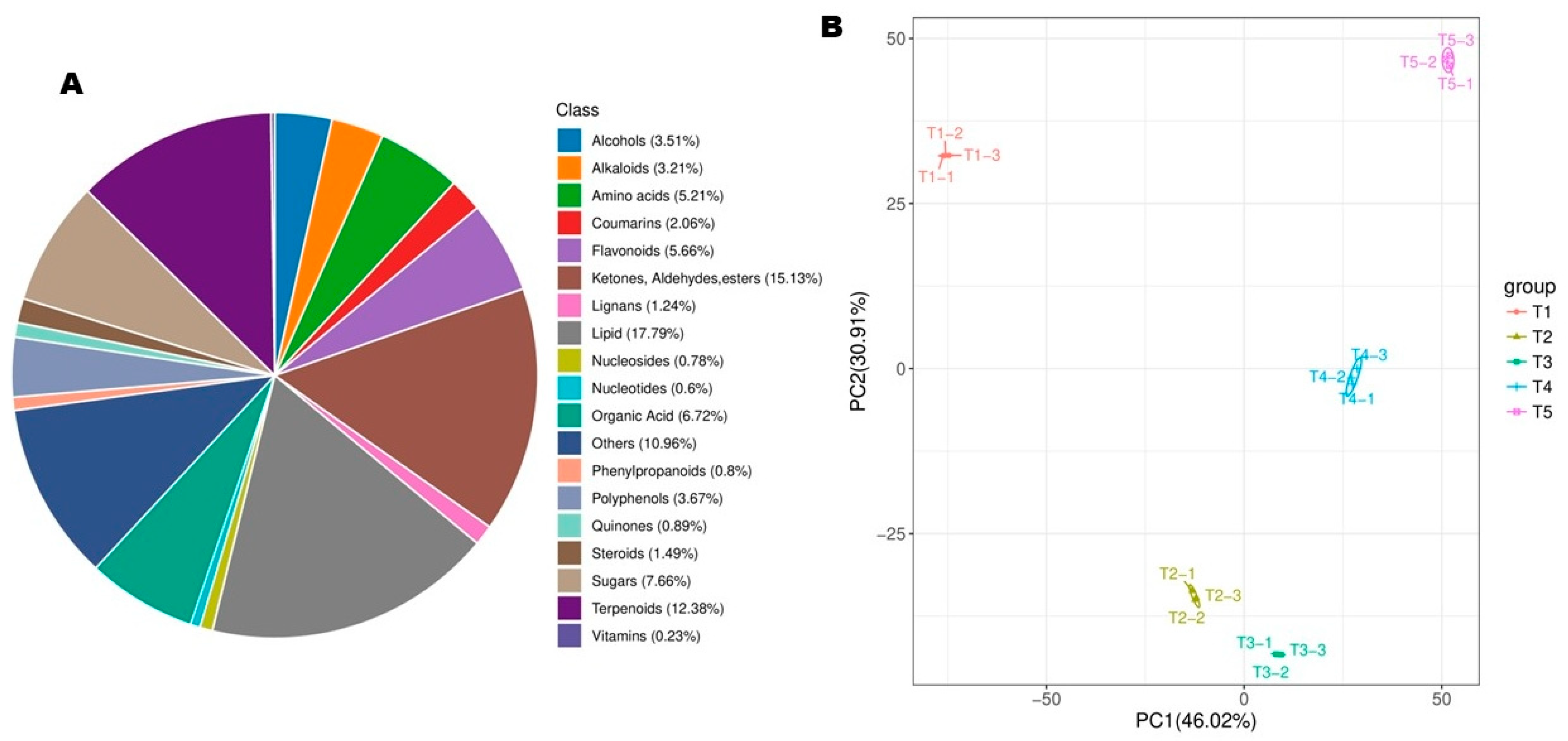
3.2. Dynamic Metabolite Profile of Developing Flowers of Michelia ‘Xin’
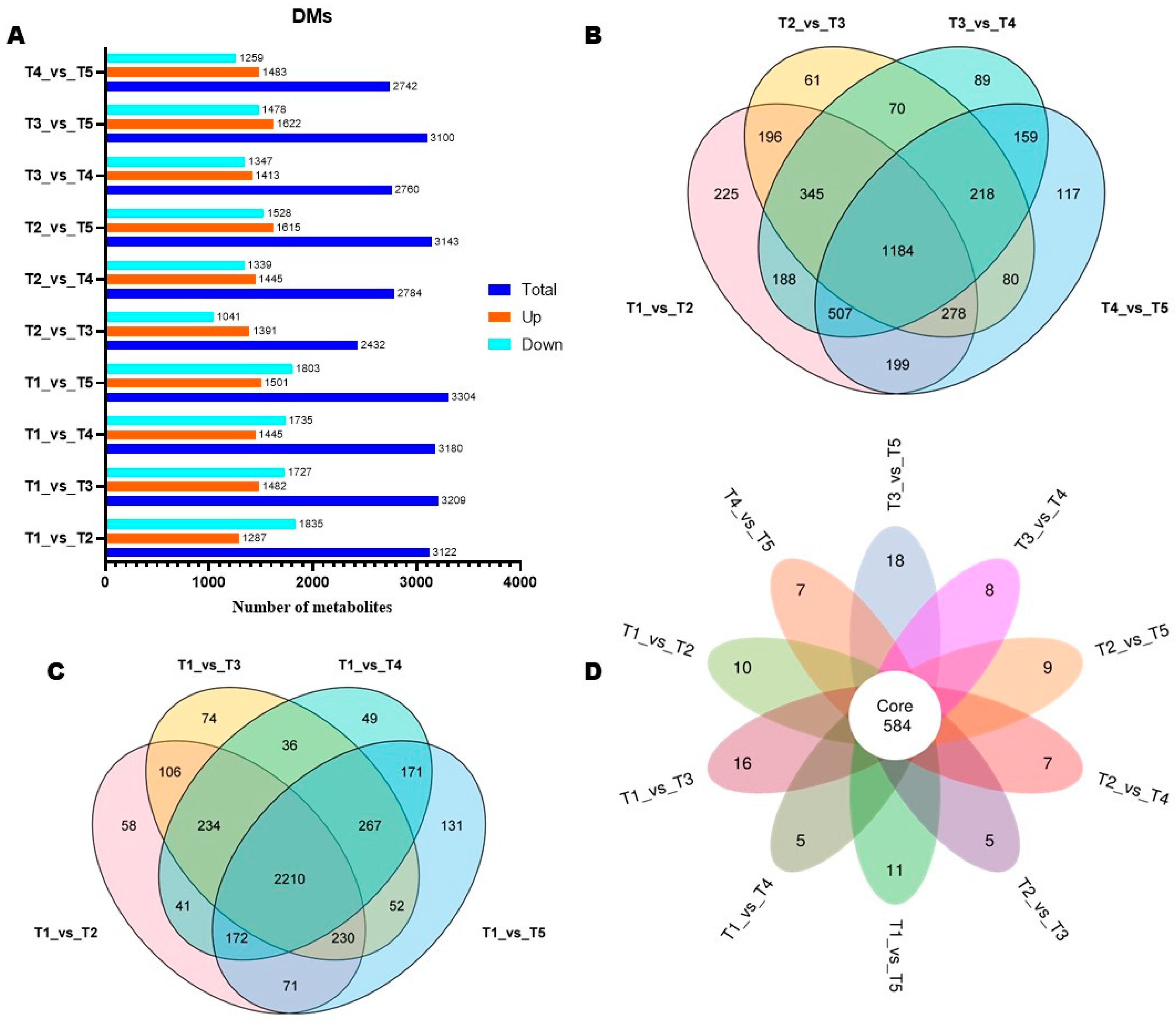
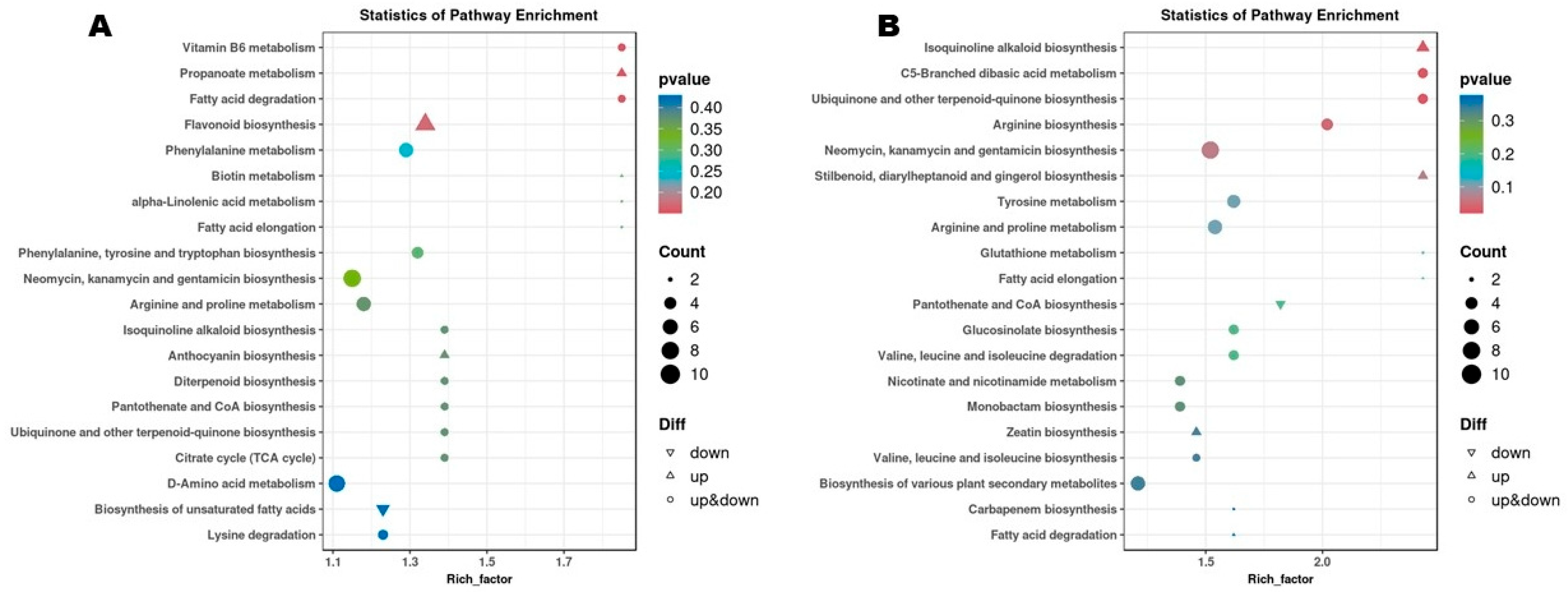
3.3. Differential Metabolites (DMs) and Pathways (DPs) Between Flowering Transition Stages
3.4. Dynamic Transcriptome Profile of Developing Flowers of Michelia ‘Xin’
3.5. Differentially Expressed Genes (DEGs) and Functional Annotation
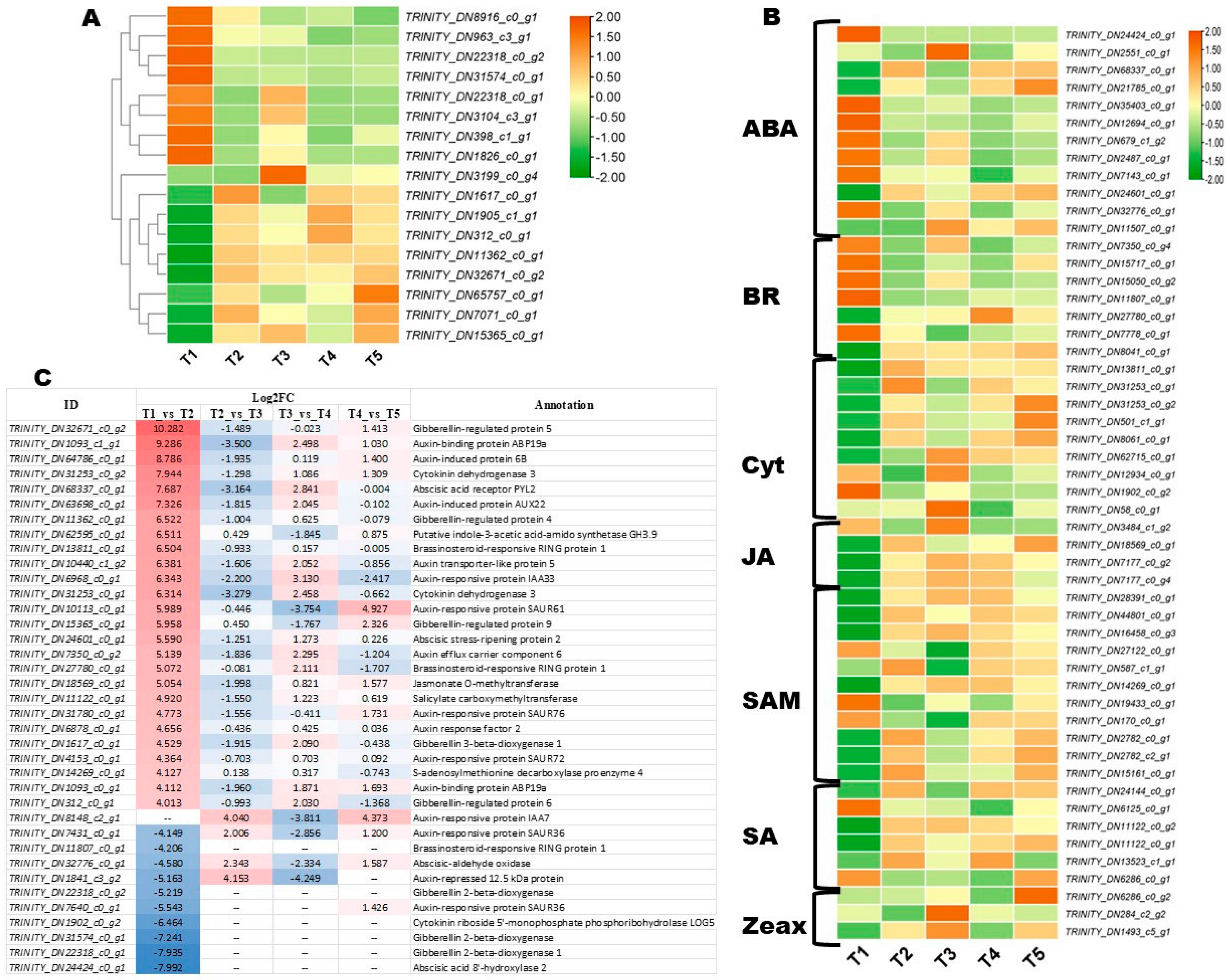
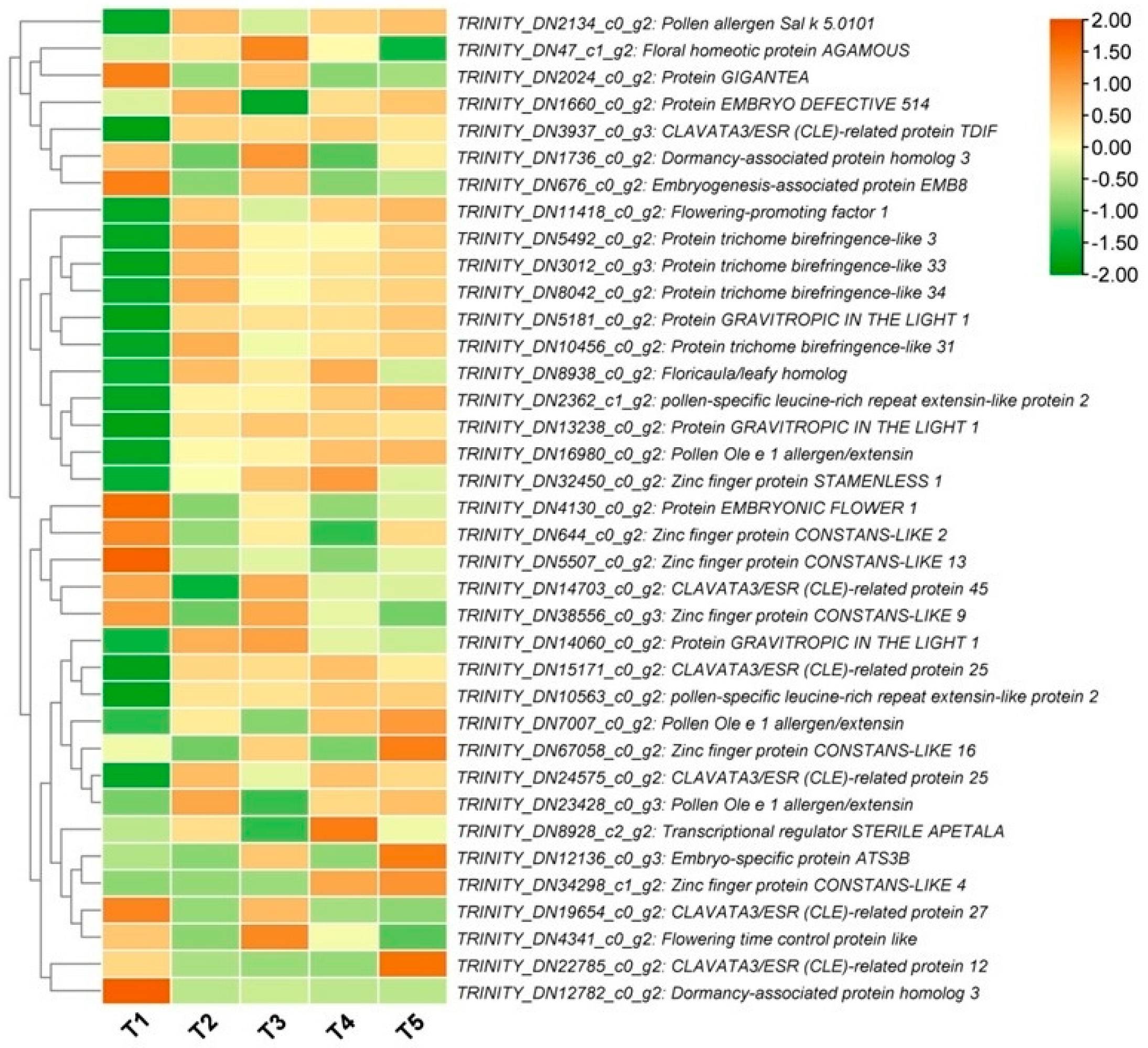
3.6. Key Phytohormone-Related Genes Involved in FBD Process in Michelia ‘Xin’
3.7. Key Transcription Factors (TFs) Regulating Flower Developmental Process in Michelia ‘Xin’
3.8. Circadian and Flowering-Related DEGs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Campos-Rivero, G.; Osorio-Montalvo, P.; Sánchez-Borges, R.; Us-Camas, R.; Duarte-Aké, F.; De-la-Peña, C. Plant Hormone Signaling in Flowering: An Epigenetic Point of View. J. Plant Physiol. 2017, 214, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Hung, F.Y.; Shih, Y.H.; Lin, P.Y.; Feng, Y.R.; Li, C.; Wu, K. WRKY63 Transcriptional Activation of COOLAIR and COLDAIR Regulates Vernalization-Induced Flowering. Plant Physiol. 2022, 190, 532–547. [Google Scholar] [CrossRef]
- Teixeira, C.; Hampton, J.; Moot, D. Phenological Development of Subterranean Clover Cultivars under Contrasting Environments. Ann. Appl. Biol. 2021, 179, 246–258. [Google Scholar] [CrossRef]
- Shrestha, R.; Gómez-Ariza, J.; Brambilla, V.; Fornara, F. Molecular Control of Seasonal Flowering in Rice, Arabidopsis and Temperate Cereals. Ann. Bot. 2014, 114, 1445–1458. [Google Scholar] [CrossRef] [PubMed]
- Quiroz, S.; Yustis, J.C.; Chávez-Hernández, E.C.; Martínez, T.; de la Paz Sanchez, M.; Garay-Arroyo, A.; Álvarez-Buylla, E.R.; García-Ponce, B. Beyond the Genetic Pathways, Flowering Regulation Complexity in Arabidopsis thaliana. Int. J. Mol. Sci. 2021, 22, 5716. [Google Scholar] [CrossRef] [PubMed]
- Kitazawa, M.S.; Fujimoto, K. A Dynamical Phyllotaxis Model to Determine Floral Organ Number. PLoS Comput. Biol. 2015, 11, e1004145. [Google Scholar] [CrossRef]
- Prenner, G.; Cardoso, D.; Zartman, C.E.; de Queiroz, L.P. Flowers of the Early-branching Papilionoid Legume Petaladenium urceoliferum Display Unique Morphological and Ontogenetic Features. Am. J. Bot. 2015, 102, 1780–1793. [Google Scholar] [CrossRef]
- Zhang, H.M.; Wang, M.J.; Hampton, J.; Rolston, P.; Lucas, R.; Ti, Z.H. Morphology of Flower Bud Differentiation and Floral Organ Specialization in Caucasian clover. Grass Forage Sci. 2024, 79, 381–391. [Google Scholar] [CrossRef]
- Huang, L.; Min, Y.; Schiessl, S.; Xiong, X.; Jan, H.U.; He, X.; Qian, W.; Guan, C.; Snowdon, R.J.; Hua, W.; et al. Integrative Analysis of GWAS and Transcriptome to Reveal Novel Loci Regulation Flowering Time in Semi-Winter Rapeseed. Plant Sci. 2021, 310, 110980. [Google Scholar] [CrossRef]
- Lee, J.; Lee, I. Regulation and Function of SOC1, a Flowering Pathway Integrator. J. Exp. Bot. 2010, 61, 2247–2254. [Google Scholar] [CrossRef]
- Sánchez-Corrales, Y.-E.; Álvarez-Buylla, E.R.; Mendoza, L. The Arabidopsis thaliana Flower Organ Specification Gene Regulatory Network Determines a Robust Differentiation Process. J. Theor. Biol. 2010, 264, 971–983. [Google Scholar] [CrossRef]
- Chávez-Hernández, E.C.; Quiroz, S.; García-Ponce, B.; Álvarez-Buylla, E.R. The Flowering Transition Pathways Converge into a Complex Gene Regulatory Network That Underlies the Phase Changes of the Shoot Apical Meristem in Arabidopsis thaliana. Front. Plant Sci. 2022, 13, 852047. [Google Scholar] [CrossRef]
- Song, H.; Duan, Z.; Huo, H.; Wang, X.; Wang, Y.; Chen, J.; Jin, L.; Lin, M. A Global Overview of Transcriptome Dynamics during the Late Stage of Flower Bud Development in Camellia oleifera. BMC Plant Biol. 2025, 25, 247. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Liu, L.; Qiang, X.; Meng, Y.; Li, Z.; Huang, F. Integrative Metabolomic and Transcriptomic Analysis Elucidates That the Mechanism of Phytohormones Regulates Floral Bud Development in Alfalfa. Plants 2024, 13, 1078. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Han, R.; Cai, K.; Guo, R.; Pei, X.; Zhao, X. Characterization of Phytohormones and Transcriptomic Profiling of the Female and Male Inflorescence Development in Manchurian Walnut (Juglans mandshurica Maxim.). Int. J. Mol. Sci. 2022, 23, 5433. [Google Scholar] [CrossRef]
- Wang, S.L.; An, H.R.; Tong, C.G.; Jang, S. Flowering and Flowering Genes: From Model Plants to Orchids. Hortic. Environ. Biotechnol. 2021, 62, 135–148. [Google Scholar] [CrossRef]
- Mishra, P.; Panigrahi, K.C. GIGANTEA—An Emerging Story. Front. Plant Sci. 2015, 6, 8. [Google Scholar] [CrossRef]
- Bond, D.M.; Dennis, E.S.; Pogson, B.J.; Finnegan, E.J. Histone Acetylation, Vernalization Insensitive 3, Flowering Locus C, and the Vernalization Response. Mol. Plant 2009, 2, 724–737. [Google Scholar] [CrossRef]
- Takada, S.; Akter, A.; Itabashi, E.; Nishida, N.; Shea, D.J.; Miyaji, N.; Mehraj, H.; Osabe, K.; Shimizu, M.; Takasaki-Yasuda, T.; et al. The Role of FRIGIDA and FLOWERING LOCUS C Genes in Flowering Time of Brassica rapa Leafy Vegetables. Sci. Rep. 2019, 9, 13843. [Google Scholar] [CrossRef] [PubMed]
- Srikanth, A.; Schmid, M. Regulation of Flowering Time: All Roads Lead to Rome. Cell. Mol. Life Sci. 2011, 68, 2013–2037. [Google Scholar] [CrossRef]
- Fan, Z.; Li, J.; Li, X.; Wu, B.; Wang, J.; Liu, Z.; Yin, H. Genome-Wide Transcriptome Profiling Provides Insights into Floral Bud Development of Summer-Flowering Camellia azalea. Sci. Rep. 2015, 5, 9729. [Google Scholar] [CrossRef]
- Qin, B.; Lu, X.; Sun, X.; Cui, J.; Deng, J.; Zhang, L. Transcriptome-Based Analysis of the Hormone Regulation Mechanism of Gender Differentiation in Juglans mandshurica Maxim. PeerJ 2021, 9, e12328. [Google Scholar] [CrossRef]
- Yuan, Z.; Zhang, D. Roles of Jasmonate Signalling in Plant Inflorescence and Flower Development. Curr. Opin. Plant Biol. 2015, 27, 44–51. [Google Scholar] [CrossRef]
- Wu, Z.; Huang, L.; Huang, F.; Lu, G.; Wei, S.; Liu, C.; Deng, H.; Liang, G. Temporal Transcriptome Analysis Provides Molecular Insights into Flower Development in Red-Flesh Pitaya. Electron. J. Biotechnol. 2022, 58, 55–69. [Google Scholar] [CrossRef]
- Cooke, J.E.K.; Eriksson, M.E.; Junttila, O. The Dynamic Nature of Bud Dormancy in Trees: Environmental Control and Molecular Mechanisms. Plant. Cell Environ. 2012, 35, 1707–1728. [Google Scholar] [CrossRef]
- Jiang, Z.; Sun, L.; Liu, X.; Liu, C.; Yin, Z. Nutritional Effect and Rhythm of Spring and Summer Flowering in Magnolia soulangeana‘Changchun’. Bull. Bot. Res. 2019, 39, 192–199. [Google Scholar]
- Sun, L.; Jiang, Z.; Ju, Y.; Zou, X.; Wan, X.; Chen, Y.; Yin, Z. A Potential Endogenous Gibberellin-Mediated Signaling Cascade Regulated Floral Transition in Magnolia × soulangeana ‘Changchun’. Mol. Genet. Genom. 2021, 296, 207–222. [Google Scholar] [CrossRef]
- Fan, L.; Chen, M.; Dong, B.; Wang, N.; Yu, Q.; Wang, X.; Xuan, L.; Wang, Y.; Zhang, S.; Shen, Y. Transcriptomic Analysis of Flower Bud Differentiation in Magnolia sinostellata. Genes 2018, 9, 212. [Google Scholar] [CrossRef]
- Chen, W.; Gong, L.; Guo, Z.; Wang, W.; Zhang, H.; Liu, X.; Yu, S.; Xiong, L.; Luoa, J. A Novel Integrated Method for Large-Scale Detection, Identification, and Quantification of Widely Targeted Metabolites: Application in the Study of Rice Metabolomics. Mol. Plant 2013, 6, 1769–1780. [Google Scholar] [CrossRef] [PubMed]
- Dossou, S.S.K.; Xu, F.; You, J.; Zhou, R.; Li, D.; Wang, L. Widely Targeted Metabolome Profiling of Different Colored Sesame (Sesamum indicum L.) Seeds Provides New Insight into Their Antioxidant Activities. Food Res. Int. 2022, 151, 110850. [Google Scholar] [CrossRef] [PubMed]
- Farag, M.A.; Khattab, A.R.; Maamoun, A.A.; Kropf, M.; Heiss, A.G. UPLC-MS Metabolome Based Classification of Lupinus and Lens Seeds: A Prospect for Phyto-Equivalency of Its Different Accessions. Food Res. Int. 2019, 115, 379–392. [Google Scholar] [CrossRef] [PubMed]
- Dossou, S.S.K.; Xu, F.; Cui, X.; Sheng, C.; Zhou, R.; You, J.; Tozo, K.; Wang, L. Comparative Metabolomics Analysis of Different Sesame (Sesamum indicum L.) Tissues Reveals a Tissue-Specific Accumulation of Metabolites. BMC Plant Biol. 2021, 21, 352. [Google Scholar] [CrossRef]
- Mu, W.; Wei, J.; Yang, T.; Fan, Y.; Cheng, L.; Yang, J.; Mu, R.; Liu, J.; Zhao, J.; Sun, W.; et al. RNA Extraction for Plant Samples Using CTAB-PBIOZOL V1. Protocols.Io 2017. [Google Scholar] [CrossRef]
- Cao, L.; Lu, X.; Wang, G.; Zhang, P.; Fu, J.; Wang, Z.; Wei, L.; Wang, T. Transcriptional Regulatory Networks in Response to Drought Stress and Rewatering in Maize (Zea mays L.). Mol. Genet. Genom. 2021, 296, 1203–1219. [Google Scholar] [CrossRef]
- Bai, M.; Zeng, W.; Chen, F.; Ji, X.; Zhuang, Z.; Jin, B.; Wang, J.; Jia, L.; Peng, Y. Transcriptome Expression Profiles Reveal Response Mechanisms to Drought and Drought-Stress Mitigation Mechanisms by Exogenous Glycine Betaine in Maize. Biotechnol. Lett. 2022, 44, 367–386. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De Novo Transcript Sequence Reconstruction from RNA-Seq Using the Trinity Platform for Reference Generation and Analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing Genome Assembly and Annotation Completeness with Single-Copy Orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript Assembly and Quantification by RNA-Seq Reveals Unannotated Transcripts and Isoform Switching during Cell Differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A Universal Tool for Annotation, Visualization and Analysis in Functional Genomics Research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Nie, T.; Jiang, Z.; Sun, L.; Chen, Y.; Li, J.; Yang, A.; Wei, Q.; Yin, Z. Reference Genes Selection for QRT-PCR Analysis in Various Flowering Transition Events of Magnolia × soulangeana ‘Changchun. ’ Sci. Hortic. 2023, 316, 112006. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-Length Transcriptome Assembly from RNA-Seq Data without a Reference Genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Wang, F.; Han, T.; Jeffrey Chen, Z. Circadian and Photoperiodic Regulation of the Vegetative to Reproductive Transition in Plants. Commun. Biol. 2024, 7, 579. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Lin, W.; Xu, Y.; Xie, B.; Yu, B.; Chen, L.; Huang, W. Flowering-Time Regulation by the Circadian Clock: From Arabidopsis to Crops. Crop J. 2024, 12, 17–27. [Google Scholar] [CrossRef]
- Chai, W.; He, X.; Wen, B.; Jiang, Y.; Zhang, Z.; Bai, R.; Zhang, X.; Xu, J.; Hou, L.; Li, M.; et al. The Molecular Mechanism of Relatively Low-Temperature-Induced Broccoli Flower Bud Differentiation Revealed by Transcriptomic Profiling. Horticulturae 2023, 9, 1353. [Google Scholar] [CrossRef]
- Komeda, Y. Genetic Regulation of Time to Flower in Arabidopsis thaliana. Annu. Rev. Plant Biol. 2004, 55, 521–535. [Google Scholar] [CrossRef]
- Luo, X.; Yin, M.; He, Y. Molecular Genetic Understanding of Photoperiodic Regulation of Flowering Time in Arabidopsis and Soybean. Int. J. Mol. Sci. 2022, 23, 466. [Google Scholar] [CrossRef]
- Putterill, J.; Robson, F.; Lee, K.; Simon, R.; Coupland, G. The CONSTANS Gene of Arabidopsis Promotes Flowering and Encodes a Protein Showing Similarities to Zinc Finger Transcription Factors. Cell 1995, 80, 847–857. [Google Scholar] [CrossRef] [PubMed]
- Samach, A.; Onouchi, H.; Gold, S.E.; Ditta, G.S.; Schwarz-Sommer, Z.; Yanofsky, M.F.; Coupland, G. Distinct Roles of CONSTANS Target Genes in Reproductive Development of Arabidopsis. Science 2000, 288, 1613–1616. [Google Scholar] [CrossRef] [PubMed]
- Sawa, M.; Kay, S.A. GIGANTEA Directly Activates Flowering Locus T in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2011, 108, 11698–11703. [Google Scholar] [CrossRef]
- Huang, G.; Ma, J.; Han, Y.; Chen, X.; Fu, Y.-F. Cloning and Expression Analysis of the Soybean CO-Like Gene GmCOL9. Plant Mol. Biol. Report. 2011, 29, 352–359. [Google Scholar] [CrossRef]
- Cheng, X.-F.; Wang, Z.-Y. Overexpression of COL9, a CONSTANS-LIKE Gene, Delays Flowering by Reducing Expression of CO and FT in Arabidopsis thaliana. Plant J. 2005, 43, 758–768. [Google Scholar] [CrossRef] [PubMed]
- Steinbach, Y. The Arabidopsis Thaliana CONSTANS-LIKE 4 (COL4)—A Modulator of Flowering Time. Front. Plant Sci. 2019, 10, 651. [Google Scholar] [CrossRef]
- Wu, W.; Zhang, Y.; Zhang, M.; Zhan, X.; Shen, X.; Yu, P.; Chen, D.; Liu, Q.; Sinumporn, S.; Hussain, K.; et al. The Rice CONSTANS-like Protein OsCOL15 Suppresses Flowering by Promoting Ghd7 and Repressing RID1. Biochem. Biophys. Res. Commun. 2018, 495, 1349–1355. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.L.; Jiang, J.F.; Ge, L.; Xu, Y.Y.; Chen, H.; Zhao, Y.; Bi, Y.R.; Wen, J.Q.; Chong, K. FPF1 Transgene Leads to Altered Flowering Time and Root Development in Rice. Plant Cell Rep. 2005, 24, 79–85. [Google Scholar] [CrossRef]
- Lee, M.B.; Shekasteband, R.; Hutton, S.F.; Lee, T.G. A Mutant Allele of the Flowering Promoting Factor 1 Gene at the Tomato BRACHYTIC Locus Reduces Plant Height with High Quality Fruit. Plant Direct 2022, 6, e422. [Google Scholar] [CrossRef]
- Wagner, D.; Sablowski, R.W.; Meyerowitz, E.M. Transcriptional Activation of APETALA1 by LEAFY. Science 1999, 285, 582–584. [Google Scholar] [CrossRef]
- Byzova, M.V.; Franken, J.; Aarts, M.G.; de Almeida-Engler, J.; Engler, G.; Mariani, C.; Van Lookeren Campagne, M.M.; Angenent, G.C. Arabidopsis STERILE APETALA, a Multifunctional Gene Regulating Inflorescence, Flower, and Ovule Development. Genes Dev. 1999, 13, 1002–1014. [Google Scholar] [CrossRef]
- Li, N.; Liu, Z.; Wang, Z.; Ru, L.; Gonzalez, N.; Baekelandt, A.; Pauwels, L.; Goossens, A.; Xu, R.; Zhu, Z.; et al. STERILE APETALA Modulates the Stability of a Repressor Protein Complex to Control Organ Size in Arabidopsis thaliana. PLoS Genet. 2018, 14, e1007218. [Google Scholar] [CrossRef]
- Nie, T.; Jiang, Z.; Sun, L.; Chen, Y.; Li, J.; Yang, A.; Yin, Z. Analysis on the Biological Basis of Stamen Abortion during the Second Flowering of Magnolia × soulangeana ‘Changchun’. Trees—Struct. Funct. 2022, 36, 1515–1528. [Google Scholar] [CrossRef]
- Liu, X.; Yuan, M.; Dang, S.; Zhou, J.; Zhang, Y. Comparative Transcriptomic Analysis of Transcription Factors and Hormones during Flower Bud Differentiation in “Red Globe” Grape under Red-blue Light. Sci. Rep. 2023, 13, 8932. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, L.; Shen, P.; Liu, X.; Zhao, R.; Zhu, J. Transcriptomic Insight into Underground Floral Differentiation in Erythronium japonicum. Biomed Res. Int. 2022, 2022, 4447472. [Google Scholar] [CrossRef]
- Hu, G.; Wang, K.; Huang, B.; Mila, I.; Frasse, P.; Maza, E.; Djari, A.; Hernould, M.; Zouine, M.; Li, Z.; et al. The Auxin-Responsive Transcription Factor SlDOF9 Regulates Inflorescence and Flower Development in Tomato. Nat. Plants 2022, 8, 419–433. [Google Scholar] [CrossRef] [PubMed]
- Krizek, B.A.; Blakley, I.C.; Ho, Y.Y.; Freese, N.; Loraine, A.E. The Arabidopsis Transcription Factor AINTEGUMENTA Orchestrates Patterning Genes and Auxin Signaling in the Establishment of Floral Growth and Form. Plant J. 2020, 103, 752–768. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, W.; Zhang, W.E.; Li, C.; Wang, R.; Pan, X.; Peng, J. Combined Transcriptional and Metabolomic Analysis of Flavonoids in the Regulation of Female Flower Bud Differentiation in Juglans sigillata Dode. BMC Plant Biol. 2025, 25, 168. [Google Scholar] [CrossRef]
- Gui, W.; Wu, P.; Wang, G.; Chen, S.; Feng, S. Systematic Identification and Prediction of Sex-Specific MADS-Box Genes in Zanthoxylum armatum during Flower Development. Sci. Hortic. 2024, 338, 113779. [Google Scholar] [CrossRef]
- Wang, P.; Wan, J.; Guo, L.; Li, Y.; Xiong, X.; Zhao, C.; Liu, Q.; Yu, J.; Xiang, L.; Liu, J.; et al. A MADS-Box Protein GhAGL8 Promotes Early Flowering and Increases Yield without Compromising Fiber Quality in Cotton. Ind. Crops Prod. 2025, 225, 120545. [Google Scholar] [CrossRef]
- Vera-Sirera, F.; Gomez, M.D.; Perez-Amador, M.A. DELLA Proteins, a Group of GRAS Transcription Regulators That Mediate Gibberellin Signaling; Elsevier Inc.: Amsterdam, The Netherlands, 2016; ISBN 9780128011270. [Google Scholar]
- Wang, P.; Zhang, Q.; Chen, Y.; Zhao, Y.; Ren, F.; Shi, H.; Wu, X. Comprehensive Identification and Analysis of DELLA Genes throughout the Plant Kingdom. BMC Plant Biol. 2020, 20, 372. [Google Scholar] [CrossRef]
- Vadassery, J.; Ballhorn, D.J.; Fleming, S.R.; Mazars, C.; Pandey, S.P.; Schmidt, A.; Schuman, M.C.; Yeh, K.-W.; Yilamujiang, A.; Mithöfer, A. Neomycin: An Effective Inhibitor of Jasmonate-Induced Reactions in Plants. J. Plant Growth Regul. 2019, 38, 713–722. [Google Scholar] [CrossRef]
- Lin, S.; Ye, M.; Li, X.; Xing, Y.; Liu, M.; Zhang, J.; Sun, X. A Novel Inhibitor of the Jasmonic Acid Signaling Pathway Represses Herbivore Resistance in Tea Plants. Hortic. Res. 2022, 9, uhab038. [Google Scholar] [CrossRef]
- He, Y.; Han, J.; Liu, R.; Ding, Y.; Wang, J.; Sun, L.; Yang, X.; Zeng, Y.; Wen, W.; Xu, J.; et al. Integrated Transcriptomic and Metabolomic Analyses of a Wax Deficient Citrus Mutant Exhibiting Jasmonic Acid-Mediated Defense against Fungal Pathogens. Hortic. Res. 2018, 5, 43. [Google Scholar] [CrossRef] [PubMed]
- Broun, P. Transcriptional Control of Flavonoid Biosynthesis: A Complex Network of Conserved Regulators Involved in Multiple Aspects of Differentiation in Arabidopsis. Curr. Opin. Plant Biol. 2005, 8, 272–279. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Dong, B.; Wang, L.; Song, Z.; Niu, L.; Li, H.; Cao, H.; Meng, D.; Fu, Y. CDPK6 Phosphorylates and Stabilizes MYB30 to Promote Hyperoside Biosynthesis That Prolongs the Duration of Full-Blooming in Okra. J. Exp. Bot. 2020, 71, 4042–4056. [Google Scholar] [CrossRef]
- Chakraborty, A.; Chaudhury, R.; Dutta, S.; Basak, M.; Dey, S.; Schäffner, A.R.; Das, M. Role of Metabolites in Flower Development and Discovery of Compounds Controlling Flowering Time. Plant Physiol. Biochem. 2022, 190, 109–118. [Google Scholar] [CrossRef]
- Wang, F.; Cai, Z.; Li, Z.; Zhang, S.; Luo, H.; Wu, Q.; Xia, H.; Guo, Y. FLOWERING LOCUS T (FT) in Photosensitive Type Chrysanthemum Accelerates Flowering in Arabidopsis. Phyton-Int. J. Exp. Bot. 2024, 93, 819–830. [Google Scholar] [CrossRef] [PubMed]




| ID | Log2FC | Annotation | |||
|---|---|---|---|---|---|
| T1_vs_T2 | T2_vs_T3 | T3_vs_T4 | T4_vs_T5 | ||
| TRINITY_DN1305_c5_g1 | 11.958 | 1.479 | 0.277 | 0.148 | Agamous-like MADS-box protein MADS9 |
| TRINITY_DN2535_c0_g1 | 11.146 | −1.720 | 1.971 | −0.600 | B3 domain-containing protein |
| TRINITY_DN9192_c0_g1 | 11.104 | 0.368 | 0.463 | 0.071 | MADS-box transcription factor 6 |
| TRINITY_DN30055_c0_g3 | 9.955 | −1.088 | 0.671 | 0.862 | NAC domain-containing protein 104 |
| TRINITY_DN18152_c0_g1 | 9.801 | −1.859 | 0.856 | 0.101 | Zinc finger CCCH domain-containing protein 9 |
| TRINITY_DN6301_c0_g1 | 9.647 | 2.300 | 0.135 | 0.398 | Agamous-like MADS-box protein MADS4 |
| TRINITY_DN4827_c0_g1 | 9.043 | −2.068 | 2.325 | −0.243 | B3 domain-containing protein Os06g0194400 |
| TRINITY_DN11681_c0_g1 | 9.042 | −0.641 | −0.207 | −0.238 | Transcription factor WER |
| TRINITY_DN604_c3_g2 | 8.766 | −4.143 | 4.401 | −3.295 | Transcription factor TRY |
| TRINITY_DN17178_c0_g1 | 8.650 | −3.142 | 3.312 | −1.467 | Transcription factor bHLH96 |
| TRINITY_DN3413_c4_g2 | 8.574 | −0.188 | 0.042 | 0.049 | MADS-box transcription factor 17 |
| TRINITY_DN16_c1_g1 | 8.535 | −1.442 | 0.530 | 1.226 | Transcription factor bHLH146 |
| TRINITY_DN8450_c0_g1 | 8.394 | −0.919 | 0.522 | 1.240 | Transcription factor PAR2 |
| TRINITY_DN4446_c0_g1 | 8.262 | −0.710 | 0.924 | 0.070 | F-box protein At1g10780 |
| TRINITY_DN22896_c0_g1 | -- | -- | -- | 8.385 | Myb-related protein 340 |
| TRINITY_DN65996_c0_g1 | 7.989 | −0.753 | −1.526 | -- | WAT1-related protein At1g21890 |
| TRINITY_DN203_c0_g1 | 7.825 | −3.287 | 1.255 | 2.045 | Transcription factor MYB1 |
| TRINITY_DN10061_c0_g1 | 7.688 | −2.343 | 3.145 | −0.731 | Basic leucine zipper 34 |
| TRINITY_DN7028_c0_g1 | 7.373 | −3.025 | -- | 1.794 | Transcription factor MYB1 |
| TRINITY_DN2070_c1_g2 | 7.296 | −1.582 | −0.365 | 1.891 | Transcription factor MYB83 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Li, D.; Ji, X.; Liu, C.; Huang, C. Dynamic Metabolome and Transcriptome Profiling Provide Molecular Insights into Floral Bud Differentiation in Michelia ‘Xin’. Biology 2025, 14, 1383. https://doi.org/10.3390/biology14101383
Chen Y, Li D, Ji X, Liu C, Huang C. Dynamic Metabolome and Transcriptome Profiling Provide Molecular Insights into Floral Bud Differentiation in Michelia ‘Xin’. Biology. 2025; 14(10):1383. https://doi.org/10.3390/biology14101383
Chicago/Turabian StyleChen, Yan, Dapeng Li, Xiaoling Ji, Caixian Liu, and Chenfei Huang. 2025. "Dynamic Metabolome and Transcriptome Profiling Provide Molecular Insights into Floral Bud Differentiation in Michelia ‘Xin’" Biology 14, no. 10: 1383. https://doi.org/10.3390/biology14101383
APA StyleChen, Y., Li, D., Ji, X., Liu, C., & Huang, C. (2025). Dynamic Metabolome and Transcriptome Profiling Provide Molecular Insights into Floral Bud Differentiation in Michelia ‘Xin’. Biology, 14(10), 1383. https://doi.org/10.3390/biology14101383




