Mitogenomic Characterization of Mining Bee Family Andrenidae (Hymenoptera: Apoidea: Anthophila) and Insights into Bee Phylogeny
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Taxon Sampling and Sequencing
2.2. Assembly, Annotation, and Composition Analyses
2.3. Phylogenetic Analysis
3. Results and Discussion
3.1. Mitogenomic Organization
3.2. Protein-Coding Genes and Evolutionary Rates
3.3. Gene Rearrangement
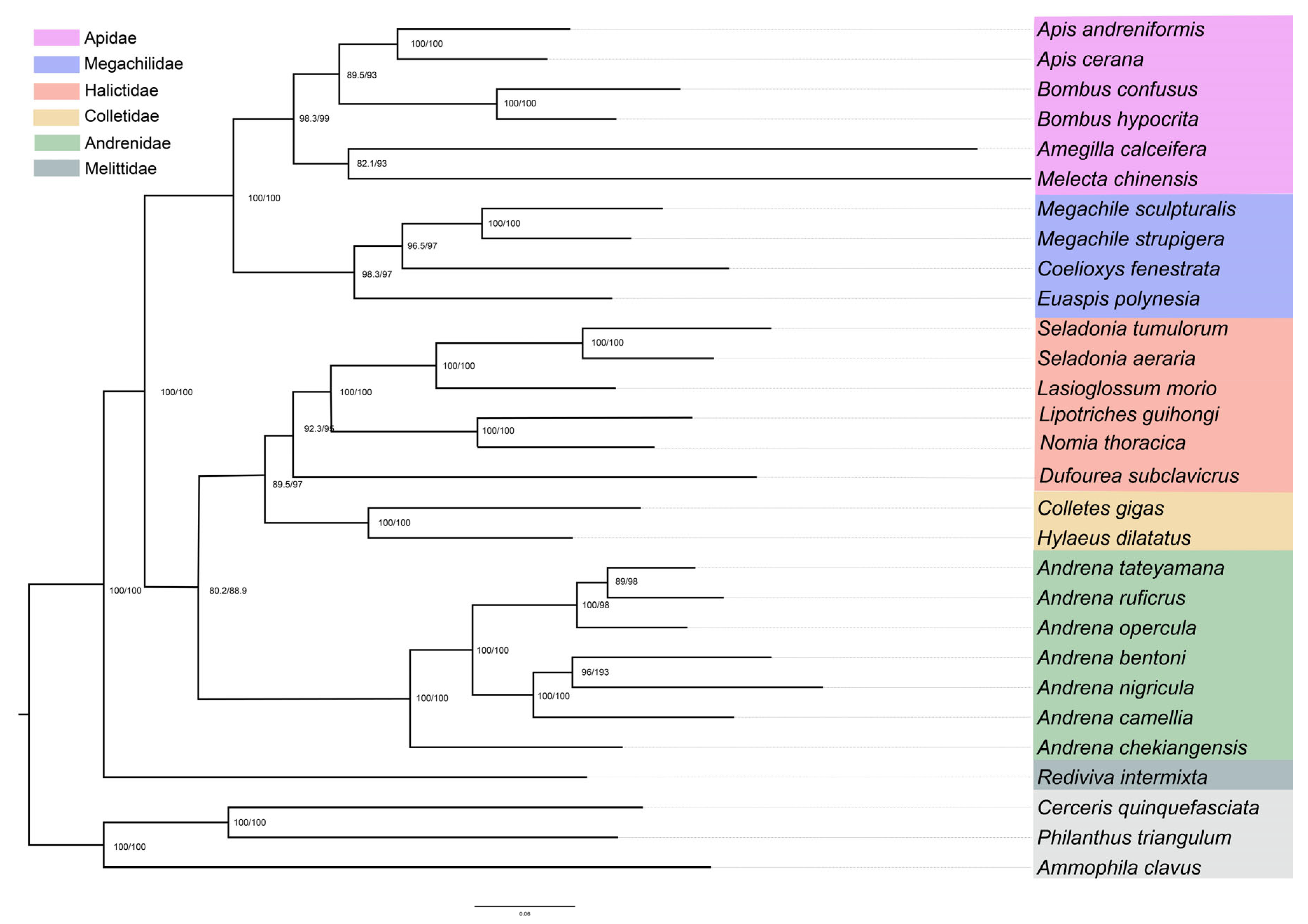
3.4. Phylogenetic Relationships
4. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ascher, J.S.; Pickering, J. Discover Life Bee Species Guide and World Checklist (Hymenoptera: Apoidea: Anthophila). Available online: http://www.discoverlife.org/mp/20q?guide=Apoidea_species (accessed on 20 August 2025).
- Klein, A.M.; Vaissière, B.E.; Cane, J.H.; Steffan-Dewenter, I.; Cunningham, S.A.; Kremen, C.; Tscharntke, T.J. Importance of pollinators in changing landscapes for world crops. Proc. R. Soc. B 2007, 274, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Ollerton, J. Pollinator diversity: Distribution, ecological function, and conservation. Annu. Rev. Ecol. Syst. 2017, 48, 353–376. [Google Scholar] [CrossRef]
- Danforth, B.N.; Minckley, R.L.; Neff, J.L.; Fawcett, F. The Solitary Bees: Biology, Evolution, Conservation; Princeton University Press: Priceton, NJ, USA, 2019. [Google Scholar]
- Michener, C.D. The Bees of the World, 2nd ed.; The John Hopkins University Press: London, UK, 2007; 953p. [Google Scholar]
- Michener, C.D. Comparative external morphology, phylogeny, and a classification of the bees (Hymenoptera). Bull. Am. Mus. Nat. Hist. 1994, 82, 151–326. [Google Scholar]
- Danforth, B.N.; Almeida, E.A. Insights into bee evolution: A tribute to Charles D. Michener. Apidologie 2008, 39, 1–2. [Google Scholar] [CrossRef]
- Danforth, B.N.; Cardinal, S.; Praz, C.; Almeida, E.A.; Michez, D. The impact of molecular data on our understanding of bee phylogeny and evolution. Annu. Rev. Entomol. 2013, 58, 57–78. [Google Scholar] [CrossRef] [PubMed]
- Danforth, B.N.; Fang, J.; Sipes, S. Analysis of family-level relationships in bees (Hymenoptera: Apiformes) using 28S and two previously unexplored nuclear genes: CAD and RNA polymerase II. Mol. Phylogenet. Evol. 2006, 39, 358–372. [Google Scholar] [CrossRef]
- Michener, C.D. The Bees of the World, 1st ed.; The John Hopkins University Press: London, UK, 2000; 913p. [Google Scholar]
- Engel, M.S. A monograph of the Baltic amber bees and evolution of the Apoidea (Hymenoptera). Bull. Am. Mus. Nat. Hist. 2001, 2001, 1–192. [Google Scholar] [CrossRef]
- Alexander, B.A.; Michener, C.D. Phylogenetic studies of the families of short-tongued bees (Hymenoptera: Apoidea). Univ. Kans. Sci. Bull. 1995, 55, 377–424. [Google Scholar]
- Branstetter, M.G.; Longino, J.T.; Ward, P.S.; Faircloth, B.C. Enriching the ant tree of life: Enhanced UCE bait set for genome-scale phylogenetics of ants and other Hymenoptera. Methods Ecol. Evol. 2017, 8, 768–776. [Google Scholar] [CrossRef]
- Sann, M.; Meusemann, K.; Niehuis, O.; Escalona, H.E.; Mokrousov, M.; Ohl, M.; Pauli, T.; Schmid-Egger, C.J. Reanalysis of the apoid wasp phylogeny with additional taxa and sequence data confirms the placement of Ammoplanidae as sister to bees. Syst. Entomol. 2021, 46, 558–569. [Google Scholar] [CrossRef]
- Sann, M.; Niehuis, O.; Peters, R.S.; Mayer, C.; Kozlov, A.; Podsiadlowski, L.; Bank, S.; Meusemann, K.; Misof, B.; Bleidorn, C.; et al. Phylogenomic analysis of Apoidea sheds new light on the sister group of bees. BMC Evol. Biol. 2018, 18, 71. [Google Scholar] [CrossRef]
- Larkin, L.L.; Neff, J.L.; Simpson, B. Phylogeny of the Callandrena subgenus of Andrena (Hymenoptera: Andrenidae) based on mitochondrial and nuclear DNA data: Polyphyly and convergent evolution. Mol. Phylogenet. Evol. 2006, 38, 330–343. [Google Scholar] [CrossRef]
- Paxton, R.; Tengö, J. Intranidal mating, emergence, and sex ratio in a communal bee Andrena jacobi Perkins 1921 (Hymenoptera: Andrenidae). J. Insect Behav. 1996, 9, 421–440. [Google Scholar] [CrossRef]
- Paxton, R.; Giovanetti, M.; Andrietti, F.; Scamoni, E.; Scanni, B. Mating in a communal bee, Andrena agilissima (Hymenoptera Andrenidae). Ethol. Ecol. Evol. 1999, 11, 371–382. [Google Scholar] [CrossRef]
- Cameron, S.L. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef] [PubMed]
- Ge, X.; Peng, L.; Vogler, A.P.; Morse, J.C.; Yang, L.; Sun, C.; Wang, B. Massive gene rearrangements of mitochondrial genomes and implications for the phylogeny of Trichoptera (Insecta). Syst. Entomol. 2022, 48, 278–295. [Google Scholar] [CrossRef]
- Lin, X.L.; Liu, Z.; Yan, L.P.; Duan, X.; Bu, W.J.; Wang, X.H.; Zheng, C.G. Mitogenomes provide new insights of evolutionary history of Boreheptagyiini and Diamesini (Diptera: Chironomidae: Diamesinae). Ecol. Evol. 2022, 12, e8957. [Google Scholar] [CrossRef]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids. Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef]
- Brown, W.N. The mitochondrial genome of animals. In Molecular Evolutionary Genetics; MacIntyre, R.J., Ed.; Plenum: New York, NY, USA, 1985; Volume 22, pp. 252–271. [Google Scholar]
- Zhang, D.; He, F.X.; Li, X.B.; Aishan, Z.; Lin, X.L. New mitogenomes of the Polypedilum generic complex (Diptera: Chironomidae): Characterization and phylogenetic implications. Insects 2023, 14, 238. [Google Scholar] [CrossRef]
- Cameron, S.L. Insect mitochondrial genomics: A decade of progress. Annu. Rev. Entomol. 2025, 70, 83–101. [Google Scholar] [CrossRef]
- Tadauchi, O.; Hirashima, Y. Synopsis of Andrena ceuandrena of Japan (Hymenoptera, andrenidae). Esakia 1984, 22, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.L.; Tadauchi, O.; Wu, Y.R. A revision of the subgenus Oreomelissa of the genus Andrena of eastern Asia (Hymenoptera, Andrenidae). Esakia 2000, 40, 41–61. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Dierckxsens, N.; Mardulyn, P.; Smits, G.J. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017, 45, e18. [Google Scholar] [CrossRef]
- Peng, Y.; Leung, H.C.; Yiu, S.M.; Chin, F.Y. IDBA–UD: A de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 2012, 28, 1420–1428. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE Online: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Shen, W.; Le, S.; Li, Y.; Hu, F. SeqKit: A cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS ONE 2016, 11, e0163962. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Branstetter, M.G.; Danforth, B.N.; Pitts, J.P.; Faircloth, B.C.; Ward, P.S.; Buffington, M.L.; Gates, M.W.; Kula, R.R.; Brady, S.G. Phylogenomic insights into the evolution of stinging wasps and the origins of ants and bees. Curr. Biol. 2017, 27, 1019–1025. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T.J.B. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Kück, P.; Longo, G.C. FASconCAT-G: Extensive functions for multiple sequence alignment preparations concerning phylogenetic studies. Fron. Zool. 2014, 11, 81. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.; Von Haeseler, A.; Jermiin, L.S.J. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Lartillot, N.; Rodrigue, N.; Stubbs, D.; Richer, J.J. PhyloBayes MPI: Phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst. Biol. 2013, 62, 611–615. [Google Scholar] [CrossRef]
- Zhang, D.; Niu, Z. Comparative analyses reveal mitogenome characteristics of Halictidae and novel rearrangement (Hymenoptera: Apoidea: Anthophila). Animals 2025, 15, 2234. [Google Scholar] [CrossRef]
- He, B.; Su, T.; Wu, Y.; Xu, J.; Huang, D. Phylogenetic analysis of the mitochondrial genomes in bees (Hymenoptera: Apoidea: Anthophila). PLoS ONE 2018, 13, e0202187. [Google Scholar] [CrossRef]
- Zheng, B.Y.; Cao, L.J.; Tang, P.; Van Achterberg, K.; Hoffmann, A.A.; Chen, H.Y.; Chen, X.X.; Wei, S.J. Gene arrangement and sequence of mitochondrial genomes yield insights into the phylogeny and evolution of bees and sphecid wasps (Hymenoptera: Apoidea). Mol. Phylogenet. Evol. 2018, 124, 1–9. [Google Scholar] [CrossRef]
- Tang, P.; Zhu, J.; Zheng, B.; Wei, S.J.; Sharkey, M.; Chen, X.X.; Vogler, A.P. Mitochondrial phylogenomics of the Hymenoptera. Mol. Phylogenet. Evol. 2019, 131, 8–18. [Google Scholar] [CrossRef]
- Wei, S.J.; Li, Q.; van Achterberg, K.; Chen, X.X. Two mitochondrial genomes from the families Bethylidae and Mutillidae: Independent rearrangement of protein-coding genes and higher-level phylogeny of the Hymenoptera. Mol. Phylogenet. Evol. 2014, 77, 1–10. [Google Scholar] [CrossRef]
- Dowton, M.; Castro, L.R.; Campbell, S.L.; Bargon, S.D.; Austin, A.D. Frequent mitochondrial gene rearrangements at the hymenopteran nad3-nad5 junction. J. Mol. Evol. 2003, 56, 517–526. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.J.; Shi, M.; Sharkey, M.J.; van Achterberg, C.; Chen, X.X. Comparative mitogenomics of Braconidae (Insecta: Hymenoptera) and the phylogenetic utility of mitochondrial genomes with special reference to Holometabolous insects. BMC Genom. 2010, 11, 371. [Google Scholar] [CrossRef] [PubMed]
- Bratic, A.; Clemente, P.; Calvo-Garrido, J.; Maffezzini, C.; Felser, A.; Wibom, R.; Wedell, A.; Freyer, C.; Wredenberg, A.J. Mitochondrial polyadenylation is a one-step process required for mRNA integrity and tRNA maturation. PLoS Genet. 2016, 12, e1006028. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Xu, X.; Jin, X.; Yin, H.; Luo, J.; Liu, G.; Zhao, Q.; Chen, Z.; Bu, W.; Gao, S.J. Using high-resolution annotation of insect mitochondrial DNA to decipher tandem repeats in the control region. RNA Biol. 2019, 16, 830–837. [Google Scholar] [CrossRef]
- Toompuu, M.; Tuomela, T.; Laine, P.; Paulin, L.; Dufour, E.; Jacobs, H.T. Polyadenylation and degradation of structurally abnormal mitochondrial tRNAs in human cells. Nucleic Acids. Res. 2018, 46, 5209–5226. [Google Scholar] [CrossRef]
- Hurst, L.D. The Ka/Ks ratio: Diagnosing the form of sequence evolution. Trends Genet. 2002, 18, 486. [Google Scholar] [CrossRef]
- Yang, Z.; Bielawski, J.P. Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. 2000, 15, 496–503. [Google Scholar] [CrossRef]
- Luo, L.; Carpenter, J.M.; Chen, B.; Li, T. First comprehensive analysis of both mitochondrial characteristics and mitogenome-based phylogenetics in the subfamily Eumeninae (Hymenoptera: Vespidae). Insects 2022, 13, 529. [Google Scholar] [CrossRef]
- Song, S.N.; Tang, P.; Wei, S.J.; Chen, X.X. Comparative and phylogenetic analysis of the mitochondrial genomes in basal hymenopterans. Sci. Rep. 2016, 6, 20972. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liu, H.; Shi, A.; Stys, P.; Zhou, X.; Cai, W. The complete mitochondrial genome and novel gene arrangement of the unique-headed bug Stenopirates sp. (Hemiptera: Enicocephalidae). PLoS ONE 2012, 7, e29419. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Shi, L.; Zhou, Y.; Xie, H.; Dai, X.; Li, R.; Chen, Y.; Wang, H. Molecular evolutionary mechanisms driving functional diversification of α-glucosidase in Lepidoptera. Sci. Rep. 2017, 7, 45787. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, K.; Chakraborty, R.; Cameron, S.L.; Sweet, A.D.; Chandra, K.; Kumar, V. Rearrangement and evolution of mitochondrial genomes in Thysanoptera (Insecta). Sci. Rep. 2020, 10, 695. [Google Scholar] [CrossRef]
- Dowton, M.; Austin, A.D. Evolutionary dynamics of a mitochondrial rearrangement “hot spot” in the Hymenoptera. Mol. Biol. Evol. 1999, 16, 298–309. [Google Scholar] [CrossRef]
- Moritz, C.; Dowling, T.E.; Brown, W.M. Evolution of animal mitochondrial DNA: Relevance for population biology and systematics. Annu. Rev. Ecol. Evol. Syst. 1987, 18, 269–292. [Google Scholar] [CrossRef]
- Lavrov, D.V.; Jeffrey, L.B.; Wesley, M.B. Complete mtDNA sequences of two millipedes suggest a new model for mitochondrial gene rearrangements: Duplication and nonrandom loss. Mol. Biol. Evol. 2002, 19, 163–169. [Google Scholar] [CrossRef]
- Dowton, M.; Campbell, N.J.H. Intramitochondrial recombination—Is it why some mitochondrial genes sleep around? Trends Ecol. Evol. 2001, 16, 269–271. [Google Scholar] [CrossRef]
- Rawlings, T.A.; Collins, T.M.; Bieler, R. Changing identities: tRNA duplication and remolding within animal mitochondrial genomes. Proc. Natl. Acad. Sci. USA 2003, 100, 15700–15705. [Google Scholar] [CrossRef]
- Clary, D.O.; Wolstenholme, D.R. The mitochondrial DNA molecule of Drosophila yakuba: Nucleotide sequence, gene organization, and genetic code. J. Mol. Evol. 1985, 22, 252–271. [Google Scholar] [CrossRef]
- Brady, S.G.; Litman, J.R.; Danforth, B.N. Rooting phylogenies using gene duplications: An empirical example from the bees (Apoidea). Mol. Phylogenet. Evol. 2011, 60, 295–304. [Google Scholar] [CrossRef]
- Kahnt, B.; Gerth, M.; Paxton, R.J.; Bleidorn, C.; Husemann, M. The complete mitochondrial genome of the endemic and highly specialized South African bee species Rediviva intermixta (Hymenoptera: Melittidae), with a comparison with other bee mitogenomes. Biol. J. Linn. Soc. 2015, 116, 940–953. [Google Scholar] [CrossRef]
- Pratlong, M.; Rancurel, C.; Pontarotti, P.; Aurelle, D.J. Monophyly of Anthozoa (Cnidaria): Why do nuclear and mitochondrial phylogenies disagree? Zool. Scr. 2017, 46, 363–371. [Google Scholar] [CrossRef]
- Fenn, J.D.; Song, H.; Cameron, S.L.; Whiting, M.F. A preliminary mitochondrial genome phylogeny of Orthoptera (Insecta) and approaches to maximizing phylogenetic signal found within mitochondrial genome data. Mol. Phylogenet. Evol. 2008, 49, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Timmermans, M.J.; Barton, C.; Haran, J.; Ahrens, D.; Culverwell, C.L.; Ollikainen, A.; Dodsworth, S.; Foster, P.G.; Bocak, L.; Vogler, A.P. Family-level sampling of mitochondrial genomes in Coleoptera: Compositional heterogeneity and phylogenetics. Genome Biol. Evol. 2016, 8, 161–175. [Google Scholar] [CrossRef] [PubMed]
- Song, F.; Li, H.; Jiang, P.; Zhou, X.; Liu, J.; Sun, C.; Vogler, A.P.; Cai, W.J. Capturing the phylogeny of Holometabola with mitochondrial genome data and Bayesian site-heterogeneous mixture models. Genome Biol. Evol. 2016, 8, 1411–1426. [Google Scholar] [CrossRef] [PubMed]
- Morgan, C.C.; Foster, P.G.; Webb, A.E.; Pisani, D.; McInerney, J.O.; O’Connell, M.J. Heterogeneous models place the root of the placental mammal phylogeny. Mol. Biol. Evol. 2013, 30, 2145–2156. [Google Scholar] [CrossRef]
- Sheffield, N.C.; Song, H.; Cameron, S.L.; Whiting, M.F. Nonstationary evolution and compositional heterogeneity in beetle mitochondrial phylogenomics. Syst. Biol. 2009, 58, 381–394. [Google Scholar] [CrossRef]
- Liu, Y.; Song, F.; Jiang, P.; Wilson, J.J.; Cai, W.; Li, H. Compositional heterogeneity in true bug mitochondrial phylogenomics. Mol. Phylogenet. Evol. 2018, 118, 135–144. [Google Scholar] [CrossRef]
- Jiang, J.; Wu, T.; Deng, J.; Peng, L. A compositional heterogeneity analysis of mitochondrial phylogenomics in chalcidoidea involving two newly sequenced mitogenomes of eupelminae (Hymenoptera: Chalcidoidea). Genes 2022, 13, 2340. [Google Scholar] [CrossRef]
- Zhang, D.; Niu, Z.Q.; Luo, A.R.; Orr, M.C.; Ferrari, R.R.; Jin, J.F.; Wu, Q.T.; Zhang, F.; Zhu, C.D. Testing the systematic status of Homalictus and Rostrohalictus with weakened cross-vein groups within Halictini (Hymenoptera: Halictidae) using low-coverage whole-genome sequencing. Insects Sci. 2022, 29, 1819–1833. [Google Scholar] [CrossRef]
- Sun, X.; Ding, Y.; Orr, M.C.; Zhang, F. Streamlining universal single-copy orthologue and ultraconserved element design: A case study in Collembola. Mol. Ecol. Resour. 2020, 20, 706–717. [Google Scholar] [CrossRef]
- Ge, X.; Peng, L.; Morse, J.C.; Wang, J.; Zang, H.; Yang, L.; Sun, C.; Wang, B. Phylogenomics resolves a 100-year-old debate regarding the evolutionary history of caddisflies (Insecta: Trichoptera). Mol. Phylogenet. Evol. 2024, 201, 108196. [Google Scholar] [CrossRef]
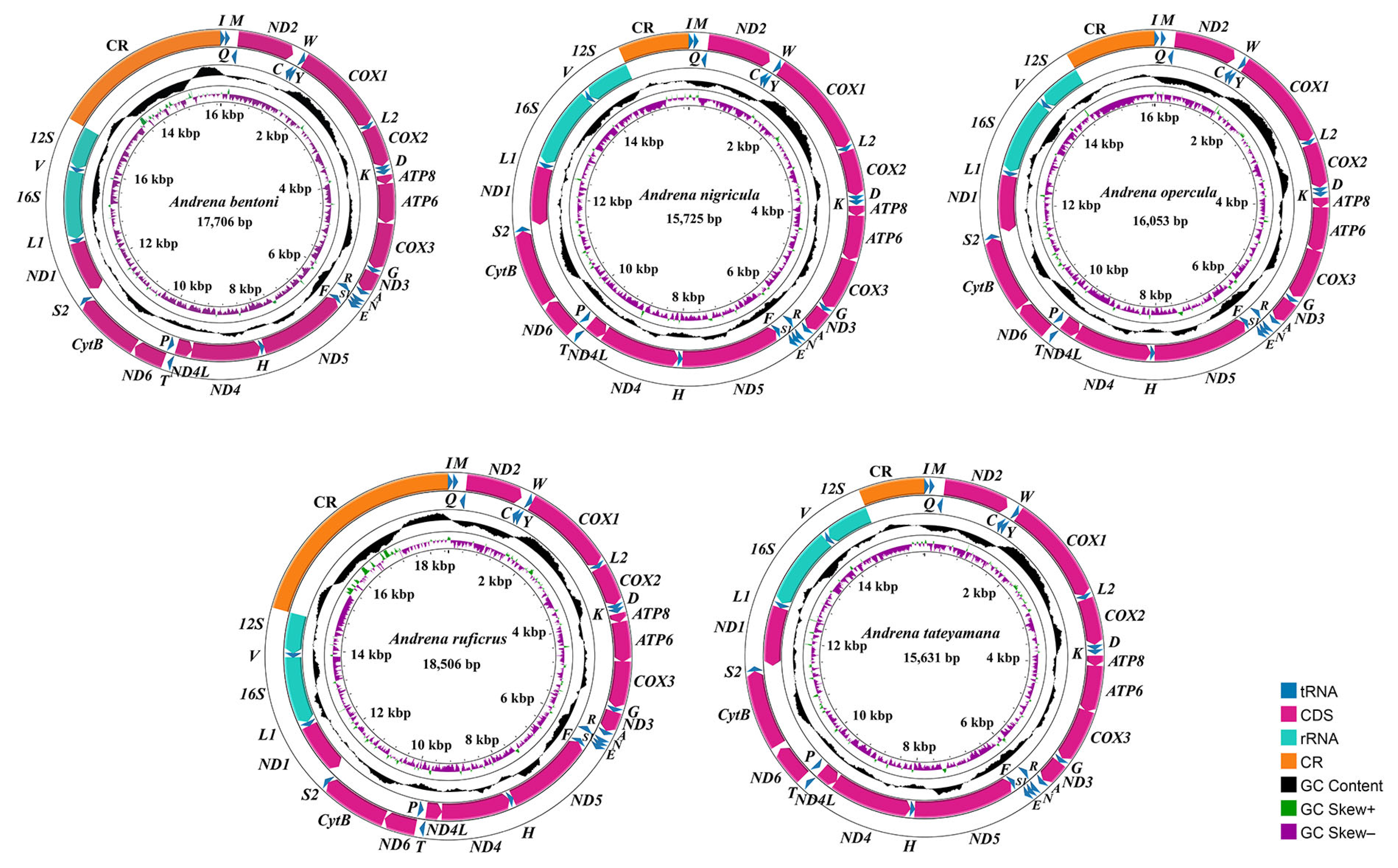
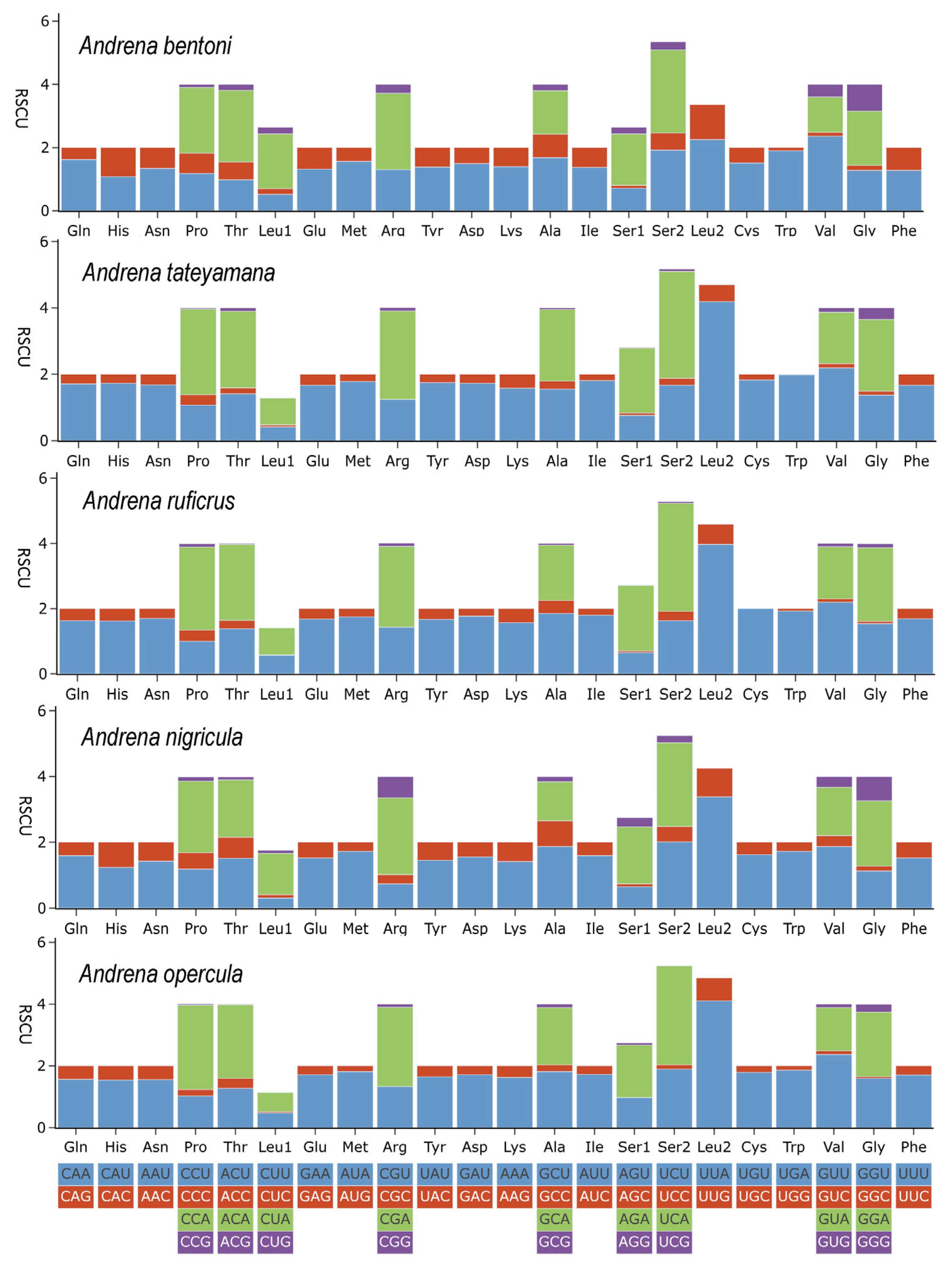

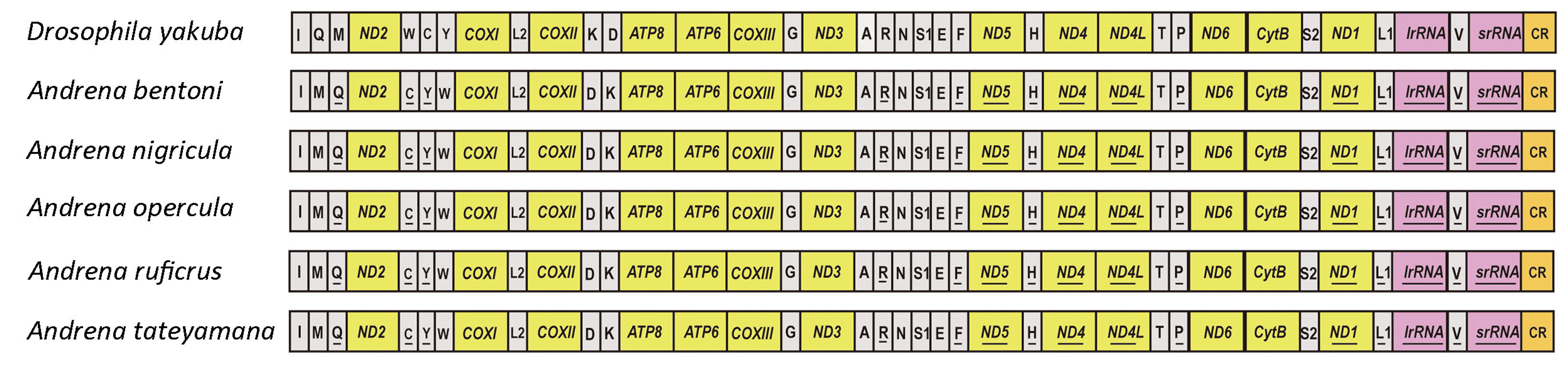
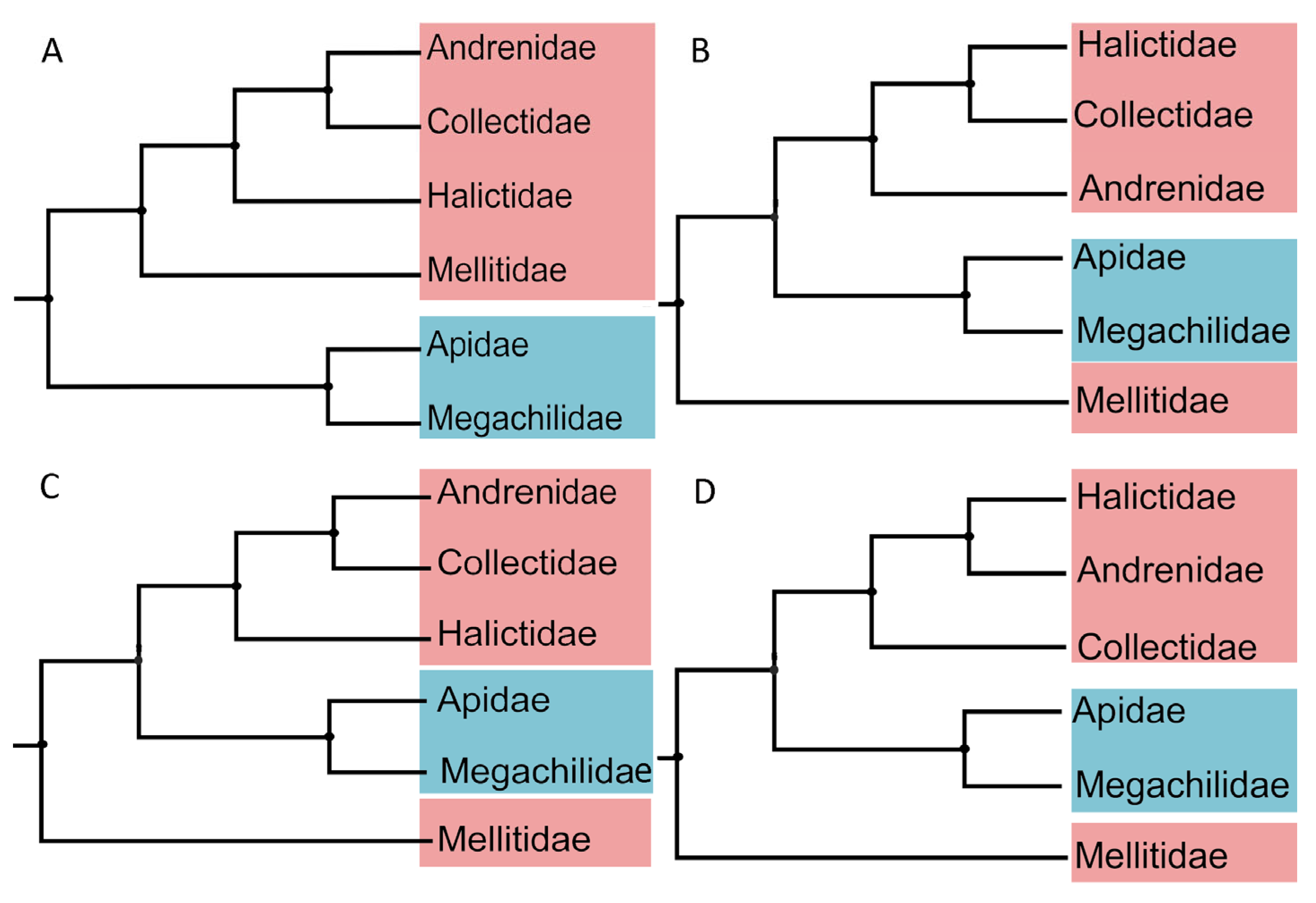
| Species | Location | Longitude and Latitude | Elevation (m) | Date | Collector | Accession Number |
|---|---|---|---|---|---|---|
| Andrena bentoni | Xizang, Yadong | E88.9330, N27.5198 | 3660 | 2023.7.9 | Qingtao Wu | PX147440 |
| Andrena nigricula | Xizang, Jilong | E85.2393, N28.5808 | 4082 | 2023.7.15 | Qingtao Wu | PX147441 |
| Andrena opercula | Xizang, Jilong | E85.3086, N28.3987 | 2858 | 2023.7.17 | Qingtao Wu | PX147442 |
| Andrena ruficrus | Xizang, Jilong | E85.4555, N28.3881 | 3265 | 2023.7.22 | Qingtao Wu | PX147443 |
| Andrena tateyamana | Xizang, Bomi | E94.8044, N30.2510 | 2262 | 2023.6.26 | Qingtao Wu | PX147444 |
| Samples | Regions | Length (bp) | A (%) | T (%) | C (%) | G (%) | A + T (%) | G + C (%) | AT-Skew | GC-Skew |
|---|---|---|---|---|---|---|---|---|---|---|
| Andrena bentoni | Whole genome | 17,706 | 42.49 | 32.11 | 17.19 | 8.14 | 74.60 | 25.32 | 0.139 | −0.357 |
| PCGs | 11,074 | 34.92 | 38.38 | 15.61 | 11.07 | 73.30 | 26.68 | −0.047 | −0.170 | |
| Site 1 | 3692 | 40.52 | 31.29 | 13.89 | 14.30 | 71.81 | 28.19 | 0.128 | 0.014 | |
| Site 2 | 3691 | 23.09 | 49.75 | 16.36 | 10.80 | 72.84 | 27.16 | −0.366 | −0.205 | |
| Site 12 | 7383 | 31.80 | 40.52 | 15.13 | 12.55 | 72.33 | 27.67 | −0.121 | −0.093 | |
| Site 3 | 3691 | 41.15 | 34.10 | 16.57 | 8.10 | 75.25 | 24.68 | 0.094 | −0.343 | |
| tRNA | 1429 | 42.20 | 39.19 | 7.56 | 11.06 | 81.39 | 18.61 | 0.037 | 0.188 | |
| l-rRNA | 1278 | 35.99 | 43.74 | 6.49 | 13.77 | 79.73 | 20.27 | −0.097 | 0.359 | |
| s-rRNA | 765 | 35.42 | 41.70 | 6.14 | 16.73 | 77.12 | 22.88 | −0.081 | 0.463 | |
| CR | 2975 | 40.40 | 35.93 | 13.78 | 9.55 | 76.33 | 23.33 | 0.059 | −0.181 | |
| Andrena nigricula | Whole genome | 15,725 | 42.52 | 34.10 | 15.80 | 7.55 | 76.62 | 23.35 | 0.110 | −0.353 |
| PCGs | 11,051 | 36.34 | 40.00 | 13.43 | 10.23 | 76.34 | 23.66 | −0.048 | −0.135 | |
| Site 1 | 3685 | 43.63 | 32.06 | 11.62 | 12.69 | 75.69 | 24.31 | 0.153 | 0.044 | |
| Site 2 | 3684 | 23.09 | 50.26 | 15.90 | 10.76 | 73.34 | 26.66 | −0.370 | −0.193 | |
| Site 12 | 7369 | 33.36 | 41.16 | 13.76 | 11.72 | 74.52 | 25.48 | −0.105 | −0.080 | |
| Site 3 | 3682 | 42.30 | 37.69 | 12.76 | 7.25 | 79.99 | 20.01 | 0.058 | −0.275 | |
| tRNA | 1424 | 40.66 | 40.66 | 8.01 | 10.67 | 81.32 | 18.68 | 0.000 | 0.142 | |
| l-rRNA | 1280 | 36.72 | 42.97 | 5.94 | 14.37 | 79.69 | 20.31 | −0.078 | 0.415 | |
| s-rRNA | 776 | 39.56 | 41.88 | 5.15 | 13.40 | 81.44 | 18.56 | −0.028 | 0.445 | |
| CR | 1032 | 42.73 | 32.75 | 18.31 | 5.72 | 75.48 | 24.03 | 0.132 | −0.524 | |
| Andrena opercula | Whole genome | 16,053 | 45.21 | 34.57 | 13.68 | 6.47 | 79.78 | 20.15 | 0.133 | −0.358 |
| PCGs | 11,071 | 37.68 | 42.02 | 11.04 | 9.26 | 79.69 | 20.31 | −0.054 | −0.088 | |
| Site 1 | 3691 | 43.53 | 34.17 | 9.72 | 12.59 | 77.69 | 22.31 | 0.120 | 0.129 | |
| Site 2 | 3690 | 23.43 | 50.34 | 15.14 | 11.09 | 73.77 | 26.23 | −0.365 | −0.155 | |
| Site 12 | 7381 | 33.48 | 42.25 | 12.43 | 11.84 | 75.73 | 24.27 | −0.116 | −0.024 | |
| Site 3 | 3690 | 46.07 | 41.54 | 8.27 | 4.11 | 87.61 | 12.39 | 0.052 | −0.336 | |
| tRNA | 1419 | 43.55 | 40.38 | 6.77 | 9.30 | 83.93 | 16.07 | 0.038 | 0.157 | |
| l-rRNA | 1268 | 36.51 | 45.58 | 5.52 | 12.38 | 82.10 | 17.90 | −0.110 | 0.383 | |
| s-rRNA | 746 | 37.40 | 43.83 | 5.63 | 13.14 | 81.23 | 18.77 | −0.079 | 0.400 | |
| CR | 1357 | 49.01 | 29.26 | 18.13 | 2.73 | 78.27 | 20.86 | 0.252 | −0.738 | |
| Andrena ruficrus | Whole genome | 18,506 | 43.82 | 36.02 | 12.98 | 7.14 | 79.84 | 20.13 | 0.098 | −0.290 |
| PCGs | 11,080 | 37.60 | 42.08 | 10.96 | 9.36 | 79.68 | 20.32 | −0.056 | −0.079 | |
| Site 1 | 3694 | 42.88 | 33.18 | 10.76 | 13.18 | 76.06 | 23.94 | 0.128 | 0.101 | |
| Site 2 | 3693 | 23.35 | 50.67 | 15.07 | 10.91 | 74.02 | 25.98 | −0.369 | −0.160 | |
| Site 12 | 7387 | 33.12 | 41.92 | 12.92 | 12.04 | 75.04 | 24.96 | −0.117 | −0.035 | |
| Site 3 | 3693 | 46.56 | 42.40 | 7.04 | 4.00 | 88.96 | 11.04 | 0.047 | −0.276 | |
| tRNA | 1423 | 43.08 | 40.34 | 6.89 | 9.70 | 83.42 | 16.58 | 0.033 | 0.169 | |
| l-rRNA | 1267 | 35.44 | 45.15 | 6.08 | 13.34 | 80.58 | 19.42 | −0.120 | 0.374 | |
| s-rRNA | 744 | 36.83 | 43.55 | 6.05 | 13.58 | 80.38 | 19.62 | −0.084 | 0.384 | |
| CR | 3834 | 42.72 | 33.72 | 16.33 | 5.55 | 76.44 | 21.88 | 0.118 | −0.493 | |
| Andrena tateyamana | Whole genome | 15,631 | 44.17 | 35.68 | 13.27 | 6.83 | 79.85 | 20.11 | 0.106 | −0.320 |
| PCGs | 11,059 | 37.47 | 42.23 | 11.05 | 9.25 | 79.70 | 20.30 | −0.060 | −0.089 | |
| Site 1 | 3687 | 42.27 | 34.03 | 10.95 | 12.74 | 76.30 | 23.70 | 0.108 | 0.075 | |
| Site 2 | 3686 | 23.08 | 50.65 | 15.07 | 11.20 | 73.74 | 26.26 | −0.374 | −0.147 | |
| Site 12 | 7373 | 32.68 | 42.34 | 13.01 | 11.97 | 75.02 | 24.98 | −0.129 | −0.042 | |
| Site 3 | 3686 | 47.04 | 42.02 | 7.13 | 3.81 | 89.06 | 10.94 | 0.056 | −0.303 | |
| tRNA | 1431 | 43.26 | 39.97 | 7.27 | 9.50 | 83.23 | 16.77 | 0.040 | 0.133 | |
| l-rRNA | 1269 | 37.27 | 44.92 | 5.67 | 12.14 | 82.19 | 17.81 | −0.093 | 0.363 | |
| s-rRNA | 743 | 37.28 | 44.41 | 4.98 | 13.32 | 81.70 | 18.30 | −0.087 | 0.456 | |
| CR | 937 | 43.65 | 37.79 | 11.95 | 7.38 | 81.44 | 19.33 | 0.072 | −0.236 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, D. Mitogenomic Characterization of Mining Bee Family Andrenidae (Hymenoptera: Apoidea: Anthophila) and Insights into Bee Phylogeny. Biology 2025, 14, 1374. https://doi.org/10.3390/biology14101374
Zhang D. Mitogenomic Characterization of Mining Bee Family Andrenidae (Hymenoptera: Apoidea: Anthophila) and Insights into Bee Phylogeny. Biology. 2025; 14(10):1374. https://doi.org/10.3390/biology14101374
Chicago/Turabian StyleZhang, Dan. 2025. "Mitogenomic Characterization of Mining Bee Family Andrenidae (Hymenoptera: Apoidea: Anthophila) and Insights into Bee Phylogeny" Biology 14, no. 10: 1374. https://doi.org/10.3390/biology14101374
APA StyleZhang, D. (2025). Mitogenomic Characterization of Mining Bee Family Andrenidae (Hymenoptera: Apoidea: Anthophila) and Insights into Bee Phylogeny. Biology, 14(10), 1374. https://doi.org/10.3390/biology14101374






