Soil Solarization Efficiently Reduces Fungal Soilborne Pathogen Populations, Promotes Lettuce Plant Growth, and Affects the Soil Bacterial Community
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Field Study Experimental Set Up
2.2. Soil Sampling and DNA Extraction from the Soil
2.3. Quantification of Soilborne Pathogens with qPCR
2.4. Disease Assessment
2.5. Plant Fresh Weight
2.6. Bacterial Microbiome Analysis
2.7. Bioinformatic Analysis
2.8. Statistical Analysis
3. Results
3.1. Standard Curves
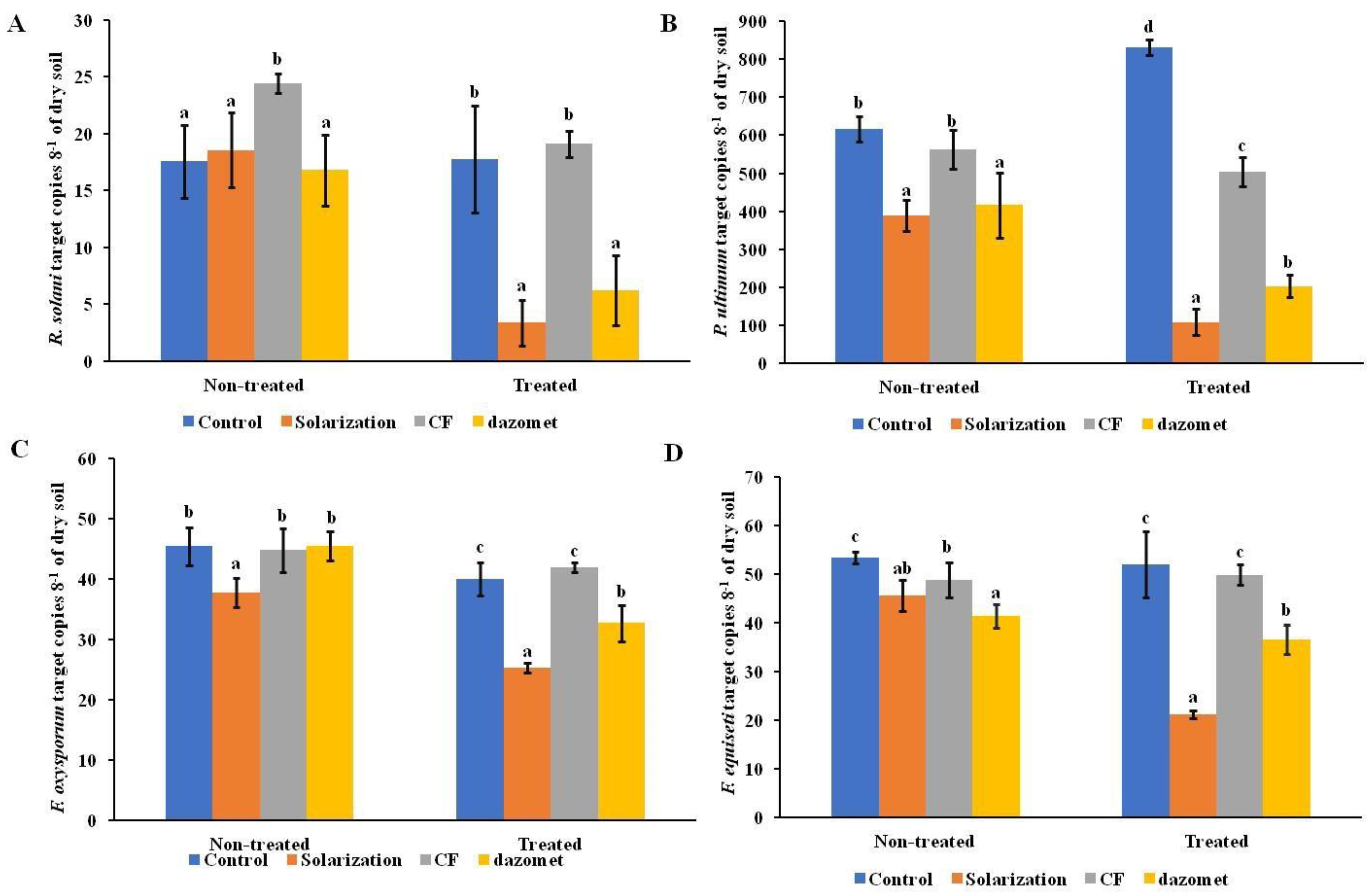
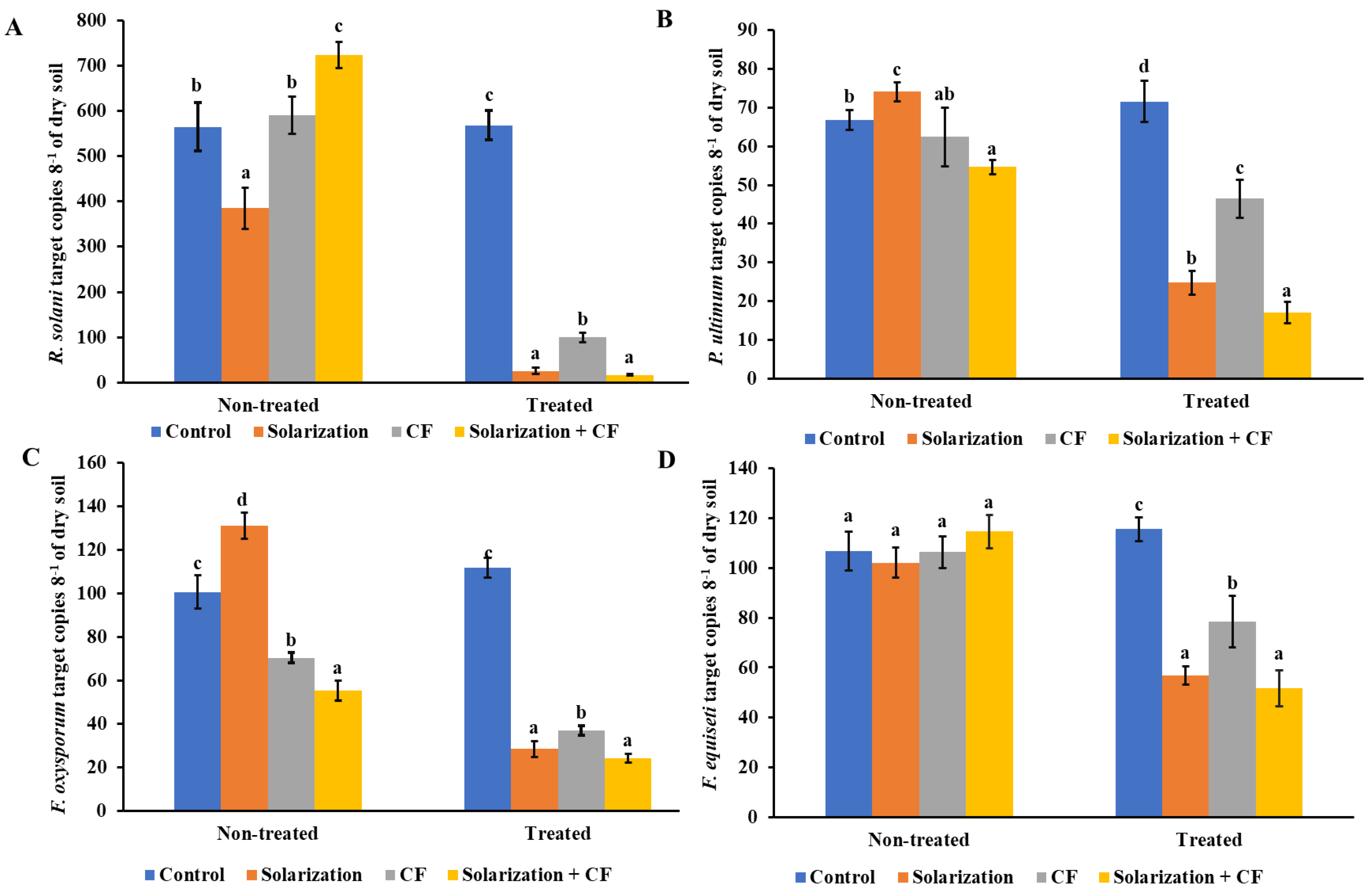
3.2. Impact of Disinfestation Treatments on Soilborne Pathogen Populations
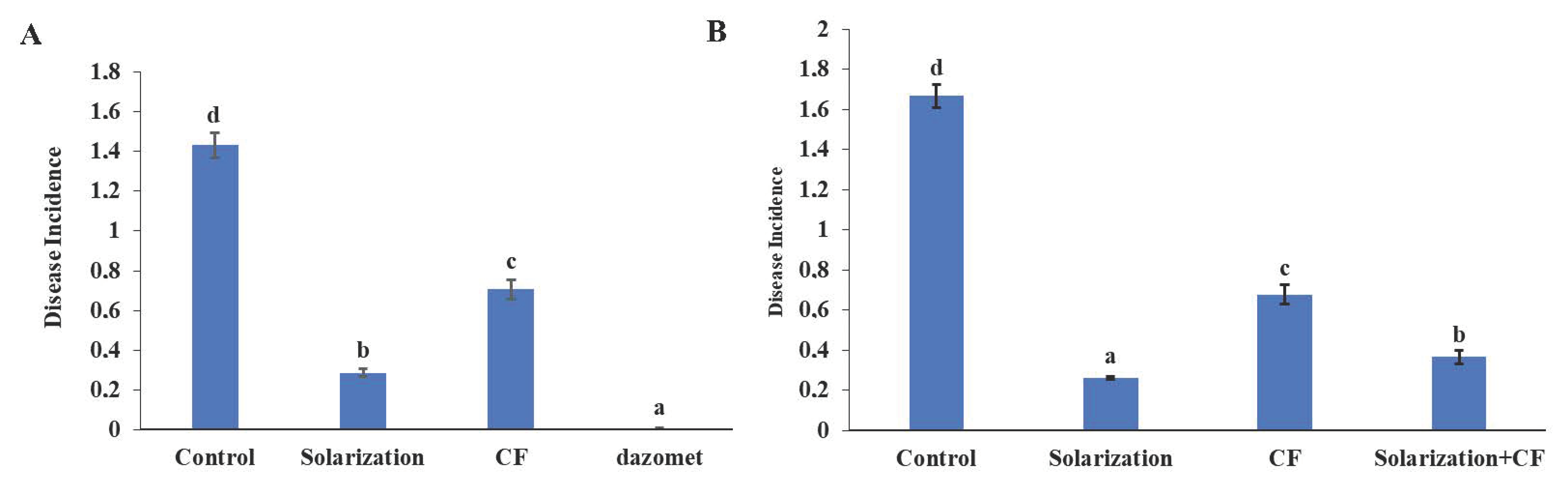
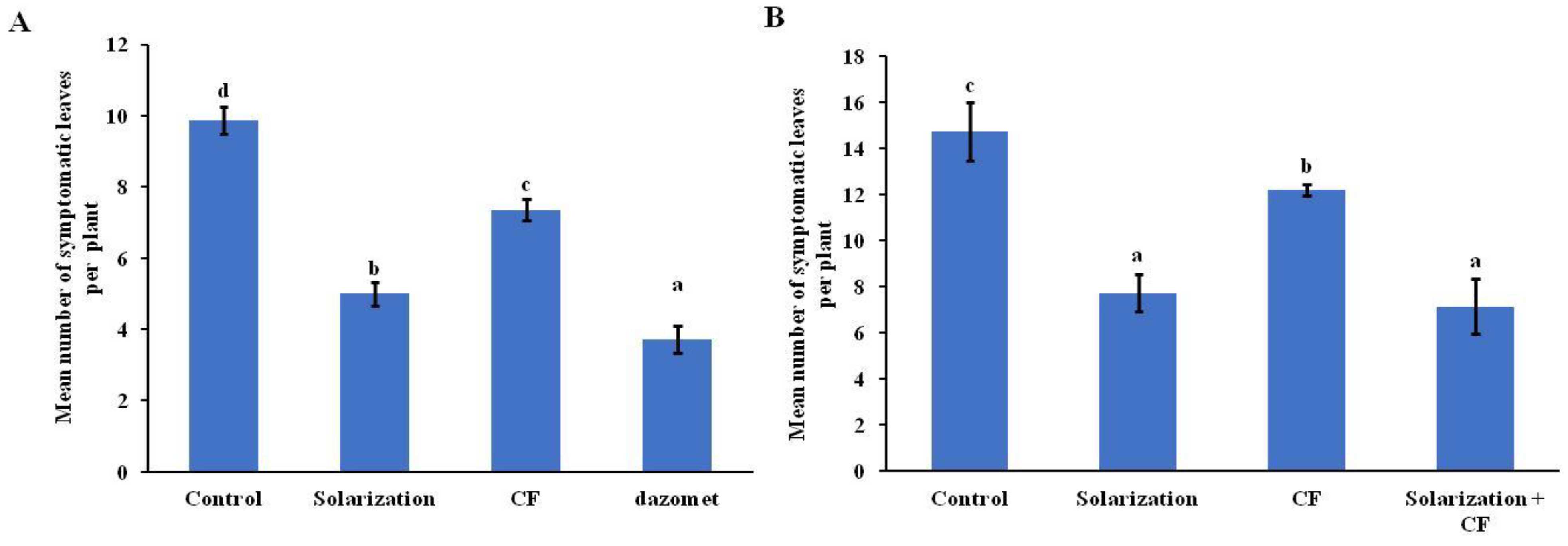
3.3. Disease Assessment
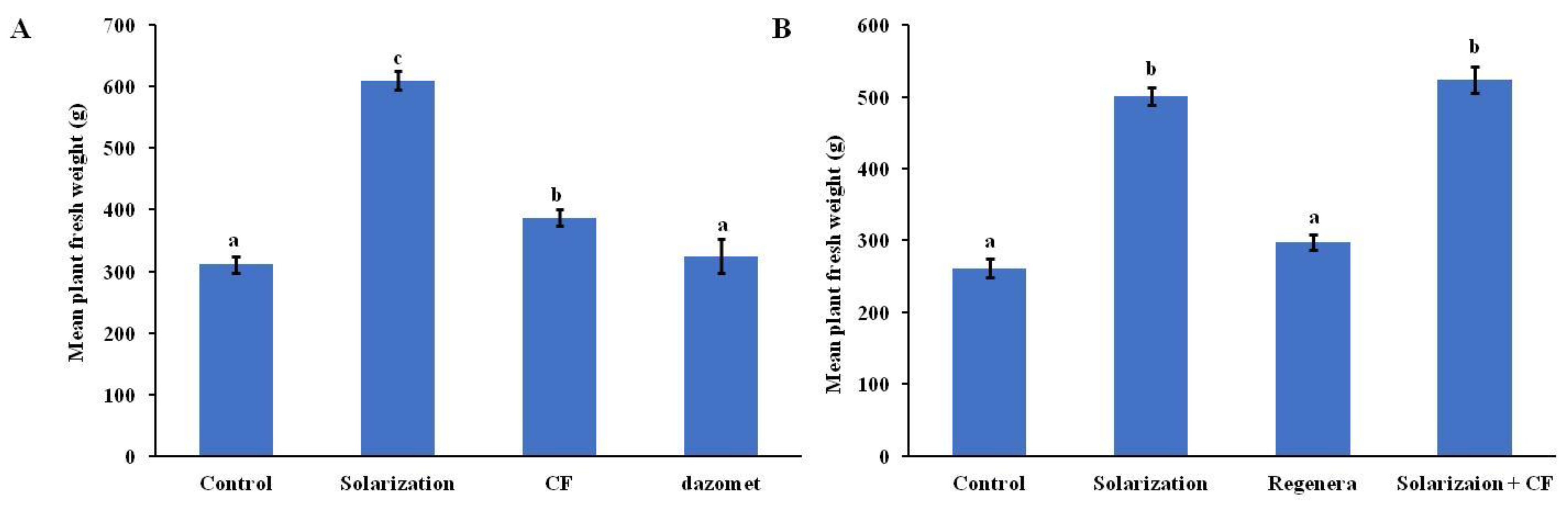
3.4. Plant Fresh Weight
3.5. Microbial Community Composition and Dynamics
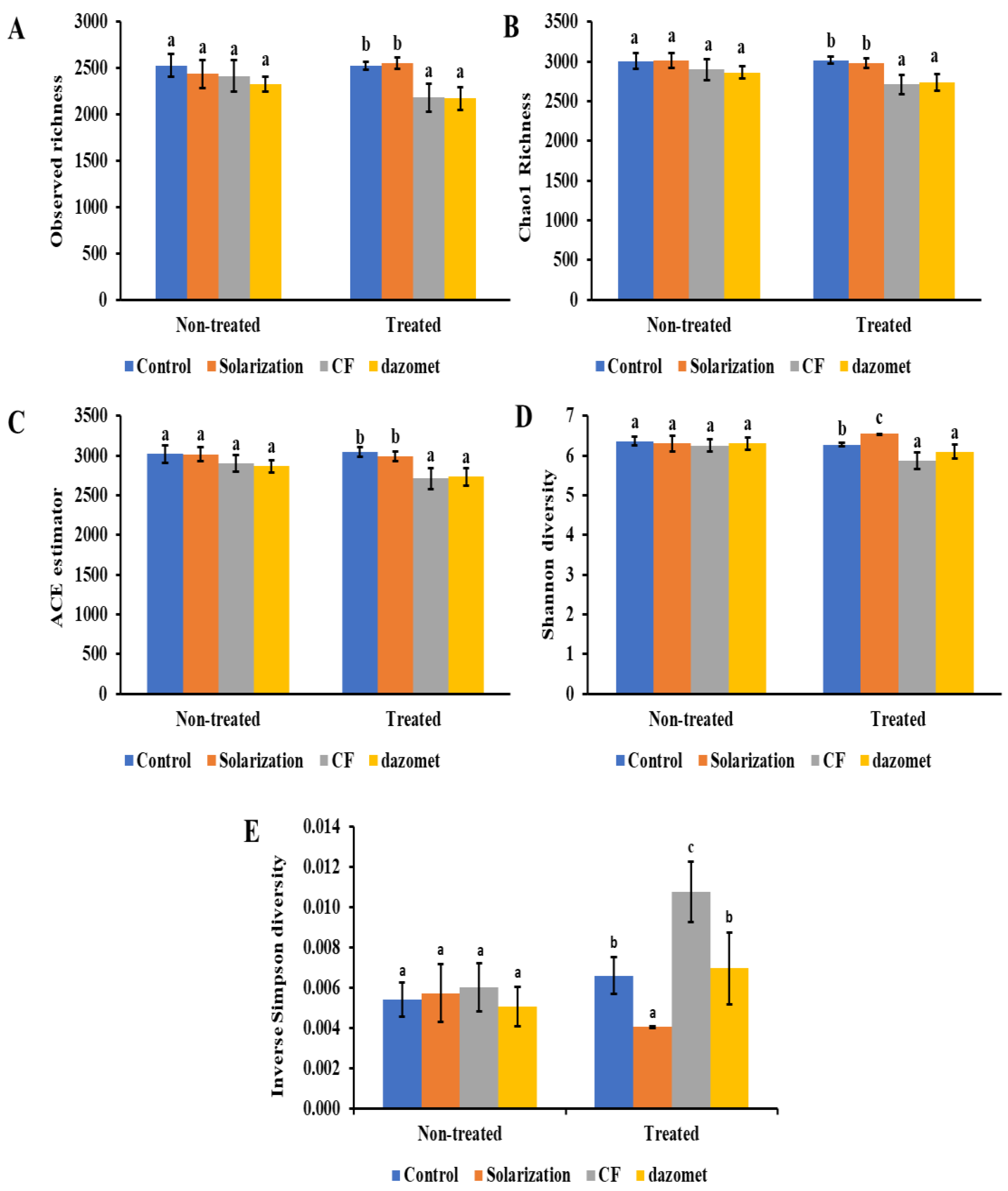
3.6. α-Diversity and β-Diversity of Soil Bacterial Communities
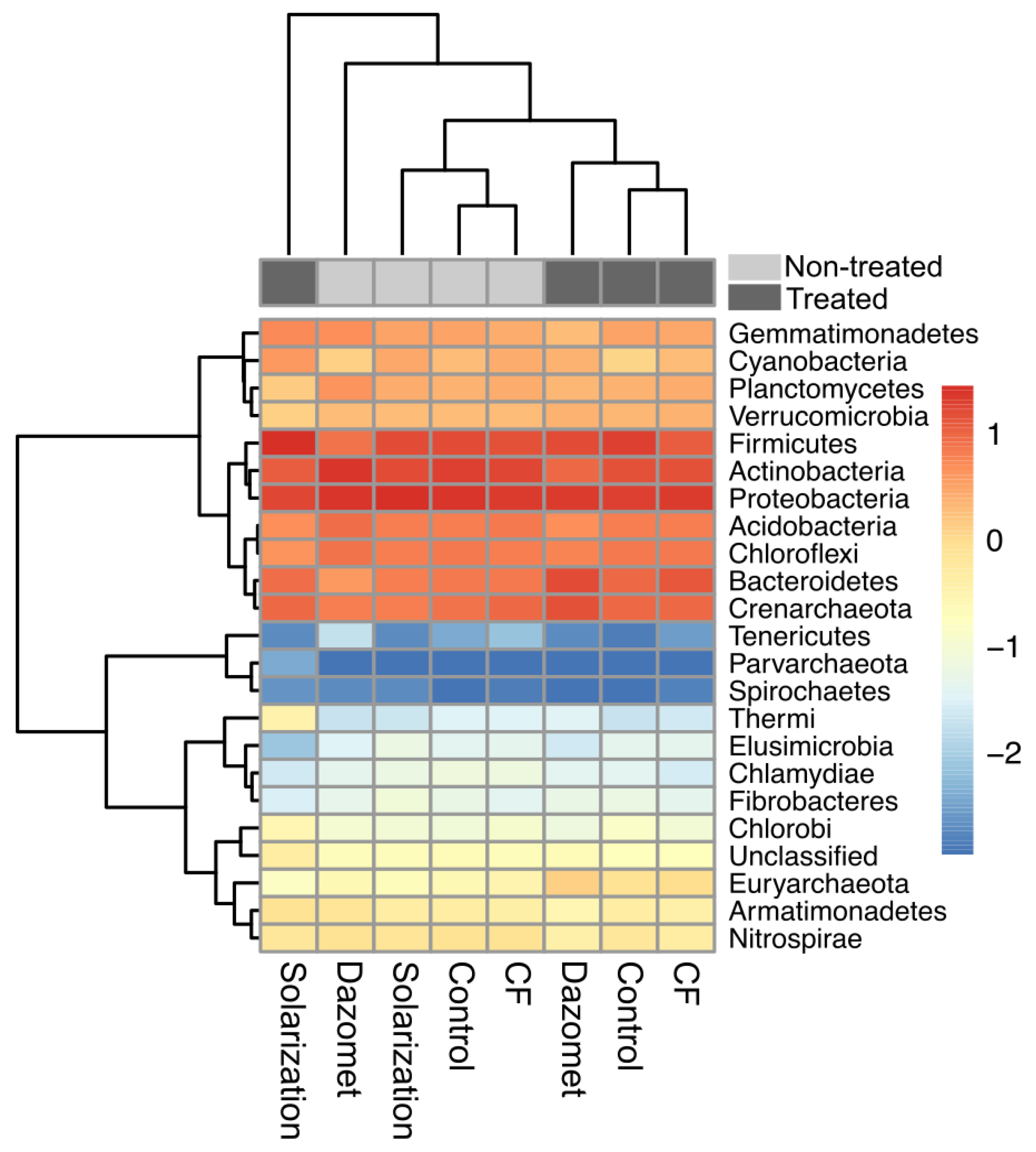
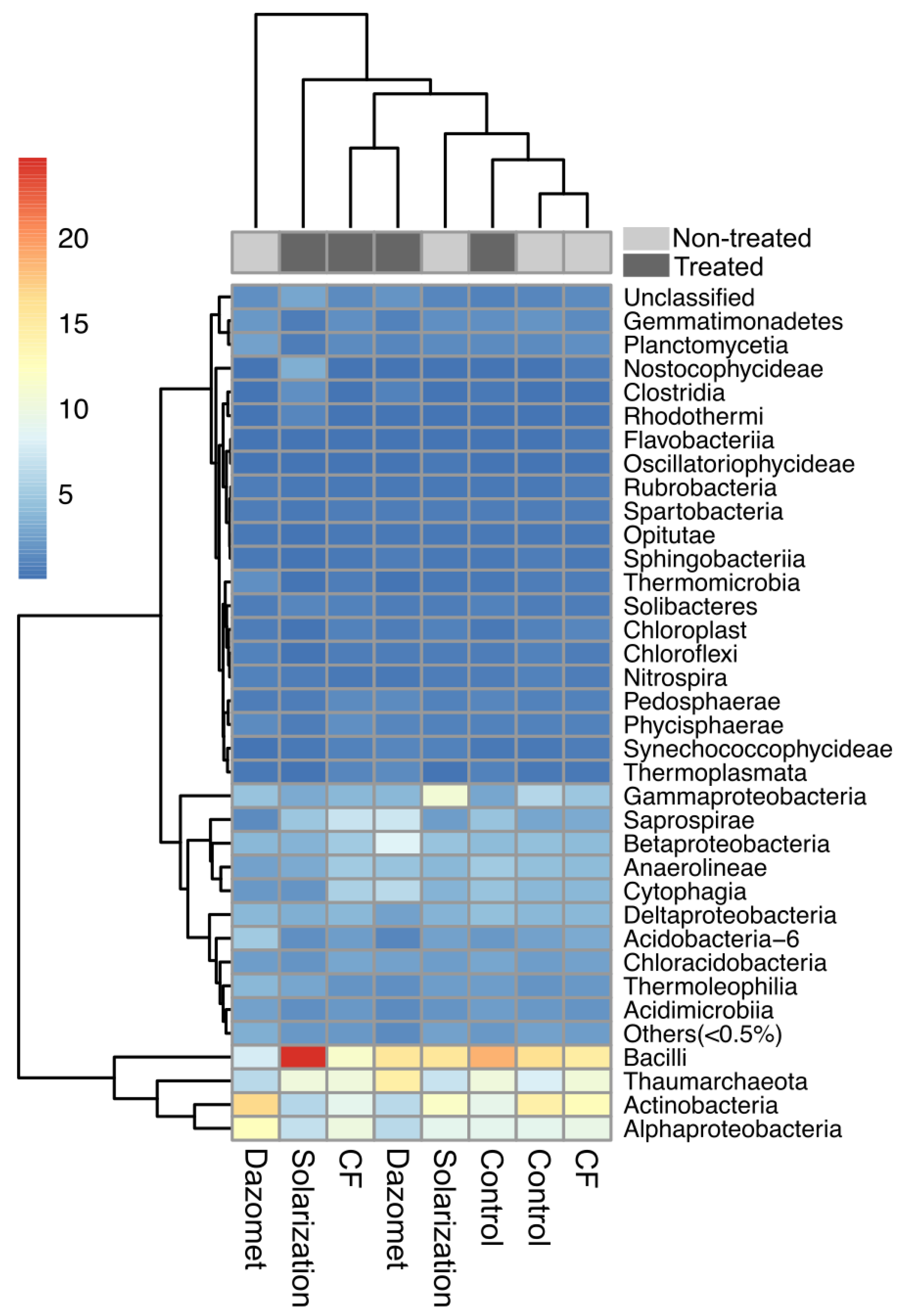
3.7. Phylum- and Class-Level Taxonomic Composition Distribution
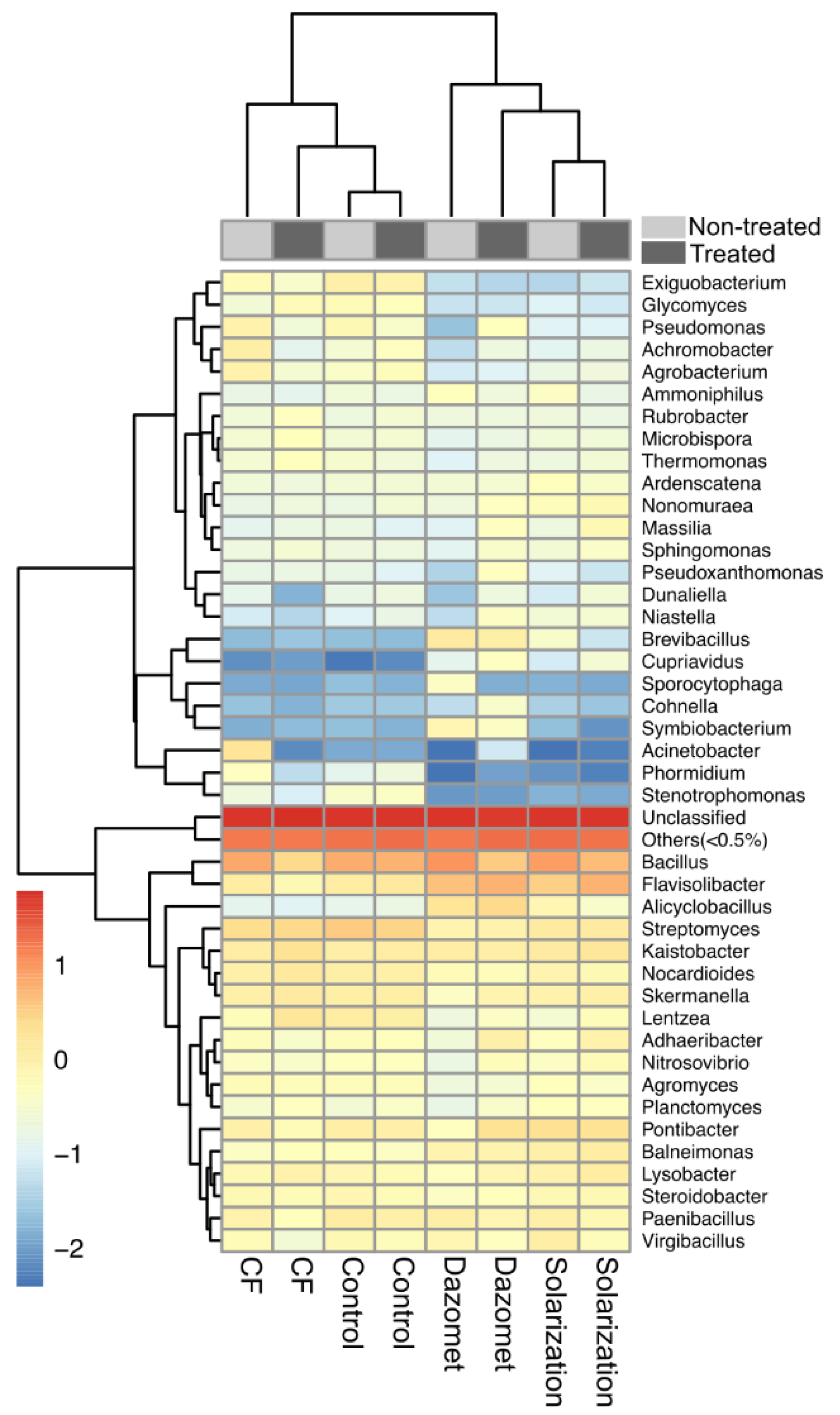
3.8. Effect of Applied Soil Treatments on Specific Bacterial Genera
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tziros, G.T.; Karaoglanidis, G.S. Molecular identification and pathogenicity of Rhizoctonia solani and Pythium spp. associated with damping-off disease on baby leafy vegetables in Greece. Plant Pathol. 2022, 71, 1381–1391. [Google Scholar] [CrossRef]
- Gullino, M.L.; Gilardi, G.; Garibaldi, A. Ready-to-eat salad crops: A plant pathogen’s heaven. Plant Dis. 2019, 10, 2153–2170. [Google Scholar] [CrossRef] [PubMed]
- Tziros, G.T.; Karaoglanidis, G.S. Identification of Fusarium oxysporum f. sp. lactucae race 1 as the causal agent of lettuce Fusarium wilt in Greece, commercial cultivars’ susceptibility, and temporal expression of defense-related genes. Microorganisms 2023, 11, 1082. [Google Scholar] [CrossRef]
- Tziros, G.T.; Samaras, A.; Karaoglanidis, G.S. Fusarium equiseti as an emerging foliar pathogen of lettuce in Greece: Identification and development of a Real-Time PCR for quantification of inoculum in soil samples. Pathogens 2002, 11, 1357. [Google Scholar] [CrossRef]
- Gamliel, A. Soil and substrate health. In Integrated Pest and Disease Management in Greenhouse Crops; Gullino, M.L., Albajes, R., Nicot, P.C., Eds.; Springer International Publishing: New York, NY, USA, 2020; Volume 9, pp. 355–384. [Google Scholar]
- Gamliel, A. Application of soil disinfestation in the view of system approach: A bottleneck or a tool for improvement. Acta Hortic. 2014, 883, 245–251. [Google Scholar] [CrossRef]
- Gamliel, A.; Katan, J. Suppression of major and minor pathogens by fluorescent pseudomonads, and in solarized and non-solarized soils. Phytopathology 1992, 83, 68–75. [Google Scholar]
- Katan, J. Diseases caused by soilborne pathogens: Biology, management and challenges. Plant Pathol. J. 2017, 99, 305–315. [Google Scholar]
- Katan, J. Integrated pest management in connection with soil disinfestation. Acta Hortic. 2015, 1044, 19–28. [Google Scholar] [CrossRef]
- Maron, P.A.; Sarr, A.; Kaisermann, A.; Lévêque, J.; Mathieu, O.; Guigue, J.; Karimi, B.; Bernard, L.; Dequiedt, S.; Terrat, S.; et al. High microbial diversity promotes soil ecosystem functioning. Appl. Environ. Microbiol. 2018, 84, e02738-17. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.K.; Ramesh, A.; Sharma, M.P.; Joshi, O.P.; Govaerts, B.; Steenwerth, K.L.; Karlen, D.L. Microbial community structure and diversity as indicators for evaluating soil quality. In Biodiversity, Biofuels, Agroforestry and Conservation Agriculture; Lichtfouse, E., Ed.; Springer: Dordrecht, The Netherland, 2011; pp. 317–358. [Google Scholar]
- Dang, S.R.; Gerik, J.S.; Tirado-Corbalá, R.; Ajwa, H. Soil microbial community structure and target organisms under different fumigation treatments. Appl. Environ. Soil Sci. 2015, 2015, 673264. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, W.; Zhang, Y.; Teng, Y.; Xu, Z. Effects of fungicide iprodione and nitrification inhibitor 3,4-dimethylpyrazole phosphate on soil enzyme and bacterial properties. Sci. Total Environ. 2017, 599, 254–263. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.T.; Hao, Y.W.; Zhu, M.Z.; Yu, S.T.; Ran, W.; Xue, C.; Ling, N.; Shen, Q.R. Characterizing differences in microbial community composition and function between Fusarium wilt diseased and healthy soils under watermelon cultivation. Plant Soil 2019, 438, 421–433. [Google Scholar] [CrossRef]
- Wei, Z.; Gu, Y.; Friman, V.P.; Kowalchuk, G.A.; Xu, Y.C.; Shen, Q.R.; Jousset, A. Initial soil microbiome composition and functioning predetermine future plant health. Sci. Adv. 2019, 5, eaaw0759. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.Q.; Liu, L.L.; Wen, T.; Zhang, J.B.; Wang, F.H.; Cai, Z.C. Changes in the soil microbial community after reductive and cucumber seedling cultivation. Appl. Microbiol. Biotechnol. 2016, 100, 5581–5593. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.L.; Huang, X.Q.; Zhang, J.B.; Cai, Z.C.; Jiang, K.; Chang, Y.Y. Deciphering the relative importance of soil and plant traits on the development of rhizosphere microbial communities. Soil Biol. Biochem. 2020, 148, 107909. [Google Scholar] [CrossRef]
- Gullino, M.L.; Garibaldi, A.; Gamliel, A.; Katan, J. Soil disinfestation: From soil treatment to soil and plant health. Plant Dis. 2022, 106, 1541–1554. [Google Scholar] [CrossRef]
- Lievens, B.; Brouwer, M.; Vanachter, A.C.; Lévesque, C.A.; Cammue, B.P.; Thomma, B.P. Quantitative assessment of phytopathogenic fungi in various substrates using a DNA macroarray. Environ. Microbiol. 2005, 7, 1698–1710. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Snindky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Spies, C.F.J.; Mazzola, M.; McLeod, A. Characterisation and detection of Pythium and Phytophthora species associated with grapevines in South Africa. Eur. J. Plant Pathol. 2011, 131, 103–119. [Google Scholar] [CrossRef]
- Li, Y.; Mao, L.; Yan, D.; Ma, T.; Shen, J.; Guo, M.; Wang, Q.; Ouyang, C.; Cao, A. Quantification of Fusarium oxysporum in fumigated soils by a newly developed real-time PCR assay to assess the efficacy of fumigants for Fusarium wilt disease in strawberry plants. Pest Manag. Sci. 2014, 70, 1669–1675. [Google Scholar] [CrossRef]
- Adesina, M.F.; Grosch, R.; Lembke, A.; Vatchev, T.D.; Smalla, K. In vitro antagonists of Rhizoctonia solani tested on lettuce: Rhizosphere competence, biocontrol efficiency and rhizosphere microbial community response. FEMS Microbiol. Ecol. 2009, 69, 62–74. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 5 January 2023).
- Douglas, W.F.; Bing, M.; Pawel, G.; Naomi, S.; Sandra, O.; Rebecca, M.B. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2014, 2, 6. [Google Scholar]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods. 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a Dual-Index Sequencing strategy and curation pipeline for analyzing amplicon sequence data on the Miseq Illumina Sequencing Platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef] [PubMed]
- Roberts, D.W. labdsv: Ordination and Multivariate Analysis for Ecology, R Package Version 1.3-1. Available online: http://ecology.msu.montana.edu/labdsv/R (accessed on 5 January 2023).
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 261–5267. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. Elegant graphics for data analysis (ggplot2). Appl. Spat. Data Anal. R 2009, 784, 785. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2010; Available online: http://www.R-project.org/ (accessed on 5 January 2023).
- Wakeham, A.J.; Pettitt, T.R. Diagnostic tests and their application in the management of soil-and water-borne oomycete pathogen species. Ann. Appl. Biol. 2017, 17, 45–67. [Google Scholar] [CrossRef]
- Ozyilmaz, U.; Benlioglu, K.; Yildiz, A.; Benlioglu, H.S. Effects of soil amendments combined with solarization on the soil microbial community in strawberry cultivation using quantitative real-time PCR. Phytoparasitica 2016, 44, 661–680. [Google Scholar] [CrossRef]
- Nakayasu, M.; Ikeda, K.; Yamazaki, S.; Aoki, Y.; Yazaki, K.; Washida, H.; Sugiyama, A. Two distinct soil disinfestations differently modify the bacterial communities in a tomato field. Agronomy 2021, 11, 1375. [Google Scholar] [CrossRef]
- Hernández-Lara, A.; Ros, M.; Cuartero, J.; Vivo, J.M.; Lozano-Pastor, P.; Pascual, J.A. Effects of solarisation combined with compost on soil pathogens and the microbial community in a spinach cropping system. Agric. Ecosyst. Environ. 2023, 346, 108359. [Google Scholar] [CrossRef]
- Zhao, H.; Zhou, T.; Xie, J.; Cheng, J.; Jiang, D.; Fu, Y. Host transcriptional response of Sclerotinia sclerotiorum induced by the mycoparasite Coniothyrium minitans. Front. Microbiol. 2020, 11, 183. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority (EFSA). Peer review of the pesticide risk assessment of the active substance Coniothyrium minitans Strain CON/M/91-08. EFSA J. 2016, 14, e04517. [Google Scholar]
- Bennett, A.J.; Leifert, C.; Whipps, J.M. Effect of combined treatment of pasteurisation and Coniothyrium minitans on sclerotia of Sclerotinia sclerotiorum in soil. Eur. J. Plant Pathol. 2005, 113, 197–209. [Google Scholar] [CrossRef]
- Elsheshtawi, M.; Elkhaky, M.T.; Sayed, S.R.; Bahkali, A.H.; Mohammed, A.A.; Gambhir, D.; Mansour, A.S.; Elgorban, A.M. Integrated control of white rot disease on beans caused by Sclerotinia sclerotiorum using Contans® and reduced fungicides application. Saudi J. Biol. Sci. 2017, 24, 405–409. [Google Scholar] [CrossRef] [PubMed]
- Nicola, L.; Turco, E.; Albanese, D.; Donati, C.; Thalheimer, M.; Pindo, M.; Insam, H.; Cavalieri, D.; Pertot, I. Fumigation with dazomet modifies soil microbiota in apple orchards affected by replant disease. Appl. Soil Ecol. 2017, 113, 71–79. [Google Scholar] [CrossRef]
- Wang, Q.; Ma, Y.; Yang, H.; Chang, Z. Effect of biofumigation and chemical fumigation on soil microbial community structure and control of pepper Phytophthora blight. World J. Microbiol. Biotechnol. 2014, 30, 507–518. [Google Scholar] [CrossRef] [PubMed]
- Gilardi, G.; Gullino, M.L.; Garibaldi, A. Soil disinfestation with dimethyl disulfide for management of Fusarium wilt on lettuce in Italy. J. Plant Dis. Prot. 2017, 124, 361–370. [Google Scholar] [CrossRef]
- Zhao, J.; Zhou, X.; Jiang, A.; Fan, J.; Lan, T.; Zhang, J.; Cai, Z. Distinct impacts of reductive soil disinfestation and chemical soil disinfestation on soil fungal communities and memberships. Appl. Microbiol. Biotechnol. 2018, 102, 7623–7634. [Google Scholar] [CrossRef]
- Bennett, R.S. Survival of Fusarium oxysporum f. sp. vasinfectum chlamydospores under solarization temperatures. Plant Dis. 2012, 96, 1564–1568. [Google Scholar] [CrossRef]
- LaMondia, J.A. Fusarium wilt of tobacco. Crop Prot. 2015, 73, 73–77. [Google Scholar] [CrossRef]
- McGovern, R.J.; Vavrina, C.S.; Noling, J.W.; Datnoff, L.A.; Yonce, H.D. Evaluation of application methods of metam sodium for management of Fusarium crown and root rot in tomato in southwest Florida. Plant Dis. 1998, 82, 919–923. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.; Li, W.; Li, Q.; Zhang, D.; Fang, W.; Li, Y.; Wang, Q.; Jin, X.; Yan, D.; Cao, A. Effects of granule size ranges on dazomet degradation and its persistence with different environmental factors. Agriculture 2022, 12, 674. [Google Scholar] [CrossRef]
- Zhang, D.; Ren, L.; Li, W.; Wang, Q.; Liu, S.; Jin, X.; Fang, W.; Yan, D.; Li, Y.; Wang, Q.; et al. The appropriate particle size of dazomet can ensure the soil fumigation effect from the source. Agriculture 2022, 12, 1832. [Google Scholar] [CrossRef]
- Scopa, A.; Candido, V.; Dumontet, S.; Pasquale, V.; Miccolis, V. Repeated solarization and long-term effects on soil microbiological parameters and agronomic traits. Crop Prot. 2009, 28, 818–824. [Google Scholar] [CrossRef]
- Matheron, M.E.; Porchas, M. Evaluation of soil solarization and flooding as management tools for Fusarium wilt of lettuce. Plant Dis. 2010, 94, 1323–1328. [Google Scholar] [CrossRef] [PubMed]
- Sinigaglia, C.; Patrício, F.R.A.; Ghini, R.; Malavolta, V.M.A.; Tessarioli Neto, J.; Freitas, S.S. Controle de Sclerotinia minor, Rhizoctonia solani e plantas daninhas em al face pela solarizaçã o do solo e sua integraçã o com controre químico. Summa Phytopathol. 2001, 27, 229–235. [Google Scholar]
- Patricio, F.R.A.; Sinigaglia, C.; Barros, B.C.; Freitas, S.S.; Neto, J.T.; Cantarella, H.; Ghini, R. Solarization and fungicides for the control of drop, bottom rot and weeds in lettuce. Crop Prot. 2006, 25, 31–38. [Google Scholar] [CrossRef]
- Pane, C.; Villecco, D.; Pentangelo, A.; Lahoz, E.; Zaccardelli, M. Integration of soil solarization with Brassica carinata seed meals amendment in a greenhouse lettuce production system. Acta Agric. Scand. Sect. B-Soil Plant Sci. 2012, 62, 291–299. [Google Scholar]
- O’Sullivan, C.A.; Belt, K.; Thatcher, L.F. Tackling control of a cosmopolitan phytopathogen: Sclerotinia. Front. Plant Sci. 2021, 12, 707509. [Google Scholar] [CrossRef]
- Kanaan, H.; Frenk, S.; Raviv, M.; Medina, S.; Minz, D. Long and short term effects of solarization on soil microbiome and agricultural production. Appl. Soil Ecol. 2018, 124, 54–61. [Google Scholar] [CrossRef]
- Hestmark, K.V.; Fernández-Bayo, J.D.; Harrold, D.R.; Randall, T.E.; Achmon, Y.; Stapleton, J.J.; Simmons, C.W.; Van der Gheynst, J.S. Compost induces the accumulation of biopesticidal organic acids during soil biosolarization. Resour. Conserv. Recycl. 2019, 143, 27–35. [Google Scholar] [CrossRef]
- Castellano-Hinojosa, A.; Boyd, N.S.; Strauss, S.L. Impact of fumigants on non-target soil microorganisms: A review. J. Hazard. Mater. 2022, 427, 128149. [Google Scholar] [CrossRef]
- Francis, I.; Holsters, M.; Vereecke, D. The Gram-positive side of plant–microbe interactions. Environ. Microbiol. 2010, 12, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Simmons, C.W.; Claypool, J.T.; Marshall, M.N.; Jabusch, L.K.; Reddy, A.P.; Simmons, B.A.; Singer, S.W.; Stapleton, J.J.; Van der Gheynst, J.S. Characterization of bacterial communities in solarized soil amended with lignocellulosic organic matter. Appl. Soil Ecol. 2014, 73, 97–104. [Google Scholar] [CrossRef]
- Wu, C.; Wang, F.; Ge, A.; Zhang, H.; Chen, G.; Deng, Y.; Yang, J.; Chen, J.; Ge, T. Enrichment of microbial taxa after the onset of wheat yellow mosaic disease. Agric Ecosyst. Environ. 2021, 322, 107651. [Google Scholar] [CrossRef]
- Trivedi, P.; Delgado-Baquerizo, M.; Trivedi, C.; Hamonts, K.; Anderson, I.C.; Singh, B.K. Keystone microbial taxa regulate the invasion of a fungal pathogen in agro-ecosystems. Soil Biol. Biochem. 2017, 111, 10–14. [Google Scholar] [CrossRef]
- Hassanisaadi, M.; Shahidi Bonjar, G.H.; Hosseinipour, A.; Abdolshahi, R.; Ait Barka, E.; Saadoun, I. Biological control of Pythium aphanidermatum, the causal agent of tomato root rot by two Streptomyces root symbionts. Agronomy 2021, 11, 846. [Google Scholar] [CrossRef]
- Benedui, A.; Ambrosini, A.; Passaglia, L.M.P. Plant growth-promoting rhizobacteria (PGPR): Their potential as antagonists and biocontrol agents. Genet. Mol. Biol. 2012, 35, 1044–1051. [Google Scholar] [CrossRef]
- He, H.; Zhang, S.; Shen, W.; Zhu, W.; Noor, I.; Liu, J.; Li, G. Benzoic acid plays a part in rhizosphere microbial composition of peach seedlings grown in replanted soil. Rhizosphere 2021, 19, 100364. [Google Scholar] [CrossRef]
- Fu, H.; Li, H.; Yin, P.; Mei, H.; Li, J.; Zhou, P.; Wang, Y.; Ma, Q.; Jeyaraj, A.; Thangaraj, K. Integrated application of rapeseed cake and green manure enhances soil nutrients and microbial communities in tea garden soil. Sustainability 2021, 13, 2967. [Google Scholar] [CrossRef]
| Treatment | Plots | |
|---|---|---|
| Non-Treated | Treated | |
| Control | 2530 ± 154.73 | 2418 ± 206.44 |
| Solarization | 2526 ± 56.96 | 2126 ± 188.25 |
| CF | 2437 ± 187.35 | 2329 ± 100.78 |
| Dazomet | 2617 ± 77.02 | 2094 ± 102.53 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tziros, G.T.; Samaras, A.; Karaoglanidis, G.S. Soil Solarization Efficiently Reduces Fungal Soilborne Pathogen Populations, Promotes Lettuce Plant Growth, and Affects the Soil Bacterial Community. Biology 2024, 13, 624. https://doi.org/10.3390/biology13080624
Tziros GT, Samaras A, Karaoglanidis GS. Soil Solarization Efficiently Reduces Fungal Soilborne Pathogen Populations, Promotes Lettuce Plant Growth, and Affects the Soil Bacterial Community. Biology. 2024; 13(8):624. https://doi.org/10.3390/biology13080624
Chicago/Turabian StyleTziros, George T., Anastasios Samaras, and George S. Karaoglanidis. 2024. "Soil Solarization Efficiently Reduces Fungal Soilborne Pathogen Populations, Promotes Lettuce Plant Growth, and Affects the Soil Bacterial Community" Biology 13, no. 8: 624. https://doi.org/10.3390/biology13080624
APA StyleTziros, G. T., Samaras, A., & Karaoglanidis, G. S. (2024). Soil Solarization Efficiently Reduces Fungal Soilborne Pathogen Populations, Promotes Lettuce Plant Growth, and Affects the Soil Bacterial Community. Biology, 13(8), 624. https://doi.org/10.3390/biology13080624







