Association between the Maternal Gut Microbiome and Macrosomia
Abstract
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Study Design
2.2. Stool Sample Processing
2.3. Metagenomic Data Processing
2.4. Diversity Analysis
2.5. Statistical Analysis
3. Results
3.1. Characteristics of the Participants
3.2. Comparison of Different Bacteria
3.3. Functional Analysis
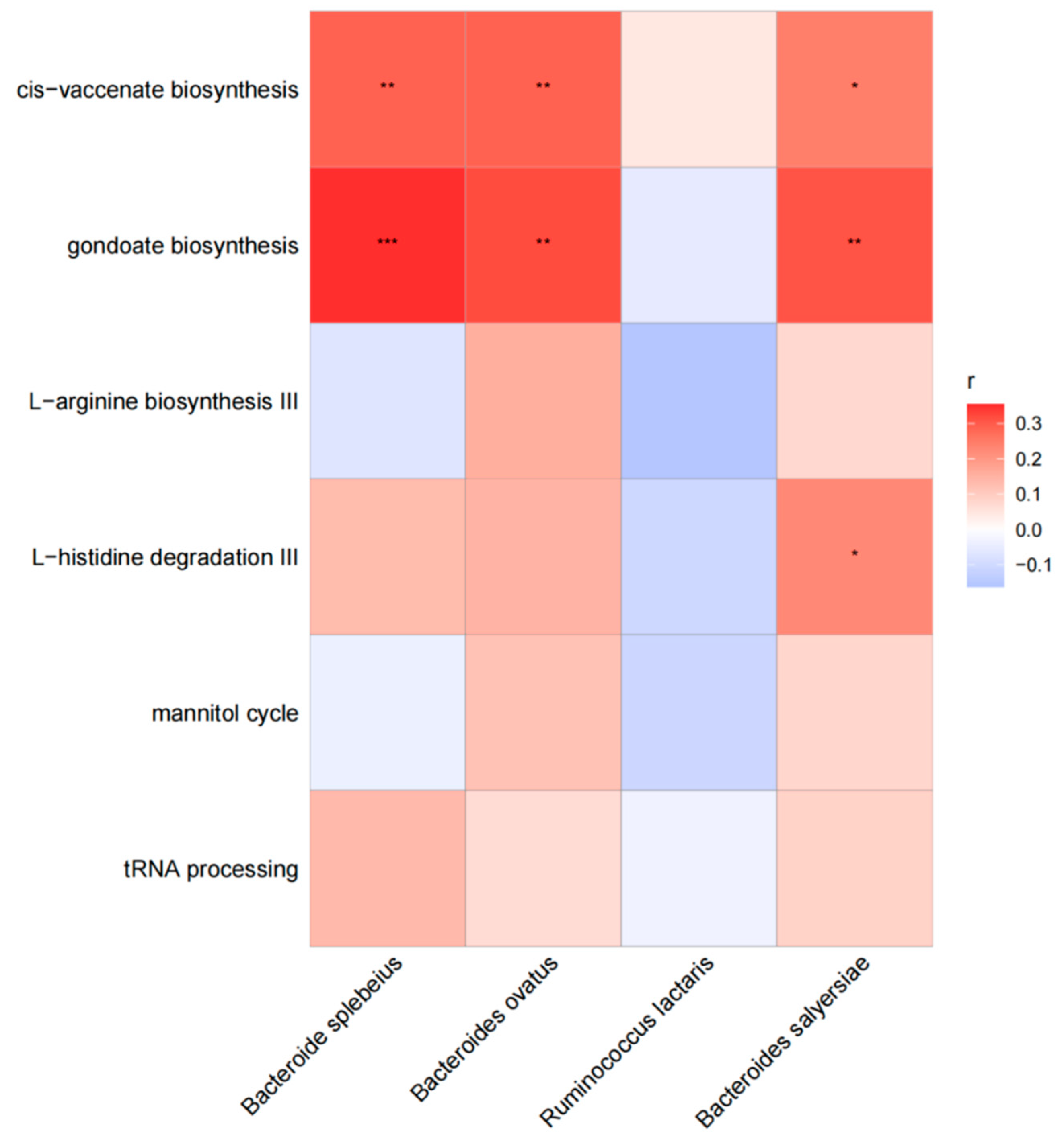
3.4. Correlation Analysis
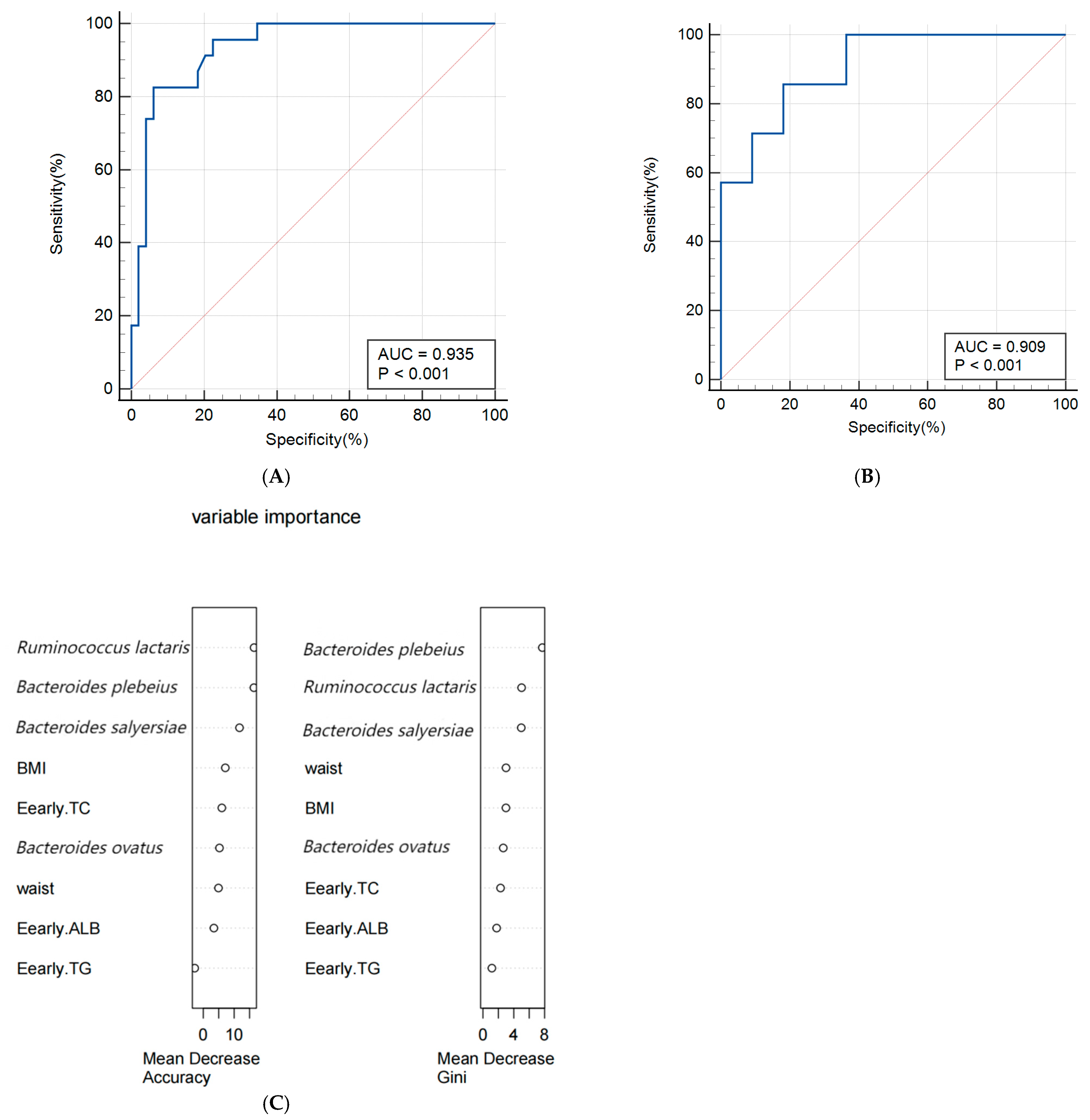
3.5. Construction of Predictive Models
4. Discussion
4.1. Gut Microbiota That Affect the Occurrence of Macrosomia
4.2. The Functional Pathways of Gut Microbiota and Their Association with Macrosomia
4.3. Correlation Analysis between Differential Species and Different Pathways
4.4. Predictive Model for Macrosomia
4.5. Strengths and Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Macrosomia: ACOG Practice Bulletin Summary, Number 216. Obstet. Gynecol. 2020, 135, 246–248. [CrossRef] [PubMed]
- Wang, D.; Hong, Y.; Zhu, L.; Wang, X.; Lv, Q.; Zhou, Q.; Ruan, M.; Chen, C. Risk factors and outcomes of macrosomia in China: A multicentric survey based on birth data. J. Matern.-Fetal Neonatal Med. 2017, 30, 623–627. [Google Scholar] [CrossRef] [PubMed]
- Araujo Júnior, E.; Peixoto, A.B.; Zamarian, A.C.; Elito Júnior, J.; Tonni, G. Macrosomia. Best Pract. Res. Clin. Obstet. Gynaecol. 2017, 38, 83–96. [Google Scholar] [CrossRef] [PubMed]
- Rossi, A.C.; Mullin, P.; Prefumo, F. Prevention, management, and outcomes of macrosomia: A systematic review of literature and meta-analysis. Obstet. Gynecol. Surv. 2013, 68, 702–709. [Google Scholar] [CrossRef] [PubMed]
- Harder, T.; Rodekamp, E.; Schellong, K.; Dudenhausen, J.W.; Plagemann, A. Birth weight and subsequent risk of type 2 diabetes: A meta-analysis. Am. J. Epidemiol. 2007, 165, 849–857. [Google Scholar] [CrossRef]
- Harder, T.; Roepke, K.; Diller, N.; Stechling, Y.; Dudenhausen, J.W.; Plagemann, A. Birth weight, early weight gain, and subsequent risk of type 1 diabetes: Systematic review and meta-analysis. Am. J. Epidemiol. 2009, 169, 1428–1436. [Google Scholar] [CrossRef] [PubMed]
- Goldenberg, R.L.; McClure, E.M.; Harrison, M.S.; Miodovnik, M. Diabetes during Pregnancy in Low- and Middle-Income Countries. Am. J. Perinatol. 2016, 33, 1227–1235. [Google Scholar] [CrossRef]
- Fekene, D.B.; Woldeyes, B.S.; Erena, M.M.; Demisse, G.A. Knowledge, uptake of preconception care and associated factors among reproductive age group women in West Shewa zone, Ethiopia, 2018. BMC Women’s Health 2020, 20, 30. [Google Scholar] [CrossRef] [PubMed]
- Arumugasaamy, N.; Rock, K.D.; Kuo, C.Y.; Bale, T.L.; Fisher, J.P. Microphysiological systems of the placental barrier. Adv. Drug Deliv. Rev. 2020, 161–162, 161–175. [Google Scholar] [CrossRef]
- Lynch, S.V.; Pedersen, O. The Human Intestinal Microbiome in Health and Disease. N. Engl. J. Med. 2016, 375, 2369–2379. [Google Scholar] [CrossRef]
- Canfora, E.E.; Jocken, J.W.; Blaak, E.E. Short-chain fatty acids in control of body weight and insulin sensitivity. Nat. Rev. Endocrinol. 2015, 11, 577–591. [Google Scholar] [CrossRef]
- Yano, J.M.; Yu, K.; Donaldson, G.P.; Shastri, G.G.; Ann, P.; Ma, L.; Nagler, C.R.; Ismagilov, R.F.; Mazmanian, S.K.; Hsiao, E.Y. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 2015, 161, 264–276. [Google Scholar] [CrossRef] [PubMed]
- Haiser, H.J.; Gootenberg, D.B.; Chatman, K.; Sirasani, G.; Balskus, E.P.; Turnbaugh, P.J. Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science 2013, 341, 295–298. [Google Scholar] [CrossRef] [PubMed]
- Fulde, M.; Hornef, M.W. Maturation of the enteric mucosal innate immune system during the postnatal period. Immunol. Rev. 2014, 260, 21–34. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zeng, S.; Egan, M.; Cherry, P.; Strain, C.; Morais, E.; Boyaval, P.; Ryan, C.A.; Dempsey, E.; Ross, R.P.; et al. Metagenomic analysis of mother-infant gut microbiome reveals global distinct and shared microbial signatures. Gut Microbes 2021, 13, 1–24. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Gordon, J.I. The core gut microbiome, energy balance and obesity. J. Physiol. 2009, 587, 4153–4158. [Google Scholar] [CrossRef]
- Gough, E.K.; Edens, T.J.; Geum, H.M.; Baharmand, I.; Gill, S.K.; Robertson, R.C.; Mutasa, K.; Ntozini, R.; Smith, L.E.; Chasekwa, B.; et al. Maternal fecal microbiome predicts gestational age, birth weight and neonatal growth in rural Zimbabwe. EBioMedicine 2021, 68, 103421. [Google Scholar] [CrossRef] [PubMed]
- Santacruz, A.; Collado, M.C.; García-Valdés, L.; Segura, M.T.; Martín-Lagos, J.A.; Anjos, T.; Martí-Romero, M.; Lopez, R.M.; Florido, J.; Campoy, C.; et al. Gut microbiota composition is associated with body weight, weight gain and biochemical parameters in pregnant women. Br. J. Nutr. 2010, 104, 83–92. [Google Scholar] [CrossRef]
- Schoenmakers, S.; Steegers-Theunissen, R.; Faas, M. The matter of the reproductive microbiome. Obstet. Med. 2019, 12, 107–115. [Google Scholar] [CrossRef]
- Edwards, S.M.; Cunningham, S.A.; Dunlop, A.L.; Corwin, E.J. The Maternal Gut Microbiome During Pregnancy. MCN Am. J. Matern. Child Nurs. 2017, 42, 310–317. [Google Scholar] [CrossRef]
- Gohir, W.; Ratcliffe, E.M.; Sloboda, D.M. Of the bugs that shape us: Maternal obesity, the gut microbiome, and long-term disease risk. Pediatr. Res. 2015, 77, 196–204. [Google Scholar] [CrossRef] [PubMed]
- Truong, D.T.; Franzosa, E.A.; Tickle, T.L.; Scholz, M.; Weingart, G.; Pasolli, E.; Tett, A.; Huttenhower, C.; Segata, N. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat. Methods 2015, 12, 902–903. [Google Scholar] [CrossRef]
- Franzosa, E.A.; McIver, L.J.; Rahnavard, G.; Thompson, L.R.; Schirmer, M.; Weingart, G.; Lipson, K.S.; Knight, R.; Caporaso, J.G.; Segata, N.; et al. Species-level functional profiling of metagenomes and metatranscriptomes. Nat. Methods 2018, 15, 962–968. [Google Scholar] [CrossRef] [PubMed]
- Del Chierico, F.; Manco, M.; Gardini, S.; Guarrasi, V.; Russo, A.; Bianchi, M.; Tortosa, V.; Quagliariello, A.; Shashaj, B.; Fintini, D.; et al. Fecal microbiota signatures of insulin resistance, inflammation, and metabolic syndrome in youth with obesity: A pilot study. Acta Diabetol. 2021, 58, 1009–1022. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Sun, J.; Liu, C.; Yu, X.; Li, H.; Zhang, W.; Li, Y.; Geng, Y.; Wang, Z. Compositional Alterations of Gut Microbiota in Patients with Diabetic Kidney Disease and Type 2 Diabetes Mellitus. Diabetes Metab. Syndr. Obes. Targets Ther. 2022, 15, 755–765. [Google Scholar] [CrossRef]
- Calderón-Pérez, L.; Gosalbes, M.J.; Yuste, S.; Valls, R.M.; Pedret, A.; Llauradó, E.; Jimenez-Hernandez, N.; Artacho, A.; Pla-Pagà, L.; Companys, J.; et al. Gut metagenomic and short chain fatty acids signature in hypertension: A cross-sectional study. Sci. Rep. 2020, 10, 6436. [Google Scholar] [CrossRef]
- Liang, X.; Zhang, S.; Zhang, D.; Hu, L.; Zhang, L.; Peng, Y.; Xu, Y.; Hou, H.; Zou, C.; Liu, X.; et al. Metagenomics-based systematic analysis reveals that gut microbiota Gd-IgA1-associated enzymes may play a key role in IgA nephropathy. Front. Mol. Biosci. 2022, 9, 970723. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.Y.; Liu, L.Y.; Jia, Y.; Li, Y.; Cai, J.N.; Shu, Y.; Tan, J.Y.; Chen, P.Y.; Li, H.W.; Cai, H.H.; et al. Correlation between gut microbiota and glucagon-like peptide-1 in patients with gestational diabetes mellitus. World J. Diabetes 2022, 13, 861–876. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Chen, L.; Zhang, D.Y.; Gan, X.P.; Cao, Y.N.; Cheng, D.C.; Liu, W.Y.; Li, F.F.; Xu, X.M.; Wang, H.K. The characteristics of intestinal flora in overweight pregnant women and the correlation with gestational diabetes mellitus. Endocr. Connect. 2021, 10, 1366–1376. [Google Scholar] [CrossRef]
- Tirosh, A.; Calay, E.S.; Tuncman, G.; Claiborn, K.C.; Inouye, K.E.; Eguchi, K.; Alcala, M.; Rathaus, M.; Hollander, K.S.; Ron, I.; et al. The short-chain fatty acid propionate increases glucagon and FABP4 production, impairing insulin action in mice and humans. Sci. Transl. Med. 2019, 11, eaav0120. [Google Scholar] [CrossRef]
- Perry, R.J.; Peng, L.; Barry, N.A.; Cline, G.W.; Zhang, D.; Cardone, R.L.; Petersen, K.F.; Kibbey, R.G.; Goodman, A.L.; Shulman, G.I. Acetate mediates a microbiome-brain-β-cell axis to promote metabolic syndrome. Nature 2016, 534, 213–217. [Google Scholar] [CrossRef] [PubMed]
- Fowden, A.L.; Hill, D.J. Intra-uterine programming of the endocrine pancreas. Br. Med. Bull. 2001, 60, 123–142. [Google Scholar] [CrossRef] [PubMed]
- Shang, M.; Wen, Z. Increased placental IGF-1/mTOR activity in macrosomia born to women with gestational diabetes. Diabetes Res. Clin. Pract. 2018, 146, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Cortez, R.V.; Taddei, C.R.; Sparvoli, L.G.; Ângelo, A.G.S.; Padilha, M.; Mattar, R.; Daher, S. Microbiome and its relation to gestational diabetes. Endocrine 2019, 64, 254–264. [Google Scholar] [CrossRef]
- Ottosson, F.; Brunkwall, L.; Ericson, U.; Nilsson, P.M.; Almgren, P.; Fernandez, C.; Melander, O.; Orho-Melander, M. Connection between BMI-Related Plasma Metabolite Profile and Gut Microbiota. J. Clin. Endocrinol. Metab. 2018, 103, 1491–1501. [Google Scholar] [CrossRef] [PubMed]
- Vieira-Silva, S.; Falony, G.; Belda, E.; Nielsen, T.; Aron-Wisnewsky, J.; Chakaroun, R.; Forslund, S.K.; Assmann, K.; Valles-Colomer, M.; Nguyen, T.T.D.; et al. Statin therapy is associated with lower prevalence of gut microbiota dysbiosis. Nature 2020, 581, 310–315. [Google Scholar] [CrossRef]
- Allin, K.H.; Tremaroli, V.; Caesar, R.; Jensen, B.A.H.; Damgaard, M.T.F.; Bahl, M.I.; Licht, T.R.; Hansen, T.H.; Nielsen, T.; Dantoft, T.M.; et al. Aberrant intestinal microbiota in individuals with prediabetes. Diabetologia 2018, 61, 810–820. [Google Scholar] [CrossRef]
- Ponzo, V.; Fedele, D.; Goitre, I.; Leone, F.; Lezo, A.; Monzeglio, C.; Finocchiaro, C.; Ghigo, E.; Bo, S. Diet-Gut Microbiota Interactions and Gestational Diabetes Mellitus (GDM). Nutrients 2019, 11, 330. [Google Scholar] [CrossRef] [PubMed]
- Felton, E.; Burrell, A.; Chaney, H.; Sami, I.; Koumbourlis, A.C.; Freishtat, R.J.; Crandall, K.A.; Hahn, A. Inflammation in children with cystic fibrosis: Contribution of bacterial production of long-chain fatty acids. Pediatr. Res. 2021, 90, 99–108. [Google Scholar] [CrossRef]
- Nakamura, M.T.; Yudell, B.E.; Loor, J.J. Regulation of energy metabolism by long-chain fatty acids. Prog. Lipid Res. 2014, 53, 124–144. [Google Scholar] [CrossRef]
- Man, W.C.; Miyazaki, M.; Chu, K.; Ntambi, J. Colocalization of SCD1 and DGAT2: Implying preference for endogenous monounsaturated fatty acids in triglyceride synthesis. J. Lipid Res. 2006, 47, 1928–1939. [Google Scholar] [CrossRef] [PubMed]
- Berk, P.D.; Verna, E.C. Nonalcoholic Fatty Liver Disease: Lipids and Insulin Resistance. Clin. Liver Dis. 2016, 20, 245–262. [Google Scholar] [CrossRef] [PubMed]
- Finucane, O.M.; Lyons, C.L.; Murphy, A.M.; Reynolds, C.M.; Klinger, R.; Healy, N.P.; Cooke, A.A.; Coll, R.C.; McAllan, L.; Nilaweera, K.N.; et al. Monounsaturated fatty acid-enriched high-fat diets impede adipose NLRP3 inflammasome-mediated IL-1β secretion and insulin resistance despite obesity. Diabetes 2015, 64, 2116–2128. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Fan, C.; Zhang, L.; Li, Y.; Hou, H.; Ma, Y.; Fan, J.; Tan, Y.; Wu, T.; Jia, S.; et al. Alterations of Gut Microbiome in Tibetan Patients with Coronary Heart Disease. Front. Cell. Infect. Microbiol. 2020, 10, 373. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Li, T.; Gao, X.; Yan, H.; Chen, J.; Huang, M.; Wang, L.; Yin, D.; Li, H.; Ma, R.; et al. Gut Microbiome Alterations in Patients with Thyroid Nodules. Front. Cell. Infect. Microbiol. 2021, 11, 643968. [Google Scholar] [CrossRef]
- Paley, E.L. Discovery of Gut Bacteria Specific to Alzheimer’s Associated Diseases is a Clue to Understanding Disease Etiology: Meta-Analysis of Population-Based Data on Human Gut Metagenomics and Metabolomics. J. Alzheimer’s Dis. JAD 2019, 72, 319–355. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Chen, X.; Hu, X.; Niu, H.; Tian, R.; Wang, H.; Pang, H.; Jiang, L.; Qiu, B.; Chen, X.; et al. Alterations in the gut microbiome and metabolism with coronary artery disease severity. Microbiome 2019, 7, 68. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.A.; Benton, T.Z.; Bennett, B.J.; Jacobs, D.R., Jr.; Lloyd-Jones, D.M.; Gross, M.D.; Carr, J.J.; Gordon-Larsen, P.; Zeisel, S.H. Microbiota-Dependent Metabolite Trimethylamine N-Oxide and Coronary Artery Calcium in the Coronary Artery Risk Development in Young Adults Study (CARDIA). J. Am. Heart Assoc. 2016, 5, e003970. [Google Scholar] [CrossRef]
- Hoang, H.H.; Padgham, S.V.; Meininger, C.J. L-arginine, tetrahydrobiopterin, nitric oxide and diabetes. Curr. Opin. Clin. Nutr. Metab. Care 2013, 16, 76–82. [Google Scholar] [CrossRef]
- Szlas, A.; Kurek, J.M.; Krejpcio, Z. The Potential of L-Arginine in Prevention and Treatment of Disturbed Carbohydrate and Lipid Metabolism-A Review. Nutrients 2022, 14, 961. [Google Scholar] [CrossRef]
- Jing, G.; Huwei, S.; Chao, C.; Lei, C.; Ping, W.; Zhongzhou, X.; Sen, Y.; Jiayuan, C.; Ruiyao, C.; Lu, L.; et al. A predictive model of macrosomic birth based upon real-world clinical data from pregnant women. BMC Pregnancy Childbirth 2022, 22, 651. [Google Scholar] [CrossRef] [PubMed]
- Weiner, E.; Mizrachi, Y.; Fainstein, N.; Elyashiv, O.; Mevorach-Zussman, N.; Bar, J.; Kovo, M. Comparison between Three Methods of Fetal Weight Estimation during the Active Stage of Labor Performed by Residents: A Prospective Cohort Study. Fetal Diagn. Ther. 2017, 42, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Mazzone, E.; Dall’Asta, A.; Kiener, A.J.O.; Carpano, M.G.; Suprani, A.; Ghi, T.; Frusca, T. Prediction of fetal macrosomia using two-dimensional and three-dimensional ultrasound. Eur. J. Obstet. Gynecol. Reprod. Biol. 2019, 243, 26–31. [Google Scholar] [CrossRef]
- Mazouni, C.; Rouzier, R.; Ledu, R.; Heckenroth, H.; Guidicelli, B.; Gamerre, M. Development and internal validation of a nomogram to predict macrosomia. Ultrasound Obstet. Gynecol. Off. J. Int. Soc. Ultrasound Obstet. Gynecol. 2007, 29, 544–549. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Zhang, Y.; Yin, Z.; Wei, L.; Lv, B.; Wu, Y. Establishment of a nomogram model to predict macrosomia in pregnant women with gestational diabetes mellitus. BMC Pregnancy Childbirth 2021, 21, 581. [Google Scholar] [CrossRef] [PubMed]


| Cases (n = 31) | Controls (n = 62) | p | |
|---|---|---|---|
| Basic characteristics and Anthropometrics indicators in early pregnancy | |||
| Age of delivery (years) | 30.58 ± 3.70 | 30.66 ± 3.83 | 0.923 |
| Waist (cm) | 82.28 ± 6.28 | 77.35 ± 7.21 | 0.002 * |
| Weight (kg) | 58.69 ± 7.60 | 52.31 ± 7.69 | <0.001 |
| Hight (cm) | 160.19 ± 4.08 | 158.52 ± 4.16 | 0.07 |
| Body Mass Index | 22.87 ± 2.88 | 20.77 ± 2.57 | 0.001 |
| SBP (mmHg) | 115.32 ± 11.41 | 115.80 ± 10.38 | 0.838 |
| DBP (mmHg) | 74.65 ± 8.34 | 75.09 ± 8.62 | 0.810 |
| History of diseases | |||
| Gestational Hypertension | 2 (6.45%) | 1 (1.61%) | 0.257 |
| Gestational Diabetes Mellitus | 0 (0.00%) | 0 (0.00) | 1.000 |
| Adverse Pregnancy | 19 (61.29%) | 38 (61.29%) | 1.000 |
| Biochemical parameters in the first trimester | |||
| HGB (g/L) | 126.67 ± 8.95 | 125.37 ± 8.44 | 0.503 |
| GLU (mmol/L) | 4.74 ± 0.31 | 4.64 ± 0.35 | 0.172 |
| ALB (g/L) | 44.79 ± 2.86 | 45.98 ± 2.58 | 0.045 |
| TG (mmol/L) | 1.76 ± 0.72 | 1.47 ± 0.46 | 0.019 |
| TC (mmol/L) | 4.89 ± 0.71 | 4.49 ± 0.71 | 0.012 |
| HDL-C (mmol/L) | 1.87 ± 0.40 | 1.95 ± 0.32 | 0.274 |
| LDL-C (mmol/L) | 2.70 ± 0.59 | 2.42 ± 0.69 | 0.060 |
| AST (U/L) | 16.40 (6.40) | 18.10 (9.00) | 0.476 |
| ALT (U/L) | 14.60 (10.50) | 17.95 (15.40) | 0.471 |
| Eating habits | |||
| Daily cereals intake (g) | 65.57 ± 26.012 | 69.29 ± 26.07 | 0.522 |
| Daily tubers intake (g) | 50.72 ± 44.64 | 45.76 ± 31.27 | 0.538 |
| Daily vegetables intake (g) | 116.85 ± 54.28 | 110.94 ± 54.70 | 0.627 |
| Daily fruits intake (g) | 164.19 ± 66.86 | 165.85 ± 55.02 | 0.900 |
| Daily meat intake (g) | 35.75 ± 30.84 | 31.88 ± 22.53 | 0.497 |
| Daily eggs intake (g) | 35.80 ± 14.73 | 29.77 ± 13.88 | 0.059 |
| Daily milk intake (mL) | 41,280.13 ± 79.27 | 25,648.51 ± 35.70 | 0.193 |
| Daily soybeans intake (g) | 48.33 ± 43.25 | 48.65 ± 42.58 | 0.973 |
| Daily peanuts intake (g) | 42.77 ± 28.87 | 28.48 ± 20.66 | 0.008 |
| Daily oil intake (g) | 10.35 ± 1.35 | 9.99 ± 1.41 | 0.256 |
| Daily salt intake (g) | 5.10 ± 1.39 | 4.69 ± 0.41 | 0.032 |
| Dail water intake (mL) | 490.28 ± 251.93 | 484.90 ± 214.29 | 0.915 |
| Mode of delivery | 0.002 | ||
| Vaginally delivery | 6 (19.40%) | 36 (58.10%) | |
| Assisted vaginal delivery | 1 (3.20%) | 2 (3.20%) | |
| Cesarean section delivery | 24 (77.40%) | 24 (38.70%) | |
| Obstetric history | |||
| Gravidity | 0.056 | ||
| 1 | 8 (25.80%) | 28 (45.16%) | |
| ≥2 | 23 (74.19%) | 34 (54.84%) | |
| Parity | 0.206 | ||
| 0 | 16 (51.61%) | 39 (62.90%) | |
| ≥1 | 15 (48.39%) | 23 (37.10%) | |
| Abortion | 0.587 | ||
| No | 12 (38.71%) | 24 (38.71%) | |
| Yes | 19 (61.29%) | 38 (61.29%) | |
| Basic characteristics of newborns | |||
| Sex | 0.518 | ||
| Male | 19 (61.30%) | 38 (61.30%) | |
| Female | 12 (38.70%) | 34 (54.80%) | |
| Birth weight (g) | 4183.87 ± 194.80 | 3005.65 ± 567.36 | <0.001 |
| Premature labor | 0 (0.00%) | 3 (4.80%) | 0.548 |
| Species | Coefficient | p | Q | Coefficient * | p * | Q * |
|---|---|---|---|---|---|---|
| Bacteroides salyersiae | −6.897 | <0.001 | <0.001 | −6.748 | <0.001 | <0.001 |
| Bacteroides plebeius | −5.210 | <0.001 | <0.001 | −5.049 | <0.001 | <0.001 |
| Ruminococcus lactaris | −4.994 | <0.001 | <0.001 | −4.880 | <0.001 | <0.001 |
| Bacteroides ovatus | −2.013 | 0.003 | 0.127 | −1.909 | 0.004 | 0.183 |
| Pathway | Coefficient | p | Q | Coefficient * | p * | Q * |
|---|---|---|---|---|---|---|
| mannitol cycle | −2.321 | <0.001 | 0.002 | −2.156 | <0.001 | 0.003 |
| L-arginine biosynthesis III | −1.363 | <0.001 | 0.011 | −1.213 | 0.001 | 0.011 |
| tRNA processing | −2.643 | <0.001 | 0.011 | −2.482 | 0.001 | 0.011 |
| L-histidine degradation III | −1.274 | <0.001 | 0.014 | −1.124 | 0.001 | 0.011 |
| gondoate biosynthesis | −0.913 | 0.002 | 0.131 | −0.763 | 0.001 | 0.121 |
| cis-vaccenate biosynthesis | −0.805 | 0.004 | 0.223 | −0.655 | 0.004 | 0.022 |
| thiamin salvage II | −0.840 | 0.005 | 0.231 | −0.689 | 0.005 | 0.258 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhong, Z.; An, R.; Ma, S.; Zhang, N.; Zhang, X.; Chen, L.; Wu, X.; Lin, H.; Xiang, T.; Tan, H.; et al. Association between the Maternal Gut Microbiome and Macrosomia. Biology 2024, 13, 570. https://doi.org/10.3390/biology13080570
Zhong Z, An R, Ma S, Zhang N, Zhang X, Chen L, Wu X, Lin H, Xiang T, Tan H, et al. Association between the Maternal Gut Microbiome and Macrosomia. Biology. 2024; 13(8):570. https://doi.org/10.3390/biology13080570
Chicago/Turabian StyleZhong, Zixin, Rongjing An, Shujuan Ma, Na Zhang, Xian Zhang, Lizhang Chen, Xinrui Wu, Huijun Lin, Tianyu Xiang, Hongzhuan Tan, and et al. 2024. "Association between the Maternal Gut Microbiome and Macrosomia" Biology 13, no. 8: 570. https://doi.org/10.3390/biology13080570
APA StyleZhong, Z., An, R., Ma, S., Zhang, N., Zhang, X., Chen, L., Wu, X., Lin, H., Xiang, T., Tan, H., & Chen, M. (2024). Association between the Maternal Gut Microbiome and Macrosomia. Biology, 13(8), 570. https://doi.org/10.3390/biology13080570






