Post-Translational Modifications (PTMs) of mutp53 and Epigenetic Changes Induced by mutp53
Abstract
Simple Summary
Abstract
1. Introduction
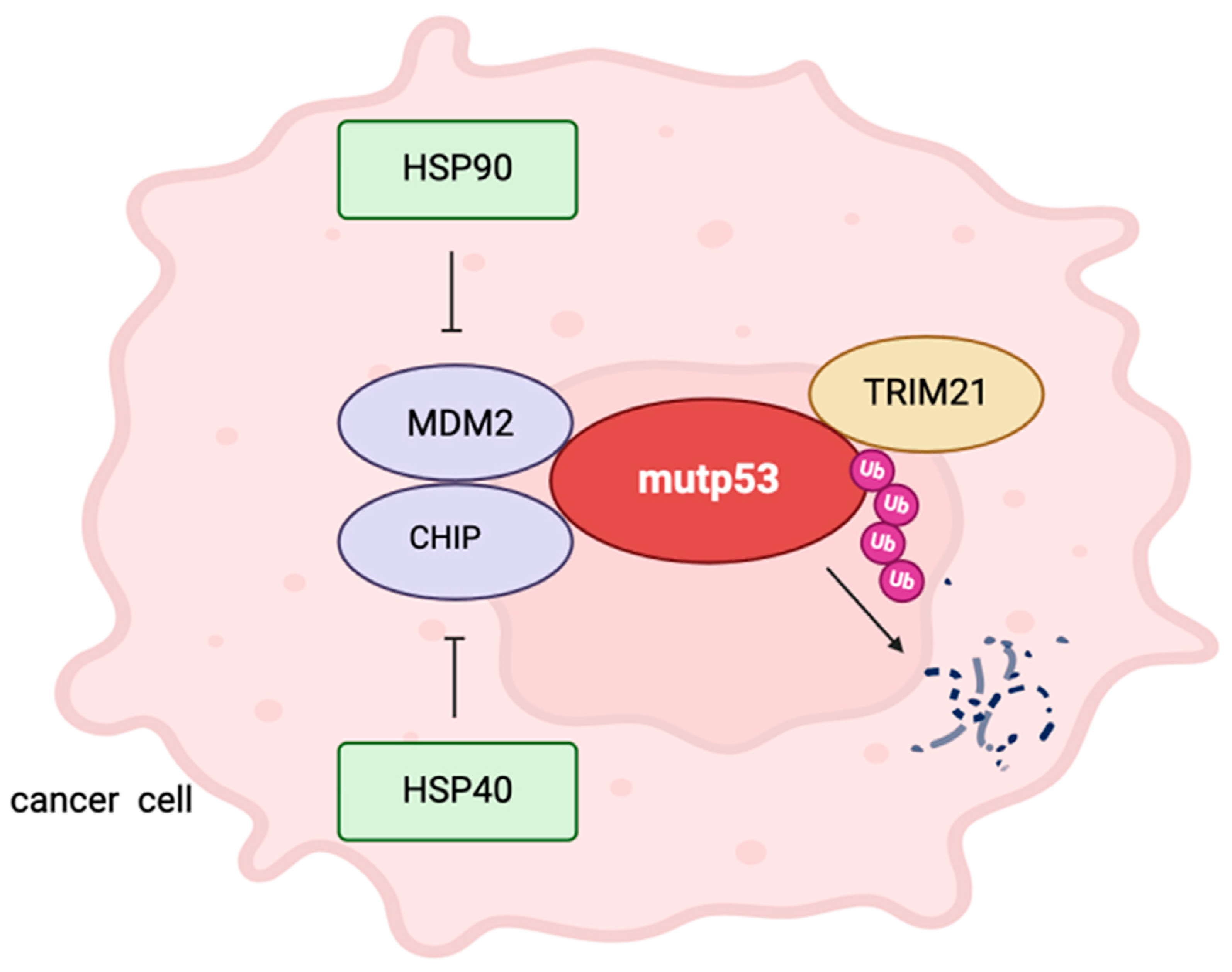
2. Mutp53, Ubiquitination and HSPs
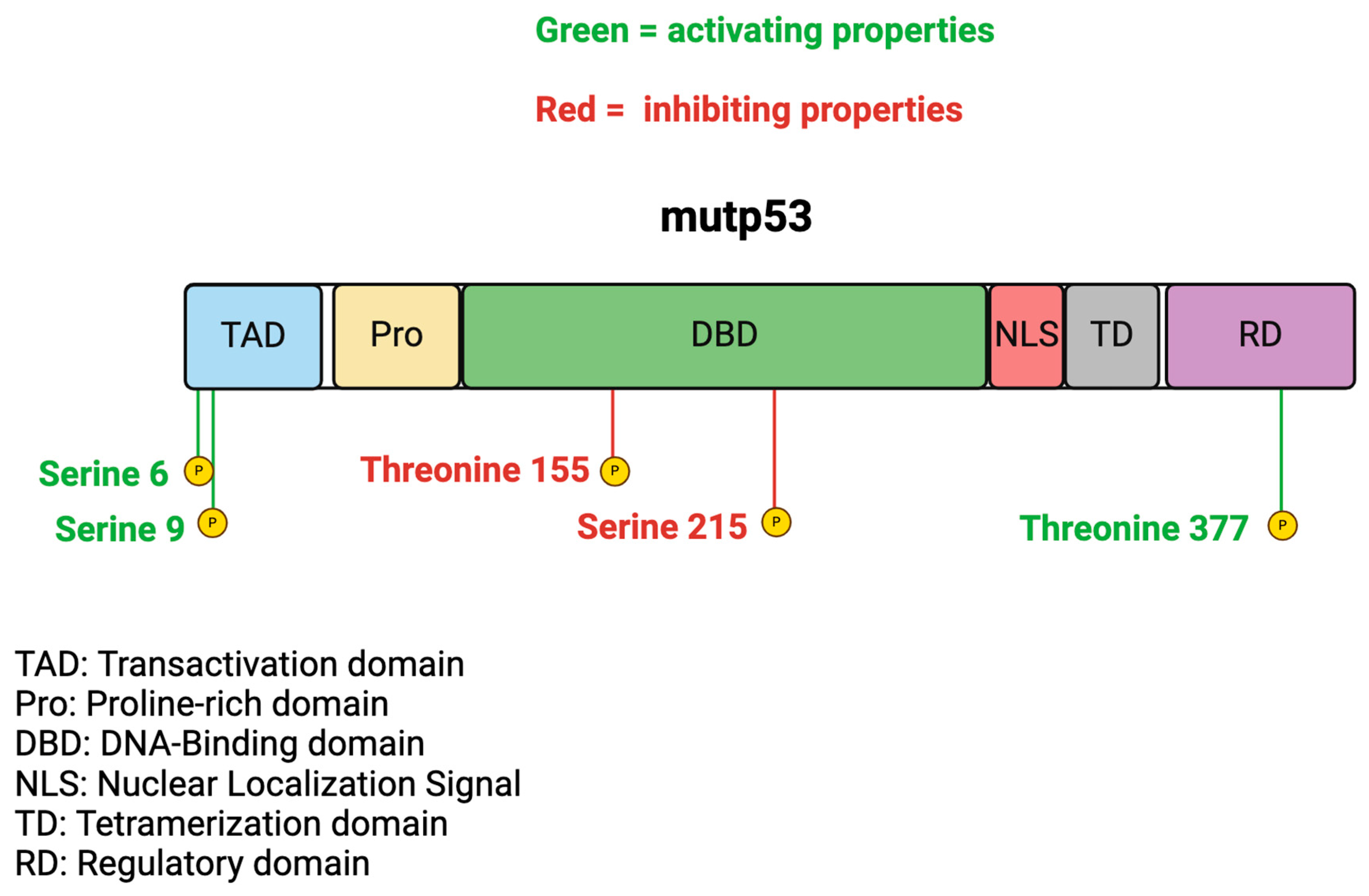
3. Mutp53 and Phosphorylation
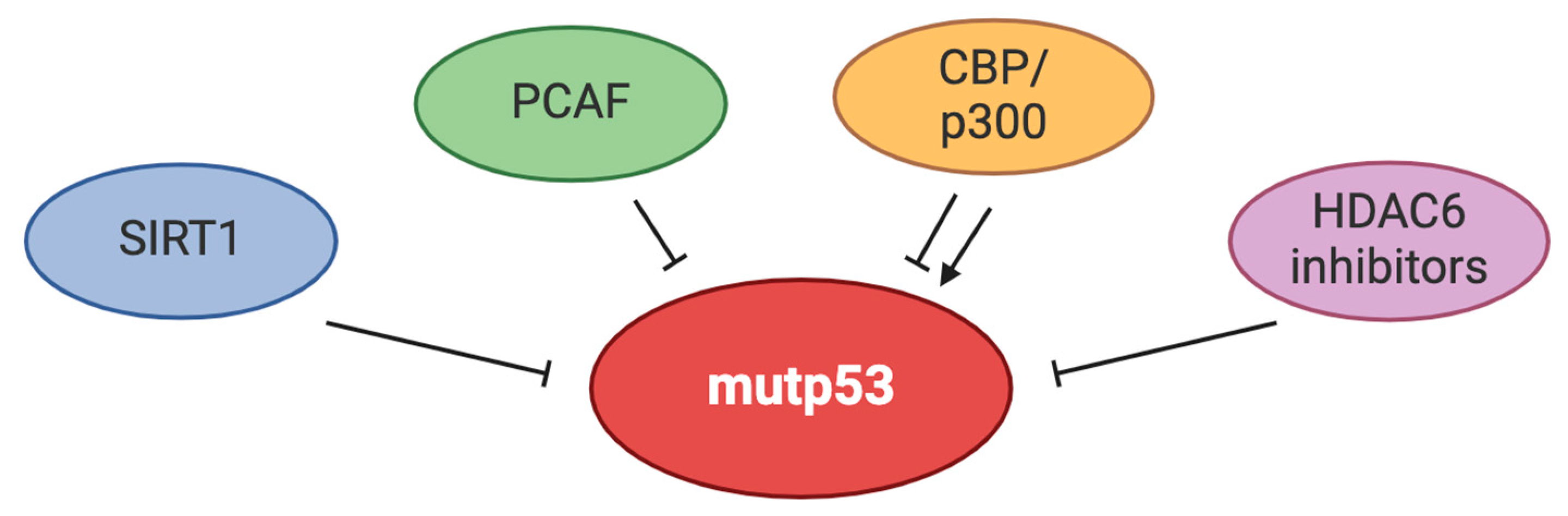
4. Mutp53 and Acetylation
5. Methylation and wt- and mutp53
6. Mutp53 and Epigenetic Changes
7. Mutp53 and miRNAs
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mantovani, F.; Collavin, L.; Del Sal, G. Mutant p53 as a guardian of the cancer cell. Cell Death Differ. 2019, 26, 199–212. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, T.; Su, W.; Dou, Z.; Zhao, D.; Jin, X.; Lei, H.; Wang, J.; Xie, X.; Cheng, B.; et al. Mutant p53 in cancer: From molecular mechanism to therapeutic modulation. Cell Death Dis. 2022, 13, 974. [Google Scholar] [CrossRef] [PubMed]
- Dolma, L.; Muller, P.A.J. GOF Mutant p53 in Cancers: A Therapeutic Challenge. Cancers 2022, 14, 5091. [Google Scholar] [CrossRef]
- Hu, J.; Cao, J.; Topatana, W.; Juengpanich, S.; Li, S.; Zhang, B.; Shen, J.; Cai, L.; Cai, X.; Chen, M. Targeting mutant p53 for cancer therapy: Direct and indirect strategies. J. Hematol. Oncol. 2021, 14, 157. [Google Scholar] [CrossRef] [PubMed]
- D’Orazi, G.; Cordani, M.; Cirone, M. Oncogenic pathways activated by pro-inflammatory cytokines promote mutant p53 stability: Clue for novel anticancer therapies. Cell Mol. Life Sci. 2021, 78, 1853–1860. [Google Scholar] [CrossRef]
- Zhao, H.; Wu, L.; Yan, G.; Chen, Y.; Zhou, M.; Wu, Y.; Li, Y. Inflammation and tumor progression: Signaling pathways and targeted intervention. Signal Transduct. Target. Ther. 2021, 6, 263. [Google Scholar] [CrossRef]
- Cirone, M. ER Stress, UPR Activation and the Inflammatory Response to Viral Infection. Viruses 2021, 13, 798. [Google Scholar] [CrossRef]
- Sicari, D.; Fantuz, M.; Bellazzo, A.; Valentino, E.; Apollonio, M.; Pontisso, I.; Di Cristino, F.; Dal Ferro, M.; Bicciato, S.; Del Sal, G.; et al. Mutant p53 improves cancer cells’ resistance to endoplasmic reticulum stress by sustaining activation of the UPR regulator ATF6. Oncogene 2019, 38, 6184–6195. [Google Scholar] [CrossRef] [PubMed]
- Ano Bom, A.P.; Rangel, L.P.; Costa, D.C.; de Oliveira, G.A.; Sanches, D.; Braga, C.A.; Gava, L.M.; Ramos, C.H.; Cepeda, A.O.; Stumbo, A.C.; et al. Mutant p53 aggregates into prion-like amyloid oligomers and fibrils: Implications for cancer. J. Biol. Chem. 2012, 287, 28152–28162. [Google Scholar] [CrossRef]
- Devine, T.; Dai, M.S. Targeting the ubiquitin-mediated proteasome degradation of p53 for cancer therapy. Curr. Pharm. Des. 2013, 19, 3248–3262. [Google Scholar] [CrossRef] [PubMed]
- Cordani, M.; Oppici, E.; Dando, I.; Butturini, E.; Dalla Pozza, E.; Nadal-Serrano, M.; Oliver, J.; Roca, P.; Mariotto, S.; Cellini, B.; et al. Mutant p53 proteins counteract autophagic mechanism sensitizing cancer cells to mTOR inhibition. Mol. Oncol. 2016, 10, 1008–1029. [Google Scholar] [CrossRef] [PubMed]
- Romeo, M.A.; Gilardini Montani, M.S.; Benedetti, R.; Santarelli, R.; D’Orazi, G.; Cirone, M. STAT3 and mutp53 Engage a Positive Feedback Loop Involving HSP90 and the Mevalonate Pathway. Front. Oncol. 2020, 10, 1102. [Google Scholar] [CrossRef] [PubMed]
- Schulz-Heddergott, R.; Stark, N.; Edmunds, S.J.; Li, J.; Conradi, L.C.; Bohnenberger, H.; Ceteci, F.; Greten, F.R.; Dobbelstein, M.; Moll, U.M. Therapeutic Ablation of Gain-of-Function Mutant p53 in Colorectal Cancer Inhibits Stat3-Mediated Tumor Growth and Invasion. Cancer Cell 2018, 34, 298–314 e297. [Google Scholar] [CrossRef]
- Benedetti, R.; Romeo, M.A.; Arena, A.; Gilardini Montani, M.S.; D’Orazi, G.; Cirone, M. ATF6 supports lysosomal function in tumor cells to enable ER stress-activated macroautophagy and CMA: Impact on mutant TP53 expression. Autophagy 2024, 1–14. [Google Scholar] [CrossRef]
- Vakifahmetoglu-Norberg, H.; Kim, M.; Xia, H.G.; Iwanicki, M.P.; Ofengeim, D.; Coloff, J.L.; Pan, L.; Ince, T.A.; Kroemer, G.; Brugge, J.S.; et al. Chaperone-mediated autophagy degrades mutant p53. Genes. Dev. 2013, 27, 1718–1730. [Google Scholar] [CrossRef]
- Gilardini Montani, M.S.; Cecere, N.; Granato, M.; Romeo, M.A.; Falcinelli, L.; Ciciarelli, U.; D’Orazi, G.; Faggioni, A.; Cirone, M. Mutant p53, Stabilized by Its Interplay with HSP90, Activates a Positive Feed-Back Loop Between NRF2 and p62 that Induces Chemo-Resistance to Apigenin in Pancreatic Cancer Cells. Cancers 2019, 11, 703. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Xiao, J.; Wang, Y.; Song, X.; Huang, L.; Ren, Z.; Kitazato, K.; Wang, Y. Posttranslational modification and beyond: Interplay between histone deacetylase 6 and heat-shock protein 90. Mol. Med. 2021, 27, 110. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Liu, T.; Rios, Z.; Mei, Q.; Lin, X.; Cao, S. Heat Shock Proteins and Cancer. Trends Pharmacol. Sci. 2017, 38, 226–256. [Google Scholar] [CrossRef]
- D’Orazi, G.; Cirone, M. Interconnected Adaptive Responses: A Way Out for Cancer Cells to Avoid Cellular Demise. Cancers 2022, 14, 2780. [Google Scholar] [CrossRef]
- Gonnella, R.; Arena, A.; Zarrella, R.; Gilardini Montani, M.S.; Santarelli, R.; Cirone, M. HSPs/STAT3 Interplay Sustains DDR and Promotes Cytokine Release by Primary Effusion Lymphoma Cells. Int. J. Mol. Sci. 2023, 24, 3933. [Google Scholar] [CrossRef]
- Gonnella, R.; Zarrella, R.; Di Crosta, M.; Benedetti, R.; Arena, A.; Santarelli, R.; Gilardini Montani, M.S.; D’Orazi, G.; Cirone, M. HSP110 Inhibition in Primary Effusion Lymphoma Cells: One Molecule, Many Pro-Survival Targets. Cancers 2023, 15, 5651. [Google Scholar] [CrossRef]
- DeHart, C.J.; Chahal, J.S.; Flint, S.J.; Perlman, D.H. Extensive post-translational modification of active and inactivated forms of endogenous p53. Mol. Cell Proteom. 2014, 13, 1–17. [Google Scholar] [CrossRef]
- Tong, Q.; Mazur, S.J.; Rincon-Arano, H.; Rothbart, S.B.; Kuznetsov, D.M.; Cui, G.; Liu, W.H.; Gete, Y.; Klein, B.J.; Jenkins, L.; et al. An acetyl-methyl switch drives a conformational change in p53. Structure 2015, 23, 322–331. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Marchenko, N.D.; Schulz, R.; Fischer, V.; Velasco-Hernandez, T.; Talos, F.; Moll, U.M. Functional inactivation of endogenous MDM2 and CHIP by HSP90 causes aberrant stabilization of mutant p53 in human cancer cells. Mol. Cancer Res. 2011, 9, 577–588. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Marchenko, N.D.; Moll, U.M. SAHA shows preferential cytotoxicity in mutant p53 cancer cells by destabilizing mutant p53 through inhibition of the HDAC6-Hsp90 chaperone axis. Cell Death Differ. 2011, 18, 1904–1913. [Google Scholar] [CrossRef] [PubMed]
- Parrales, A.; Ranjan, A.; Iyer, S.V.; Padhye, S.; Weir, S.J.; Roy, A.; Iwakuma, T. DNAJA1 controls the fate of misfolded mutant p53 through the mevalonate pathway. Nat. Cell Biol. 2016, 18, 1233–1243. [Google Scholar] [CrossRef]
- Romeo, M.A.; Gilardini Montani, M.S.; Arena, A.; Benedetti, R.; D’Orazi, G.; Cirone, M. c-Myc Sustains Pancreatic Cancer Cell Survival and mutp53 Stability through the Mevalonate Pathway. Biomedicines 2022, 10, 2489. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, C.; Xu, D.; Zhang, T.; Chang, C.Y.; Wang, J.; Liu, J.; Zhang, L.; Haffty, B.G.; Zong, W.X.; et al. The ubiquitin ligase TRIM21 regulates mutant p53 accumulation and gain of function in cancer. J. Clin. Invest. 2023, 133, e164354. [Google Scholar] [CrossRef]
- Yogosawa, S.; Yoshida, K. Tumor suppressive role for kinases phosphorylating p53 in DNA damage-induced apoptosis. Cancer Sci. 2018, 109, 3376–3382. [Google Scholar] [CrossRef]
- D’Orazi, G.; Cecchinelli, B.; Bruno, T.; Manni, I.; Higashimoto, Y.; Saito, S.; Gostissa, M.; Coen, S.; Marchetti, A.; Del Sal, G.; et al. Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nat. Cell Biol. 2002, 4, 11–19. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, J.; Xu, D.; Zhang, T.; Hu, W.; Feng, Z. Gain-of-function mutant p53 in cancer progression and therapy. J. Mol. Cell Biol. 2020, 12, 674–687. [Google Scholar] [CrossRef]
- Yue, X.; Zhao, Y.; Xu, Y.; Zheng, M.; Feng, Z.; Hu, W. Mutant p53 in Cancer: Accumulation, Gain-of-Function, and Therapy. J. Mol. Biol. 2017, 429, 1595–1606. [Google Scholar] [CrossRef] [PubMed]
- Gujral, P.; Mahajan, V.; Lissaman, A.C.; Ponnampalam, A.P. Histone acetylation and the role of histone deacetylases in normal cyclic endometrium. Reprod. Biol. Endocrinol. 2020, 18, 84. [Google Scholar] [CrossRef]
- Ryu, H.W.; Shin, D.H.; Lee, D.H.; Choi, J.; Han, G.; Lee, K.Y.; Kwon, S.H. HDAC6 deacetylates p53 at lysines 381/382 and differentially coordinates p53-induced apoptosis. Cancer Lett. 2017, 391, 162–171. [Google Scholar] [CrossRef]
- Jethwa, A.; Slabicki, M.; Hullein, J.; Jentzsch, M.; Dalal, V.; Rabe, S.; Wagner, L.; Walther, T.; Klapper, W.; Project, M.N.; et al. TRRAP is essential for regulating the accumulation of mutant and wild-type p53 in lymphoma. Blood 2018, 131, 2789–2802. [Google Scholar] [CrossRef] [PubMed]
- McMahon, S.B.; Van Buskirk, H.A.; Dugan, K.A.; Copeland, T.D.; Cole, M.D. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell 1998, 94, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Vervoorts, J.; Luscher-Firzlaff, J.M.; Rottmann, S.; Lilischkis, R.; Walsemann, G.; Dohmann, K.; Austen, M.; Luscher, B. Stimulation of c-MYC transcriptional activity and acetylation by recruitment of the cofactor CBP. EMBO Rep. 2003, 4, 484–490. [Google Scholar] [CrossRef]
- Ionov, Y.; Matsui, S.; Cowell, J.K. A role for p300/CREB binding protein genes in promoting cancer progression in colon cancer cell lines with microsatellite instability. Proc. Natl. Acad. Sci. USA 2004, 101, 1273–1278. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Qian, W.; Yang, Z.; Zhang, Z.; Sun, P.; Wan, Q.; Yin, Y.; Hu, Y.; Gong, L.; Zhang, B.; et al. Acetylation halts missense mutant p53 aggregation and rescues tumor suppression in non-small cell lung cancers. iScience 2023, 26, 107003. [Google Scholar] [CrossRef]
- Stojanovic, N.; Hassan, Z.; Wirth, M.; Wenzel, P.; Beyer, M.; Schafer, C.; Brand, P.; Kroemer, A.; Stauber, R.H.; Schmid, R.M.; et al. HDAC1 and HDAC2 integrate the expression of p53 mutants in pancreatic cancer. Oncogene 2017, 36, 1804–1815. [Google Scholar] [CrossRef]
- Sakaguchi, K.; Herrera, J.E.; Saito, S.; Miki, T.; Bustin, M.; Vassilev, A.; Anderson, C.W.; Appella, E. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes. Dev. 1998, 12, 2831–2841. [Google Scholar] [CrossRef] [PubMed]
- Kaypee, S.; Sahadevan, S.A.; Patil, S.; Ghosh, P.; Roy, N.S.; Roy, S.; Kundu, T.K. Mutant and Wild-Type Tumor Suppressor p53 Induces p300 Autoacetylation. iScience 2018, 4, 260–272. [Google Scholar] [CrossRef]
- Kawai, H.; Nie, L.; Wiederschain, D.; Yuan, Z.M. Dual role of p300 in the regulation of p53 stability. J. Biol. Chem. 2001, 276, 45928–45932. [Google Scholar] [CrossRef]
- Knowell, A.E.; Patel, D.; Morton, D.J.; Sharma, P.; Glymph, S.; Chaudhary, J. Id4 dependent acetylation restores mutant-p53 transcriptional activity. Mol. Cancer 2013, 12, 161. [Google Scholar] [CrossRef] [PubMed]
- Perez, R.E.; Knights, C.D.; Sahu, G.; Catania, J.; Kolukula, V.K.; Stoler, D.; Graessmann, A.; Ogryzko, V.; Pishvaian, M.; Albanese, C.; et al. Restoration of DNA-binding and growth-suppressive activity of mutant forms of p53 via a PCAF-mediated acetylation pathway. J. Cell Physiol. 2010, 225, 394–405. [Google Scholar] [CrossRef]
- Kirilyuk, A.; Shimoji, M.; Catania, J.; Sahu, G.; Pattabiraman, N.; Giordano, A.; Albanese, C.; Mocchetti, I.; Toretsky, J.A.; Uversky, V.N.; et al. An intrinsically disordered region of the acetyltransferase p300 with similarity to prion-like domains plays a role in aggregation. PLoS ONE 2012, 7, e48243. [Google Scholar] [CrossRef]
- Carafa, V.; Altucci, L.; Nebbioso, A. Dual Tumor Suppressor and Tumor Promoter Action of Sirtuins in Determining Malignant Phenotype. Front. Pharmacol. 2019, 10, 38. [Google Scholar] [CrossRef]
- Yi, Y.W.; Kang, H.J.; Kim, H.J.; Kong, Y.; Brown, M.L.; Bae, I. Targeting mutant p53 by a SIRT1 activator YK-3-237 inhibits the proliferation of triple-negative breast cancer cells. Oncotarget 2013, 4, 984–994. [Google Scholar] [CrossRef] [PubMed]
- Jambhekar, A.; Dhall, A.; Shi, Y. Roles and regulation of histone methylation in animal development. Nat. Rev. Mol. Cell Biol. 2019, 20, 625–641. [Google Scholar] [CrossRef]
- Chuikov, S.; Kurash, J.K.; Wilson, J.R.; Xiao, B.; Justin, N.; Ivanov, G.S.; McKinney, K.; Tempst, P.; Prives, C.; Gamblin, S.J.; et al. Regulation of p53 activity through lysine methylation. Nature 2004, 432, 353–360. [Google Scholar] [CrossRef]
- Ivanov, G.S.; Ivanova, T.; Kurash, J.; Ivanov, A.; Chuikov, S.; Gizatullin, F.; Herrera-Medina, E.M.; Rauscher, F., 3rd; Reinberg, D.; Barlev, N.A. Methylation-acetylation interplay activates p53 in response to DNA damage. Mol. Cell Biol. 2007, 27, 6756–6769. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, D.; Zhao, Y.; Tu, B.; Zheng, Z.; Wang, L.; Wang, H.; Gu, W.; Roeder, R.G.; Zhu, W.G. Methyltransferase Set7/9 regulates p53 activity by interacting with Sirtuin 1 (SIRT1). Proc. Natl. Acad. Sci. USA 2011, 108, 1925–1930. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Perez-Burgos, L.; Placek, B.J.; Sengupta, R.; Richter, M.; Dorsey, J.A.; Kubicek, S.; Opravil, S.; Jenuwein, T.; Berger, S.L. Repression of p53 activity by Smyd2-mediated methylation. Nature 2006, 444, 629–632. [Google Scholar] [CrossRef]
- Shi, X.; Kachirskaia, I.; Yamaguchi, H.; West, L.E.; Wen, H.; Wang, E.W.; Dutta, S.; Appella, E.; Gozani, O. Modulation of p53 function by SET8-mediated methylation at lysine 382. Mol. Cell 2007, 27, 636–646. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Sengupta, R.; Espejo, A.B.; Lee, M.G.; Dorsey, J.A.; Richter, M.; Opravil, S.; Shiekhattar, R.; Bedford, M.T.; Jenuwein, T.; et al. p53 is regulated by the lysine demethylase LSD1. Nature 2007, 449, 105–108. [Google Scholar] [CrossRef]
- Lee, D.H.; Kim, G.W.; Yoo, J.; Lee, S.W.; Jeon, Y.H.; Kim, S.Y.; Kang, H.G.; Kim, D.H.; Chun, K.H.; Choi, J.; et al. Histone demethylase KDM4C controls tumorigenesis of glioblastoma by epigenetically regulating p53 and c-Myc. Cell Death Dis. 2021, 12, 89. [Google Scholar] [CrossRef]
- Nagasaka, M.; Tsuzuki, K.; Ozeki, Y.; Tokugawa, M.; Ohoka, N.; Inoue, Y.; Hayashi, H. Lysine-Specific Demethylase 1 (LSD1/KDM1A) Is a Novel Target Gene of c-Myc. Biol. Pharm. Bull. 2019, 42, 481–488. [Google Scholar] [CrossRef]
- Lu, Y.; Chan, Y.T.; Tan, H.Y.; Li, S.; Wang, N.; Feng, Y. Epigenetic regulation in human cancer: The potential role of epi-drug in cancer therapy. Mol. Cancer 2020, 19, 79. [Google Scholar] [CrossRef]
- Mantovani, F.; Walerych, D.; Sal, G.D. Targeting mutant p53 in cancer: A long road to precision therapy. FEBS J. 2017, 284, 837–850. [Google Scholar] [CrossRef]
- Arena, A.; Gilardini Montani, M.S.; Romeo, M.A.; Benedetti, R.; Gaeta, A.; Cirone, M. DNA damage triggers an interplay between wtp53 and c-Myc affecting lymphoma cell proliferation and Kaposi sarcoma herpesvirus replication. Biochim. Biophys. Acta Mol. Cell Res. 2022, 1869, 119168. [Google Scholar] [CrossRef]
- Tang, X.; Milyavsky, M.; Shats, I.; Erez, N.; Goldfinger, N.; Rotter, V. Activated p53 suppresses the histone methyltransferase EZH2 gene. Oncogene 2004, 23, 5759–5769. [Google Scholar] [CrossRef] [PubMed]
- Versemann, L.; Patil, S.; Steuber, B.; Zhang, Z.; Kopp, W.; Krawczyk, H.E.; Kaulfuss, S.; Wollnik, B.; Strobel, P.; Neesse, A.; et al. TP53-Status-Dependent Oncogenic EZH2 Activity in Pancreatic Cancer. Cancers 2022, 14, 3451. [Google Scholar] [CrossRef]
- Zhao, Y.; Ding, L.; Wang, D.; Ye, Z.; He, Y.; Ma, L.; Zhu, R.; Pan, Y.; Wu, Q.; Pang, K.; et al. EZH2 cooperates with gain-of-function p53 mutants to promote cancer growth and metastasis. EMBO J. 2019, 38, e99599. [Google Scholar] [CrossRef]
- Jiang, F.Z.; He, Y.Y.; Wang, H.H.; Zhang, H.L.; Zhang, J.; Yan, X.F.; Wang, X.J.; Che, Q.; Ke, J.Q.; Chen, Z.; et al. Mutant p53 induces EZH2 expression and promotes epithelial-mesenchymal transition by disrupting p68-Drosha complex assembly and attenuating miR-26a processing. Oncotarget 2015, 6, 44660–44674. [Google Scholar] [CrossRef]
- Kuser-Abali, G.; Gong, L.; Yan, J.; Liu, Q.; Zeng, W.; Williamson, A.; Lim, C.B.; Molloy, M.E.; Little, J.B.; Huang, L.; et al. An EZH2-mediated epigenetic mechanism behind p53-dependent tissue sensitivity to DNA damage. Proc. Natl. Acad. Sci. USA 2018, 115, 3452–3457. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Sammons, M.A.; Donahue, G.; Dou, Z.; Vedadi, M.; Getlik, M.; Barsyte-Lovejoy, D.; Al-awar, R.; Katona, B.W.; Shilatifard, A.; et al. Gain-of-function p53 mutants co-opt chromatin pathways to drive cancer growth. Nature 2015, 525, 206–211. [Google Scholar] [CrossRef] [PubMed]
- Xu, A.; Liu, M.; Huang, M.F.; Zhang, Y.; Hu, R.; Gingold, J.A.; Liu, Y.; Zhu, D.; Chien, C.S.; Wang, W.C.; et al. Rewired m(6)A epitranscriptomic networks link mutant p53 to neoplastic transformation. Nat. Commun. 2023, 14, 1694. [Google Scholar] [CrossRef]
- Rahnamoun, H.; Hong, J.; Sun, Z.; Lee, J.; Lu, H.; Lauberth, S.M. Mutant p53 regulates enhancer-associated H3K4 monomethylation through interactions with the methyltransferase MLL4. J. Biol. Chem. 2018, 293, 13234–13246. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Tsai, M.H.; Shiao, Y.H.; Chen, L.H.; Wei, M.L.; Lv, X.; Gius, D.; Little, J.B.; Mitchell, J.B.; Chuang, E.Y. DNA (cytosine-5)-methyltransferase 1 as a mediator of mutant p53-determined p16(ink4A) down-regulation. J. Biomed. Sci. 2008, 15, 163–168. [Google Scholar] [CrossRef]
- Wang, Q.; Xiong, F.; Wu, G.; Liu, W.; Chen, J.; Wang, B.; Chen, Y. Gene body methylation in cancer: Molecular mechanisms and clinical applications. Clin. Epigenetics 2022, 14, 154. [Google Scholar] [CrossRef]
- Gurtner, A.; Falcone, E.; Garibaldi, F.; Piaggio, G. Dysregulation of microRNA biogenesis in cancer: The impact of mutant p53 on Drosha complex activity. J. Exp. Clin. Cancer Res. 2016, 35, 45. [Google Scholar] [CrossRef] [PubMed]
- Garibaldi, F.; Falcone, E.; Trisciuoglio, D.; Colombo, T.; Lisek, K.; Walerych, D.; Del Sal, G.; Paci, P.; Bossi, G.; Piaggio, G.; et al. Mutant p53 inhibits miRNA biogenesis by interfering with the microprocessor complex. Oncogene 2016, 35, 3760–3770. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Cheng, B.; Miao, L.; Mei, Y.; Wu, M. Mutant p53-R273H gains new function in sustained activation of EGFR signaling via suppressing miR-27a expression. Cell Death Dis. 2013, 4, e574. [Google Scholar] [CrossRef] [PubMed]
- Madrigal, T.; Hernandez-Monge, J.; Herrera, L.A.; Gonzalez-De la Rosa, C.H.; Dominguez-Gomez, G.; Candelaria, M.; Luna-Maldonado, F.; Calderon Gonzalez, K.G.; Diaz-Chavez, J. Regulation of miRNAs Expression by Mutant p53 Gain of Function in Cancer. Front. Cell Dev. Biol. 2021, 9, 695723. [Google Scholar] [CrossRef]
- Donzelli, S.; Fontemaggi, G.; Fazi, F.; Di Agostino, S.; Padula, F.; Biagioni, F.; Muti, P.; Strano, S.; Blandino, G. MicroRNA-128-2 targets the transcriptional repressor E2F5 enhancing mutant p53 gain of function. Cell Death Differ. 2012, 19, 1038–1048. [Google Scholar] [CrossRef]
- Neilsen, P.M.; Noll, J.E.; Mattiske, S.; Bracken, C.P.; Gregory, P.A.; Schulz, R.B.; Lim, S.P.; Kumar, R.; Suetani, R.J.; Goodall, G.J.; et al. Mutant p53 drives invasion in breast tumors through up-regulation of miR-155. Oncogene 2013, 32, 2992–3000. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Benedetti, R.; Di Crosta, M.; D’Orazi, G.; Cirone, M. Post-Translational Modifications (PTMs) of mutp53 and Epigenetic Changes Induced by mutp53. Biology 2024, 13, 508. https://doi.org/10.3390/biology13070508
Benedetti R, Di Crosta M, D’Orazi G, Cirone M. Post-Translational Modifications (PTMs) of mutp53 and Epigenetic Changes Induced by mutp53. Biology. 2024; 13(7):508. https://doi.org/10.3390/biology13070508
Chicago/Turabian StyleBenedetti, Rossella, Michele Di Crosta, Gabriella D’Orazi, and Mara Cirone. 2024. "Post-Translational Modifications (PTMs) of mutp53 and Epigenetic Changes Induced by mutp53" Biology 13, no. 7: 508. https://doi.org/10.3390/biology13070508
APA StyleBenedetti, R., Di Crosta, M., D’Orazi, G., & Cirone, M. (2024). Post-Translational Modifications (PTMs) of mutp53 and Epigenetic Changes Induced by mutp53. Biology, 13(7), 508. https://doi.org/10.3390/biology13070508




