Simple Summary
Natural microorganism-derived products are an essential source of valuable pharmaceuticals and agrichemicals. Streptomyces spp. are the most environmentally abundant bacteria that are capable of producing various valuable natural products exhibiting significant biological activity to be used in such industries as medicine, environmental science, food industry, and agriculture. However, many natural products among Streptomyces bacteria remain unstudied. Because of the rapid increase in antimicrobial and pesticide resistance, it is currently extremely topical to develop novel antibiotics, antiviral agents, agrichemicals, and other substances. The modern methods of molecular biology allow one to detect antimicrobial resistance gene clusters in the bacterial cell. Our study detected antibiotics and terpenes in cells of investigated bacteria; these compounds are of significant interest for the plant-growing sector.
Abstract
This work aimed to study the genome organization and the metabolic potential of Streptomyces carpaticus strain SCPM-O-B-9993, a promising plant-protecting and plant-stimulating strain isolated from brown semi-desert soils with very high salinity. The strain genome contains a linear chromosome 5,968,715 bp long and has no plasmids. The genome contains 5331 coding sequences among which 2139 (40.1%) are functionally annotated. Biosynthetic gene clusters (BGCs) of secondary metabolites exhibiting antimicrobial properties (ohmyungsamycin, pellasoren, naringenin, and ansamycin) were identified in the genome. The most efficient period of SCPM-O-B-9993 strain cultivation was 72 h: during this period, the culture went from the exponential to the stationary growth phase as well as exhibited excellent phytostimulatory properties and antiviral activity against the cucumber mosaic virus in tomatoes under laboratory conditions. The Streptomyces carpaticus SCPM-OB-9993 strain is a biotechnologically promising producer of secondary metabolites exhibiting antiviral and phytostimulatory properties.
1. Introduction
Actinobacteria are characterized by high ecological plasticity, labile enzymatic systems, and a powerful and complex secondary metabolism [1,2].
According to the research into abundance and species diversity of Actinobacteria, Streptomyces spp. constitute 80–95% of all soil-dwelling actinobacteria. The overwhelming majority (70–80%) of all the known biologically active microbial secondary metabolites are produced by Streptomyces spp. [3].
The genus Streptomyces includes spore-forming, filamentous, and Gram-positive Actinobacteria [4,5]. Members of the genus Streptomyces are able to survive under adverse environmental conditions, while retaining metabolic activity for a long time, and degrade natural and synthetic substances as they possess enzymes with a wide substrate specificity. These bacteria produce a great variety of chemical components (polyketides, peptides, macrolides, indoles, aminoglycosides, terpenes, etc.) [6,7,8] through which they exert regulatory effects on the plant and control development of phytopathogens [9].
Members of the Streptomyces genus can affect phytopathogens either directly by producing antibiotics, siderophores, hydrolytic, or detoxifying enzymes, or indirectly by stimulating host plant growth through synthesis of phytohormones, increasing their resistance to diseases, forming mechanisms of induced and/or acquired system resistance, or simply by competing with phytopathogens for available nutrients [10,11].
Functions of the genes annotated in the genomes are currently poorly understood and require further comprehensive studies [12]. Using in silico genome analysis methods, Streptomyces genomes have been found to contain 25–70 biologically active compounds, but only a small fraction of these compounds are synthesized in the laboratory using culturing methods [13]. Modern sequencing techniques pose serious computational challenges because of short lengths of the sequenced fragments and large data volumes, which especially affect the functional annotation of the genomes of Streptomyces strains, since the latter contain many proteins with repeats, multiblock structures such as polyketide synthases, non-ribosomal peptide synthases (NRPSs), and serine threonine kinases [14,15]. The functional genes of Streptomyces are currently being intensively studied [16,17]. Thus, the genome of the S. clavuligerus strain contains many biosynthetic gene clusters (BGCs) of secondary metabolites such as staurosporine [18], moenomycin [19], terpenes, pentalenes, phytoenes, siderophores, and lantibiotics [20].
Given the high degree of polymorphism of Streptomyces, it is undoubtedly important, from a scientific and practical point of view, to study the level of specificity and biological activity of S. carpaticus strain SCPM-O-B-9993 [21]. The S. carpaticus SCPM-O-B-9993 strain was isolated from brown semi-desert soils with very high salinity in the Astrakhan Region of the Russian Federation. The laboratory studies conducted previously demonstrated that this strain exhibits phytostimulatory, antifungal, antioxidant, insecticidal, and antiviral activities, thus being of interest for the plant-growing industry [22]. This work aimed to investigate the genome organization and metabolic potential of Streptomyces carpaticus strain SCPM-O-B-9993, a promising plant-protecting and plant-stimulating strain.
2. Materials and Methods
2.1. Strain Cultivation
The S. carpaticus SCPM-O-B-9993 strain was cultured in starch casein agar at 28 °C. The composition of starch casein agar (g/L distilled water) was as follows: soluble starch, 10.0; casein, 0.3; KNO3, 2.0; K2HPO4, 2.0; MgSO4·7H2O, 0.05; NaCl, 2.0; CaCO3, 0.02; FeSO4·7H2O, 0.01, agar, 20.0. For the experiment, a 1 cm2 fragment of 7-day strain colonies with aerial and substrate mycelium was reinoculated from solid starch casein agar to 1 L of liquid growth medium of the same composition. The SCPM-O-B-9993 strain was cultured during 168 h at 28 °C under constant stirring in a shaker (80 rpm). During 7-day cultivation, an aliquot of the culture medium with cells (suspension) was used to inoculate solid starch casein agar (0.1 mL) every 24 h for calculating colony-forming units (CFU) and testing culture purity; cell count was estimated by measuring optical density of the suspension at 440 nm and used to determine phytostimulatory and antiviral activities. Strain cells were examined using an Amplival microscope (Zeiss, Oberkochen, Germany).
2.2. Phytotoxicity Assessment
Phytotoxicity of the SCPM-O-B-9993 strain suspension was studied by the wet cell method using seeds of Ducat cress (Lepidium sativum). The seeds were pre-sterilized in 70% ethanol during 3 min and then washed with distilled water several times. Each Petri dish was bottomed with a circle made of filter paper; 30 cress seeds were placed on it, and then 5 mL of the suspension was added (distilled water was used as a control). The dishes were exposed to 20 °C under natural light conditions for 3 days. The experiment was performed in two replicates. Germination was then calculated; root length and stem height of seedlings were measured using a ruler.
The results were analyzed using the conventional mathematical statistics methods in the Excel (2023) and Biostat (https://www.analystsoft.com/en/products/biostat/) software.
2.3. Studying Antiviral Activity
Tomato plants (Solanum lycopersicum) at the growth stage of 3–4 true leaves were infected with the cucumber mosaic virus (CMV) by placing the inoculum onto the upper surface of leaf laminae. A CMV isolate derived from open-ground tomato plants (F1 Adonis bred in Russia) was used as material for infection.
Seven days after treatment with the CMV inoculum, tomato plants were sprayed with a suspension of the strain. Control plants were sprayed with sterile distilled water (K−) and pharmaiodine (Scientific Production Center Farmbiomed, Moscow, Russia) (K+). An aqueous solution of pharmaiodine was prepared immediately before use: 3–5 g/10 L of water. Each experimental and control group consisted of ten plants. The experiment was performed in two replicates. In all the experiments, the plants were watered to the root zone with settled tap water in the morning. Throughout the entire experiment, the plants were subjected to two treatments with bacterial suspensions with a 7-day interval. Three days after the second spraying, immunochromatography tests were conducted to detect infection by the phytovirus using ImmunoStrip CMV (Cucumber Mosaic Virus) Test Kit Flashkits® (Agdia Inc., Elkhart, IN, USA) consisting of a microtiter strip impregnated with an alkaline enzyme and bilaterally coated with anti-phytovirus antibodies and a buffer-filled bag for sample extraction. For diagnostics, a leaf portion (0.15 g) of each plant was placed into the lower part of the bag containing phosphate-buffered saline (0.2 M, pH 7.4), and a plant tissue sample was ground between mesh pads. An immunostrip was submerged into the resulting suspension until the mark “sample” and left in a vertical position for 30 min to further process the analysis results. Staining of the band whose mobility corresponded to the positive control suggested that plants had been infected. Plants infected by the virus were characterized by leaf deformation, a yellow-green mosaic pattern, and significant growth delay.
2.4. Genome Analysis
The strain genome was sequenced and completely assembled [22]. The genome was annotated with the NCBI Prokaryotic Genome Annotation Pipeline (PGAP) version 4.6 [23], Prokka [24], and RAST [25]. The whole-genome tree was built using the TYGS web service [https://tygs.dsmz.de/ (accessed on 1 May 2023)]. The genomic map was built using the Proksee.ca web service (accessed on 1 May 2023) [26]. The average nucleotide identity (ANI) value was calculated using the Ezbiocloud service (accessed on 1 May 2023) [27]; the DNA–DNA hybridization (DDH) value, using the Genome-to-Genome distance calculator (Formula (1) (i.e., GBDP formula d0)) [28]. Chromosome alignment was performed using Mauve software, ver. 2.4.0 (21 December 2014) [29]. Pangenome analysis and searching for unique genes were performed using OrthoVenn service (https://orthovenn3.bioinfotoolkits.net/, accessed on 28 February 2024) [30]. Searching for gene clusters responsible for secondary metabolite production was conducted using the AntiSmash service (https://antismash.secondarymetabolites.org/, accessed on 28 February 2024) [31]. Functional analysis based on metabolic and regulatory pathways was carried out using KEGG (https://www.kegg.jp/blastkoala/, accessed on 28 February 2024) [32]. SNP searches were performed using Nucmer from the Mummer v. 4.0.0 package [33].
3. Results
3.1. Morphological Features of the Strain
The S. carpaticus SCPM-O-B-9993 strain has dark brown aerial mycelium and cherry-red substrate mycelium (Figure S1). The optimal growth temperature is 28 °C; the optimal pH is 7.0–7.1 [22]. Spores are straight or twisted, and short. Spores are oval or globular with a dense shell, sized 0.5–1.0 × 1.0–1.1 µm.
3.2. Evaluation of Productivity as well as the Phytostimulatory and Antiviral Properties of the Strain
During the first 12 h of growth, the strain is in the lag phase when the cells are adapting to the nutrient medium. The cells and mycelium enter the exponential growth phase by 12–24 h. At this time, strain SCPM-O-B-9993 actively produces metabolites, their cells multiply, and their numbers increase to 2.0 × 109 CFU/mL. Starting with 48 h of exposure, the culture begins to enter the stationary growth phase and remains in it for five days. Abundance does not decrease even on day 7. The optical density changes in accordance with strain biomass growth (Figure 1).
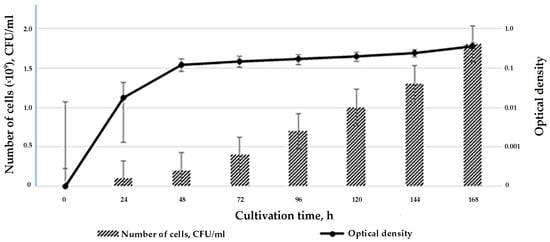
Figure 1.
The growth curve of S. carpaticus strain SCPM-O-B-9993 when cultured on liquid starch–casein medium.
When studying the effect of strain SCPM-O-B-9993 suspension on cress seeds, a toxic effect on germination of 4-day and 7-day cultures and positive control pharmaiodine was detected (Table S1). A toxic effect was observed in germination of less than 30% of seeds. The concentration of cells, spores, mycelium, and the number of metabolites in the suspension cultivated under laboratory conditions in liquid nutrient medium increased with cultivation duration, which increased the toxicity of the suspension. However, it should be taken into account that bacteria entering natural conditions in plant treatments, at a concentration of 109 CFU/mL, are distributed in soil or on plants in lower quantities than when they are under laboratory flask conditions due to lack of substrate, environmental factors, etc. In addition, when microorganisms enter the soil, a microbial pool is formed, which further leaves the number of cells no more than 104 CFU/mL.
The longest root (23.89 mm) was found for treatment with a 3-day suspension of the strain. The cultivation period of the strain influenced cress growth; 1-, 2-, 6-, and 7-day cultures exhibited an inhibitory effect on root and shoot growth. It is quite natural that 6- and 7-day-old cultures inhibited plant growth because of the accumulation of metabolites at this stage.
The data indicate that S. carpaticus strain SCPM-O-B-9993 exhibits an antiviral activity against CMV on tomatoes under laboratory conditions relative to positive and negative controls (Table 1, Figure S2). When comparing the antagonistic properties of the strain suspension by hours of cultivation, it was found that 3-day suspensions of the strain exhibited the maximum antiviral activity: all the treated plants were symptom-free of the virus. Hence, 3-day cultivation of the strain is optimal, since the suspension showed effective phytostimulatory properties, while having an inhibitory effect on CMV. The study of secondary metabolites of the 3-day suspension of the strain showed the presence of alcohols, aldehydes, hydrocarbons, esters, sulfates, and other groups of low-molecular-weight organic compounds with high biological activity [21].

Table 1.
Determination of antiviral activity of the suspension of strain S. carpaticus SCPM-O-B-9993 in laboratory conditions on tomatoes inoculated with CMV (each experimental group included ten plants).
Suspensions of the strain after 24 and 48 h of cultivation were completely ineffective against the phytovirus. It was found that 4-day and 5-day suspensions of the strain continued to exhibit an antiviral activity, but it was insignificantly lower than that of 3-day suspensions. The antiviral activity of the 6- and 7-day suspensions of the strain was approximately 50%. Ara et al. [34] found out that symptoms of the tobacco mosaic virus in Datura plant treated with Streptomyces strains decreased after 7-day incubation due to the effect of bioactive metabolites of the strains.
3.3. Taxonomic Positioning of the Strain SCPM-O-B-9993
The genomes of Streptomyces strains include large unknown BGCs, making them a potential repository for discovering biotechnologically valuable compounds. Diverse molecules exhibiting an antibacterial activity encoded in the genome exist in the “silent” repressed state. Therefore, there arises a need for using genome editing and metagenomic analysis methods to identify new biosynthetic clusters of antibiotics and alter expression of the respective genes, which would potentially enable synthesis of novel molecules exhibiting an antibacterial activity [35].
Strain SCPM-O-B-9993 is the only representative of the S. carpaticus species whose genome has been completely sequenced [22]. The genome assembly consisted of a 5,968,715 bp long single linear chromosome with 72.84% GC composition (Figure 2). No plasmids were detected. During genome annotation and analysis, 5206 protein-coding sequences, 60 tRNA sequences, 15 rRNA sequences (5–5S, 5–16S, and 5–23S), and eight CRISPR loci were identified.
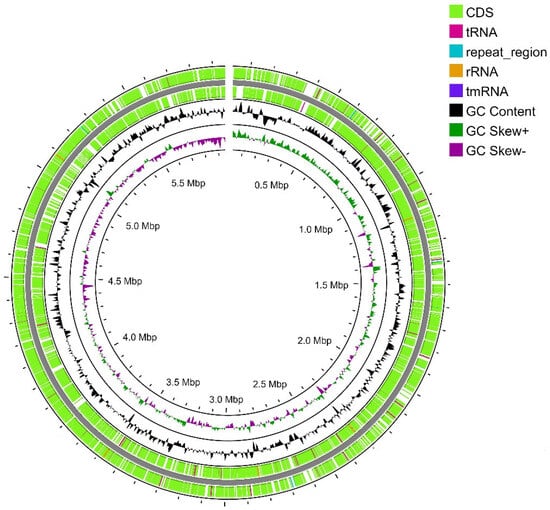
Figure 2.
Map of the linear (gap on the top of the figure) chromosome of strain SCPM-O-B-9993. From outside to the center: all CDS and RNA genes on the forward strand, all CDS and RNA genes on the reverse strand, GC content, and GC skew.
A BLAST search for housekeeping gene sequences (gyrB, rpoB, trpB, recA) was performed to determine its closest relatives among Streptomyces strains with completely sequenced genomes (Table S2). Streptomyces harbinensis NA02264, Streptomyces xiamenensis 318 and Streptomyces sp. XC2026 were found to be most closely related to strain SCPM-O-B-9993 (Table 2).

Table 2.
Genome parameters of strain SCPM-O-B-9993 and its closest relatives.
Strains SCPM-O-B-9993 and NA02264 are deposited in the Genbank database as representatives of different species. However, the ANI and DDH values show that these strains are very likely to belong to the same species (Table 3). Hence, the threshold for species assignment by DDH is 70%, while strains SCPM-O-B-9993 and NA02264 have DDH values >90%.

Table 3.
The ANI and DDH values for strain SCPM-O-B-9993 and its closest relatives.
The genome of the type strain S. carpaticus has not been sequenced yet (July 2023). However, the genome of the type strain S. harbinensis CGMCC4.7047T (FPAB000000000000.1) has been sequenced, which makes it possible to calculate the main parameters of species membership (Table 4).

Table 4.
The ANI and DDH values for strains SCPM-O-B-9993 and Streptomyces harbinensis NA02264 compared to those of the type strain of S. harbinensis.
Considering that the ANI and DDH values of strain SCPM-O-B-9993 compared to the type strain of S. harbinensis exceed the thresholds for species assignment (ANI > 95% and DDH > 70%), we assume that strain SCPM-O-B-9993 is probably a representative of the species S. harbinensis, but the final taxonomic position of the strain will be possible only after the genome sequence of the type strain of S. carpaticus appears in the Genbank database.
The phylogenetic tree (Figure 3) demonstrates that the strain SCPM-O-B-9993 is also close to S. harbinensis (Streptomyces gingkonis species is not validated, so we do not consider it).
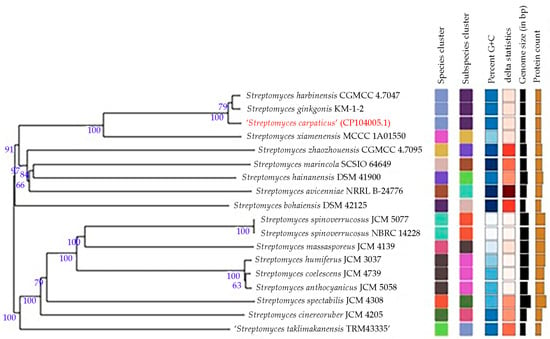
Figure 3.
Whole-genome tree demonstrating the position of strain SCPM-O-B-9993 within the genus Streptomyces.
Alignment of the genomes of Streptomyces carpaticus strain SCPM-O-B-9993 and Streptomyces harbinensis strain NA02264 (Figure 4) relative to each other shows that the main blocks (marked with one color) retain a similar arrangement on the chromosomes and in general, the structures of the genomes are very similar. Only a few displacements of gene blocks can be noted (marked with vertical bars).
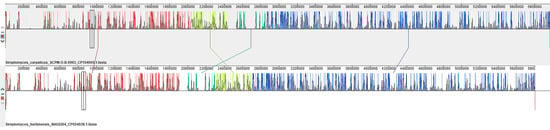
Figure 4.
Mauve visualization of locally collinear blocks identified between chromosomes of Streptomyces carpaticus SCPM-O-B-9993 and Streptomyces harbinensis NA02264. Vertical bars demarcate interchromosomal boundaries.
In the S. harbinensis NA02264 genome relative to the S. carpaticus SCPM-O-B-9993 genome, 20,153 single nucleotide substitutions (SNPs) were found, which are evenly dispersed throughout the genome. Single nucleotide substitutions accounted for 0.34% of the total genome length.
3.4. Analysis of the Genome of Streptomyces carpaticus Strain SCPM-O-B-9993 and Its Closest Relatives
We analyzed the genome of the strains listed in Table 1 as the closest relatives of strain SCPM-O-B-9993 (Figure 5A), and separately, the pangenome of the pair Streptomyces carpaticus SCPM-O-B-9993 and Streptomyces harbinensis NA02264 (Figure 5B) as the strains closest to each other among the Streptomyces whose genomes were sequenced. It was of interest to identify whether these two strains, whose genomes are nearly identical in terms of structure, have genes that are unique relative to each other.
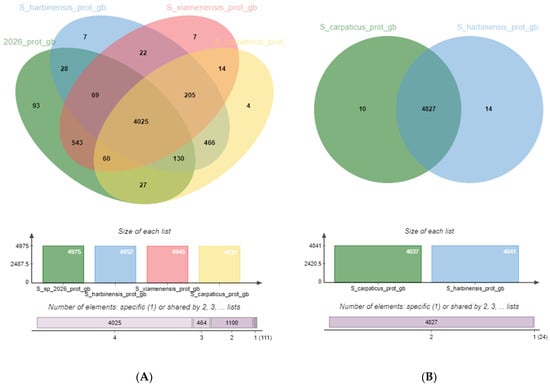
Figure 5.
Venn diagram depicting the genome of S. carpaticus SCPM-O-B-9993 and its relatives: (A) Streptomyces harbinensis NA02264, Streptomyces xiamenensis 318, Streptomyces sp. XC2026; (B) Streptomyces harbinensis NA02264 as the closest relative.
The genome of four Streptomyces strains is represented by 5700 orthologous clusters (OCs) (clusters of genes in different species that originated by vertical descent from a single gene in the last common ancestor), of which 4025 (70.6%) are core. Expectedly, the SCPM-O-B-9993/NA02264 pair had the maximum (of all pairs with strain SCPM-O-B-9993) number of OCs unique to the pair: 466 are characteristic only of this pair and absent in the other strains (Table S3).
The genome of the Streptomyces carpaticus SCPM-O-B-9993 and Streptomyces harbinensis NA02264 pair is represented by 4851 clusters, of which 4827 (99.4%) are core. Despite the close relatedness, each strain has unique CDSs, but most of them cannot be classified at this time.
3.5. Functional Annotation of Streptomyces carpaticus Strain SCPM-O-B-9993
The genome of strain SCPM-O-B-9993 contains 5331 coding sequences, of which 2139 (40.1%) are functionally annotated (Figure 6).
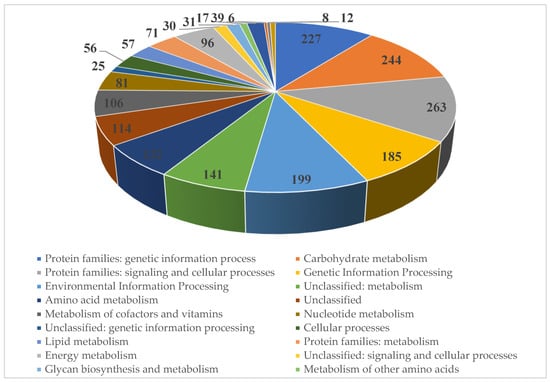
Figure 6.
KEGG function classification of the Streptomyces carpaticus strain SCPM-O-B-9993.
Since the strain SCPM-O-B-9993 is a biotechnologically promising producer of secondary metabolites, the genetic organization of secondary metabolite production clusters was analyzed.
Antibiotic biosynthesis by microorganisms plays an adaptive role; it is an inherited feature of microorganisms and is regulated by specialized genes. 3-Amino-5-hydroxybenzoic acid (AHBA) is the starting unit in the biosynthesis of ansamycin antibiotics by Streptomyces bacteria [36]. The genome of strain SCPM-O-B-9993 contains a sequence of genes whose products control the pathway of AHBA biosynthesis from UDP glucose, consisting of ten reactions. The terpenoid biosynthesis pathway in the genome of strain SCPM-O-B-9993 involves production of key metabolites such as isopentenyl pyrophosphate, geranyl-PP, farnesyl-PP, and geranyl-geranyl-PP, which are precursors of many secondary metabolites in Streptomyces.
We found the following highly conserved secondary metabolite production clusters in the strain genome (>80% structural similarity to similar clusters in other strains) (Table 5).

Table 5.
Production clusters of secondary metabolites in strain SCPM-O-B-9993.
The organization of the coelibactin gene cluster in strain SCPM-O-B-9993 is completely identical to that of S. harbinensis (Figure S3e). Differences between them are present mainly at the level of single nucleotide substitutions (SNPs), except for a few extended regions (Table S4).
The antimicrobial properties of the strain may be due to production of ohmyungsamycin, pellasoren, and naringenin. Ohmyungsamycins are cyclic peptides first isolated from marine Streptomyces [37,38]. They are synthesized by a non-ribosomal peptide synthetase. Kim et al. [39] reported their activity against Mycobacterium tuberculosis and cancer cells in humans, with OMS A showing greater activity than OMS B. Considering that strain SCPM-O-B-9993 possesses the ohmyungsamycin BGC, it can be considered promising in medicine as well. There are some structural differences among the ohmyungsamycin BGC in SPM-O-B-9993 and NA02264 strains. In the NA02264 genome, in this region there resides (there are no in SCPM-O-B-9993) IS110 family transposase and a region embedded between the hypothetical protein and α/β hydrolase, having a length of 8393 bp and containing several genes both annotated (tyrosine-type recombinase, NAD-dependent epimerase/dehydratase family protein) and hypothetical.
Previously, naringenin biosynthesis was believed to be characteristic only of plants [40,41]. Naringenin is a flavonoid whose biosynthesis has been repeatedly reported in citrus trees (lemons, oranges, etc) and tomatoes [42]. Álvarez-Álvarez et al. [43] first showed its biosynthesis in Streptomyces clavuligerus. Naringenin exhibits anti-inflammatory, chemoprotective, and antitumor properties [44,45]. The naringenin BGC is completely structurally similar to that of the strain Streptomyces narbonensis NA02264.
Sorangium cellulosum is the best-known producer of pellasoren [46]. However, the gene cluster for biosynthesis of this compound is found in the genomes of Streptomyces species, in particular in the type strain S. harbinensis (Figure S3). Nothing is known about the antimicrobial properties of pellasoren, this compound was reported to exhibit cytotoxic effects against cancer cells [47,48]. Pellasorene BGC in the SCPM-O-B-9993 and NA02264 strains have the following structural difference: the type I polyketide synthase gene is present between fatty acyl-AMP ligase and SDR family NAD(P)-dependent oxidoreductase genes in the SCPM-O-B-9993 strain, having coordinates 543.262–544.999 (1737 bp). The NA02264 strain does not contain it.
The detected BGCs in SCPM-O-B-9993 strain have a strong potential for being used in plant protection, since they exhibit antimicrobial properties.
Investigation of the component composition of suspension and extracts (water-alcohol, methanol, and hexane) of S. carpaticus strain SCPM-O-B-9993 showed the presence of secondary metabolites: alcohols, aldehydes, hydrocarbons, esters, sulfates, and other groups of low-molecular-weight organic compounds (LMCs). Alcohols and esters prevailed among LMCs for all the extraction variants. The identified LMCs have valuable properties from the agricultural point of view: antiviral, antimicrobial and antitumor (ethyl 5-(pyridin-4-yl)-1H-pyrazol-3-carboxylate); bactericidal, fungicidal, and antiseptic properties (1,2-hexanediol); and insectoacaricidal ones (isopropyl myristate). 1-Dodecanol is a component of pheromones, sex attractants and surfactants for controlling insect pests [21].
Annotation of the Streptomyces genomes shows that different BGCs are present. Thus, the genome of the Streptomyces tendae strain UTMC 3329 shows the presence of clusters of the genes encoding polyketides, ribosomally and non-ribosomally synthesized peptides. Various genes have been discovered in the xenobiotic degradation pathway and heavy metal resistance [49]. An analysis of S. avermitilis genome revealed the feasibility of polyketide synthesis [50]. Genome mining and manipulations with the genome, and with antibiotic BGC in particular, have a significant potential for identifying novel molecules exhibiting an antibacterial activity [51].
4. Conclusions
The studies revealed clusters of biosynthesis of secondary metabolites of the strain SCPM-O-B-9993: terpenoids and ansamycin antibiotics. Under laboratory conditions, the strain exhibited phytostimulatory and antiviral properties. The most effective cultivation period of the strain was 72 h, during which the culture went from exponential to stationary growth phase with the production of a spectrum of bioactive metabolites.
There are alternatives to genome analysis, such as metabolomic analysis (coupled to dereplication of known compounds) to assess the presence of potentially new compounds in any Streptomyces culture. The Streptomyces carpaticus SCPM-OB-9993 strain is a biotechnologically promising producer of secondary metabolites exhibiting antiviral and phytostimulatory properties.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biology13060388/s1, Figure S1. Emergence of colonies of S. carpaticus strain SCPM-O-B-9993 on potato agar on cultivation day 14; Table S1. Phytotoxicity of S. carpaticus strain SCPM-O-B-9993; Figure S2. Manifestation of symptoms of the cucumber mosaic virus on day 7 of exposure after the first treatment: (A) control; (B) the experiment (exposure to the strain); Table S2. Search for strains most related to strain SCPM-O-B-9993 using marker genes (housekeeping genes); Table S3. Distribution of coding sequences unique to the SCPM-O-B-9993/NA02264 pair by biological processes; Figure S3. Schemes of genetic organization of highly conserved clusters of secondary metabolite biosynthesis; Table S4. Regions in conserved clusters of secondary metabolite biosynthesis containing extended mismatched regions.
Author Contributions
Conceptualization, Y.B.; methodology, Y.B., Y.D. and L.G.; software, Y.D.; validation, Y.B. and A.B.; formal analysis, Y.D. and A.B.; investigation, L.S. and L.G.; resources, A.B.; data curation, Y.B.; writing—original draft preparation, Y.D. and Y.B.; writing—review and editing, A.B.; visualization, Y.D.; supervision, Y.B.; project administration, Y.B. and L.G.; funding acquisition, A.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Ministry of Science and Higher Education of the Russian Federation, grant number 075-15-2019-1671 (agreement dated 31 October 2019).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data on genome sequence of the strain S. carpaticus strain SCPM-O-B-9993 can be found in the Genbank database under the accession number CP104005.1 (BioProject PRJNA269675, BioSample SAMN30493425).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Chang, T.-L.; Huang, T.-W.; Wang, Y.-X.; Liu, C.-P.; Kirby, R.; Chu, C.-M.; Huang, C.-H. An actinobacterial isolate, Streptomyces sp. YX44, produces broad-spectrum antibiotics that strongly inhibit Staphylococcus aureus. Microorganisms 2021, 9, 630. [Google Scholar] [CrossRef] [PubMed]
- Veselá, A.B.; Pelantová, H.; Šulc, M.; Macková, M.; Lovecká, P.; Thimová, M.; Pasquarelli, F.; Pičmanová, M.; Pátek, M.; Bhalla, T.C.; et al. Biotransformation of benzonitrile herbicides via the nitrile hydratase–amidase pathway in rhodococci. J. Ind. Microbiol. Biotechnol. 2012, 39, 1811–1819. [Google Scholar] [CrossRef] [PubMed]
- Alam, K.; Mazumder, A.; Sikdar, S.; Zhao, Y.M.; Hao, J.; Song, C.; Wang, Y.; Sarkar, R.; Islam, S.; Zhang, Y.; et al. Streptomyces: The biofactory of secondary metabolites. Front. Microbiol. 2022, 13, 968053. [Google Scholar] [CrossRef] [PubMed]
- Khadayat, K.; Sherpa, D.D.; Malla, K.P.; Shrestha, S.; Rana, N.; Marasini, B.P.; Khanal, S.; Rayamajhee, B.; Bhattarai, B.R.; Parajuli, N. Molecular identification and antimicrobial potential of Streptomyces species from nepalese soil. Int. J. Microbiol. 2020, 2020, 8817467. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.; Hwang, S.; Kim, J.; Cho, S.; Palsson, B.; Cho, B.-K. Mini review: Genome mining approaches for the identification of secondary metabolite biosynthetic gene clusters in Streptomyces. Comput. Struct. Biotechnol. J. 2020, 18, 1548–1556. [Google Scholar] [CrossRef] [PubMed]
- Lapaz, M.I.; Cisneros, E.J.; Pianzzola, M.J.; Francis, I.M. Exploring the exceptional properties of Streptomyces: A hands-on discovery of natural products. Am. Biol. Teach. 2019, 81, 658–664. [Google Scholar] [CrossRef]
- Tarkka, M.; Hampp, R. Secondary Metabolites of Soil Streptomycetes in Biotic Interactions; Springer: Berlin/Heidelberg, Germany, 2008; pp. 107–126. [Google Scholar] [CrossRef]
- Xia, H.; Li, X.; Li, Z.; Zhan, X.; Mao, X.; Li, Y. The Application of regulatory cascades in Streptomyces: Yield enhancement and metabolite mining. Front. Microbiol. 2020, 11, 508962. [Google Scholar] [CrossRef]
- Shirokikh, I.G.; Ashikhmina, T.Y. Actinobacteria in protecting the environment from industrial pollution. Theor. Appl. Ecol. 2022, 4, 14–21. [Google Scholar] [CrossRef]
- Hamedi, J.; Mohammadipanah, F. Biotechnological application and taxonomical distribution of plant growth promoting actinobacteria. J. Ind. Microbiol. Biotechnol. 2015, 42, 157–171. [Google Scholar] [CrossRef]
- Vurukonda, S.S.; Giovanardi, D.; Stefani, E. Plant growth promoting and biocontrol activity of Streptomyces spp. as endophytes. Int. J. Mol. Sci. 2018, 19, 952. [Google Scholar] [CrossRef]
- Wright, G.D. Environmental and clinical antibiotic resistomes, same only different. Curr. Opin. Microbiol. 2019, 51, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Belknap, K.C.; Park, C.J.; Barth, B.M.; Andam, C.P. Genome mining of biosynthetic and chemotherapeutic gene clusters in Streptomyces bacteria. Sci. Rep. 2020, 10, 2003. [Google Scholar] [CrossRef] [PubMed]
- Klassen, J.L.; Currie, C.R. Gene fragmentation in bacterial draft genomes: Extent, consequences and mitigation. BMC Genom. 2012, 13, 14. [Google Scholar] [CrossRef] [PubMed]
- Semenkov, I.N.; Shelyakin, P.V.; Nikolaeva, D.D.; Tutukina, M.N.; Sharapova, A.V.; Lednev, S.A.; Sarana, Y.V.; Gelfand, M.S.; Krechetov, P.P.; Koroleva, T.V. Data on the temporal changes in soil properties and microbiome composition after a jet-fuel contamination during the pot and field experiments. Data Brief 2023, 46, 108860. [Google Scholar] [CrossRef]
- Kalkreuter, E.; Pan, G.; Cepeda, A.J.; Shen, B. Targeting bacterial genomes for natural product discovery. Trends Pharmacol. Sci. 2020, 41, 13–26. [Google Scholar] [CrossRef] [PubMed]
- Caicedo-Montoya, C.; Manzo-Ruiz, M.; Ríos-Estepa, R. Pan-Genome of the genus Streptomyces and prioritization of biosynthetic gene clusters with potential to produce antibiotic compounds. Front. Microbiol. 2021, 12, 677558. [Google Scholar] [CrossRef] [PubMed]
- Salas, J.A.; Méndez, C. Indolocarbazole antitumour compounds by combinatorial biosynthesis. Curr. Opin. Chem. Biol. 2009, 13, 152–160. [Google Scholar] [CrossRef] [PubMed]
- Ostash, B.; Doud, E.H.; Lin, C.; Ostash, I.; Perlstein, D.L.; Fuse, S.; Wolpert, M.; Kahne, D.; Walker, S. Complete characterization of the seventeen step moenomycin biosynthetic pathway. Biochemistry 2009, 48, 8830–8841. [Google Scholar] [CrossRef] [PubMed]
- Song, J.Y.; Jeong, H.; Yu, D.S.; Fischbach, M.A.; Park, H.-S.; Kim, J.J.; Seo, J.-S.; Jensen, S.E.; Oh, T.K.; Lee, K.J.; et al. Draft genome sequence of Streptomyces clavuligerus NRRL 3585, a producer of diverse secondary metabolites. J. Bacteriol. 2010, 192, 6317–6318. [Google Scholar] [CrossRef]
- Bataeva, Y.V.; Grigoryan, L.N.; Kurashov, E.A.; Krylova, J.V.; Fedorova, E.V.; Iavid, E.J.; Khodonovich, V.V.; Yakovleva, L.V. Study of metabolites of Streptomyces carpaticus RCAM04697 for the creation of environmentally friendly plant protection products. Theor. Appl. Ecol. 2021, 3, 172–178. [Google Scholar] [CrossRef]
- Bataeva, Y.V.; Grigoryan, L.N.; Bogun, A.G.; Kislichkina, A.A.; Platonov, M.E.; Kurashov, E.A.; Krylova, J.V.; Fedorenko, A.G.; Andreeva, M.P. Biological Activity and Composition of Metabolites of Potential Agricultural Application from Streptomyces carpaticus K-11 RCAM04697 (SCPM-O-B-9993). Microbiology 2023, 92, 459–467. [Google Scholar] [CrossRef]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.-H.; Ha, S.; Lim, J.; Kwon, S.; Chun, J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 2017, 110, 1281–1286. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Darling, A.C.E.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Dong, Z.; Fang, L.; Luo, Y.; Wei, Z.; Guo, H.; Zhang, G.; Gu, Y.Q.; Coleman-Derr, D.; Xia, Q.; et al. OrthoVenn2: A web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 2019, 47, W52–W58. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef]
- Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Marçais, G.; Delcher, A.L.; Phillippy, A.M.; Coston, R.; Salzberg, S.L.; Zimin, A. MUMmer4: A fast and versatile genome alignment system. PLoS Comput. Biol. 2018, 14, e1005944. [Google Scholar] [CrossRef] [PubMed]
- Ara, I.; Bukhari, N.A.; Aref, N.M.; Shinwari, M.A.; Bakir, M.A. Antiviral activities of streptomycetes against tobacco mosaic virus (TMV) in Datura plant: Evaluation of different organic compounds in their metabolites. Afr. J. Biotechnol. 2012, 11, 2130–2138. [Google Scholar] [CrossRef]
- Onaka, H. Novel antibiotic screening methods to awaken silent or cryptic secondary metabolic pathways in actinomycetes. J. Antibiot. 2017, 70, 865–870. [Google Scholar] [CrossRef]
- Kim, C.-G.; Yu, T.-W.; Fryhle, C.B.; Handa, S.; Floss, H.G. 3-Amino-5-hydroxybenzoic Acid Synthase, the Terminal Enzyme in the Formation of the Precursor of mC7N Units in Rifamycin and Related Antibiotics. J. Biol. Chem. 1998, 273, 6030–6040. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.S.; Shin, Y.-H.; Lee, H.-M.; Kim, J.K.; Choe, J.H.; Jang, J.-C.; Um, S.; Jin, H.S.; Komatsu, M.; Cha, G.-H.; et al. Ohmyungsamycins promote antimicrobial responses through autophagy activation via AMP-activated protein kinase pathway. Sci. Rep. 2017, 7, 3431. [Google Scholar] [CrossRef] [PubMed]
- Um, S.; Choi, T.J.; Kim, H.; Kim, B.Y.; Kim, S.-H.; Lee, S.K.; Oh, K.-B.; Shin, J.; Oh, D.-C. Ohmyungsamycins A and B: Cytotoxic and antimicrobial cyclic peptides produced by Streptomyces sp. from a volcanic island. J. Org. Chem. 2013, 78, 12321–12329. [Google Scholar] [CrossRef]
- Kim, E.; Du, Y.E.; Ban, Y.H.; Shin, Y.-H.; Oh, D.-C.; Yoon, Y.J. Enhanced ohmyungsamycin a production via adenylation domain engineering and optimization of culture conditions. Front. Microbiol. 2021, 12, 626881. [Google Scholar] [CrossRef]
- An, J.; Kim, S.H.; Bahk, S.; Vuong, U.T.; Nguyen, N.T.; Do, H.L.; Kim, S.H.; Chung, W.S. Naringenin induces pathogen resistance against Pseudomonas syringae through the activation of NPR1 in Arabidopsis. Front. Plant Sci. 2021, 12, 672552. [Google Scholar] [CrossRef]
- Din, S.; Hamid, S.; Yaseen, A.; Yatoo, A.M.; Ali, S.; Shamim, K.; Mahdi, W.A.; Alshehri, S.; Rehman, M.U.; Shah, W.A. Isolation and characterization of flavonoid naringenin and evaluation of cytotoxic and biological efficacy of water lilly (Nymphaea mexicana Zucc.). Plants 2022, 11, 3588. [Google Scholar] [CrossRef]
- Wu, L.-H.; Lin, C.; Lin, H.-Y.; Liu, Y.-S.; Wu, C.Y.-J.; Tsai, C.-F.; Chang, P.-C.; Yeh, W.-L.; Lu, D.-Y. Naringenin suppresses neuroinflammatory responses through inducing suppressor of cytokine signaling 3 expression. Mol. Neurobiol. 2016, 53, 1080–1091. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Álvarez, R.; Botas, A.; Albillos, S.M.; Rumbero, A.; Martín, J.F.; Liras, P. Molecular genetics of naringenin biosynthesis, a typical plant secondary metabolite produced by Streptomyces clavuligerus. Microb. Cell Factories 2015, 14, 178. [Google Scholar] [CrossRef] [PubMed]
- Cavia-Saiz, M.; Busto, M.D.; Pilar-Izquierdo, M.C.; Ortega, N.; Perez-Mateos, M.; Muñiz, P. Antioxidant properties, radical scavenging activity and biomolecule protection capacity of flavonoid naringenin and its glycoside naringin: A comparative study. J. Sci. Food Agric. 2010, 90, 1238–1244. [Google Scholar] [CrossRef] [PubMed]
- Jagetia, A.; Jagetia, G.C.; Jha, S. Naringin, a grapefruit flavanone, protects V79 cells against the bleomycin-induced genotoxicity and decline in survival. J. Appl. Toxicol. 2007, 27, 122–132. [Google Scholar] [CrossRef] [PubMed]
- Jahns, C.; Hoffmann, T.; Müller, S.; Gerth, K.; Washausen, P.; Höfle, G.; Reichenbach, H.; Kalesse, M.; Müller, R. Pellasoren: Structure elucidation, biosynthesis, and total synthesis of a cytotoxic secondary metabolite from Sorangium cellulosum. Angew. Chem. Int. Ed. 2012, 51, 5239–5243. [Google Scholar] [CrossRef] [PubMed]
- Abbassi-Ghanavati, M.; Alexander, J.; McIntire, D.; Savani, R.; Leveno, K. Neonatal effects of magnesium sulfate given to the mother. Am. J. Perinatol. 2012, 29, 795–800. [Google Scholar] [CrossRef]
- Jaita, S.; Phakhodee, W.; Chairungsi, N.; Pattarawarapan, M. Mechanochemical synthesis of primary amides from carboxylic acids using TCT/NH4SCN. Tetrahedron Lett. 2018, 59, 3571–3573. [Google Scholar] [CrossRef]
- Eftekharivash, L.; Hamedi, J. Genome sequence and annotation of Streptomyces tendae UTMC 3329, acid and alkaline tolerant actinobacterium. Iran. J. Microbiol 2020, 12, 343–352. [Google Scholar] [CrossRef] [PubMed]
- Korolev, S.A.; Zverkov, O.A.; Seliverstov, A.V.; Lyubetsky, V.A. Ribosome reinitiation at leader peptides increases translation of bacterial proteins. Biol. Direct 2016, 11, 20. [Google Scholar] [CrossRef]
- Ziemert, N.; Jensen, P.R. Phylogenetic approaches to natural product structure prediction. Meth Enzym. 2012, 517, 161–182. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).