Role of TFEB in Huntington’s Disease
Abstract
Simple Summary
Abstract
1. Huntington’s Disease
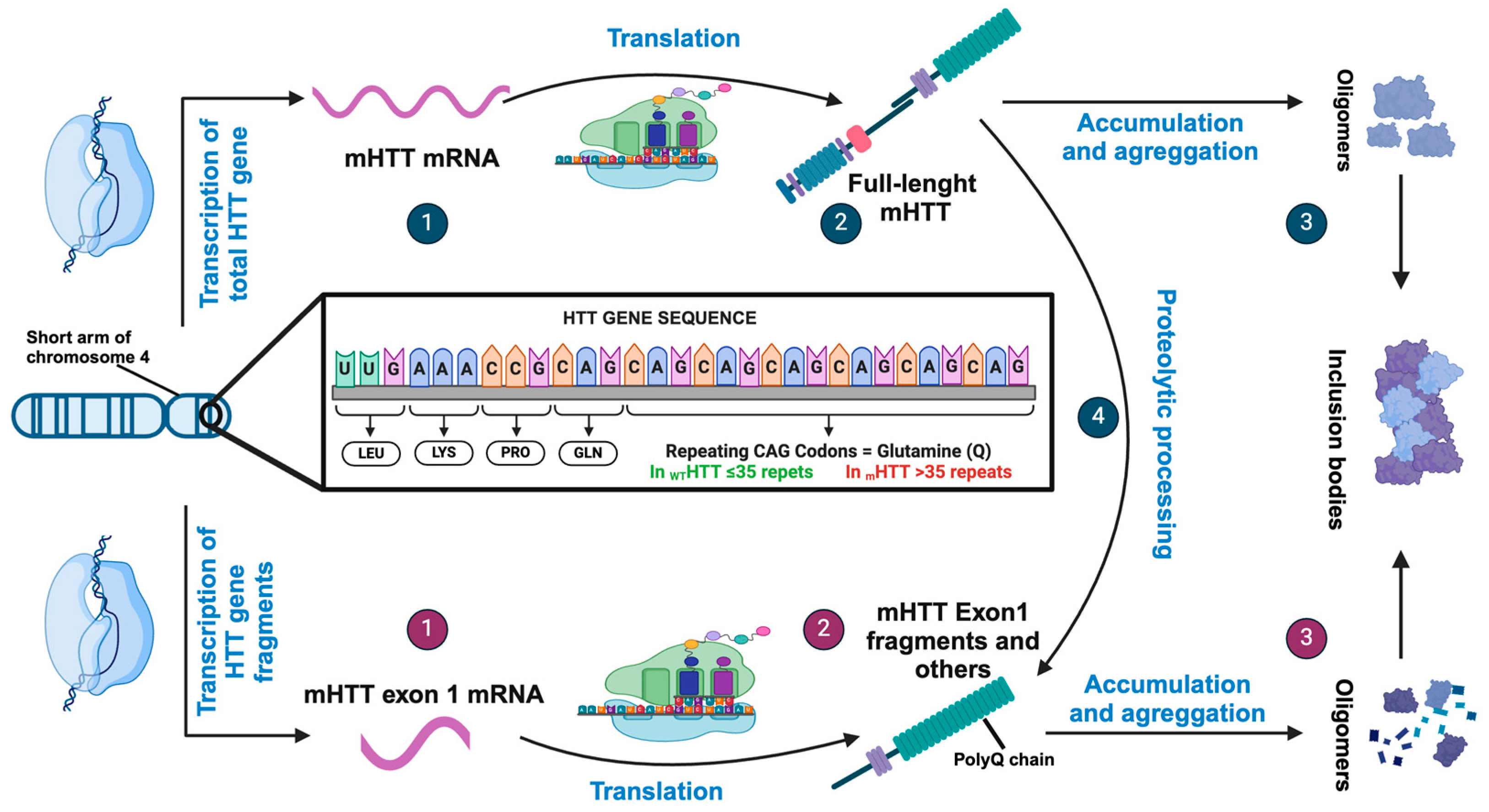
HTT and mHTT
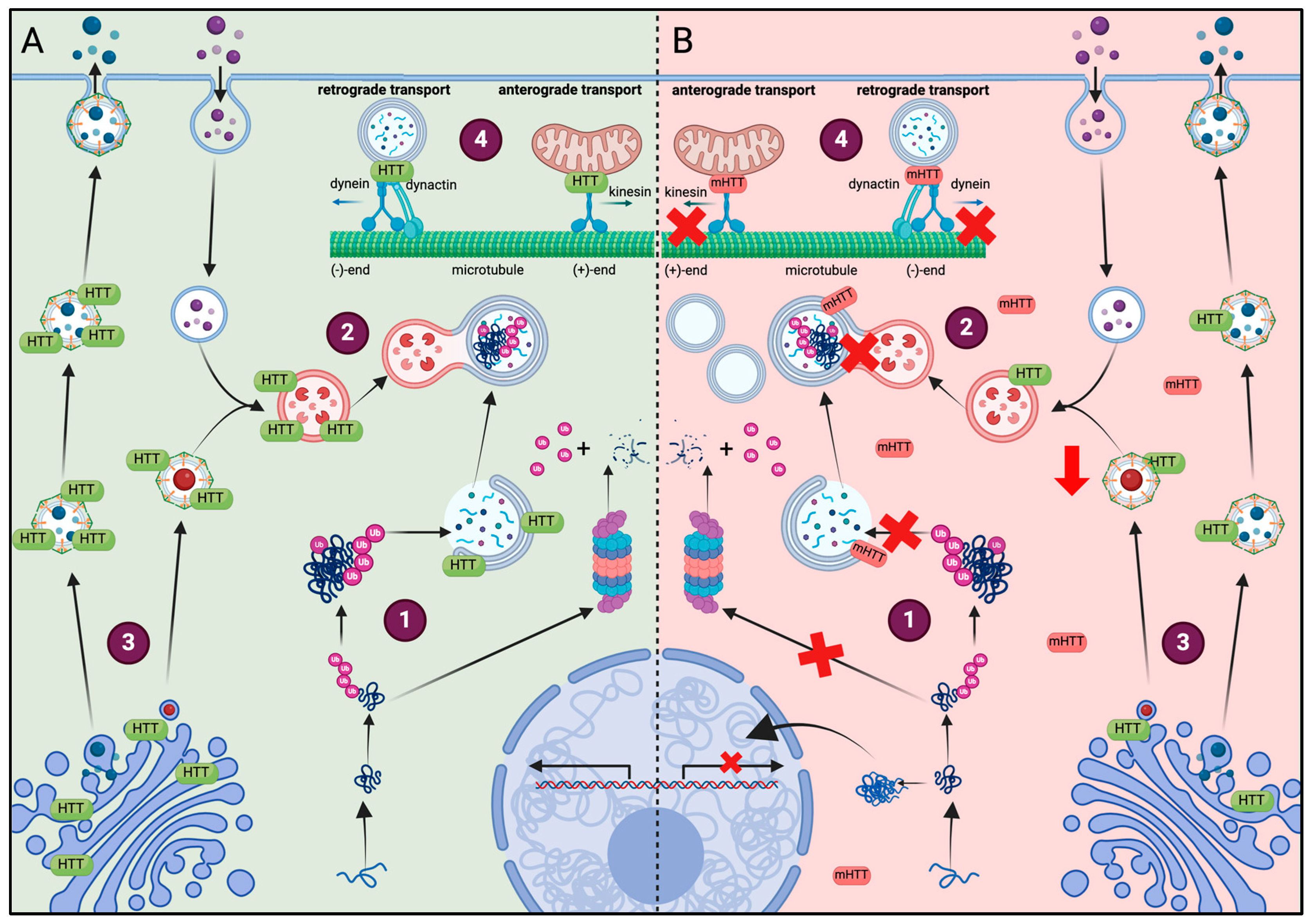
2. Alteration in HD
3. TFEB
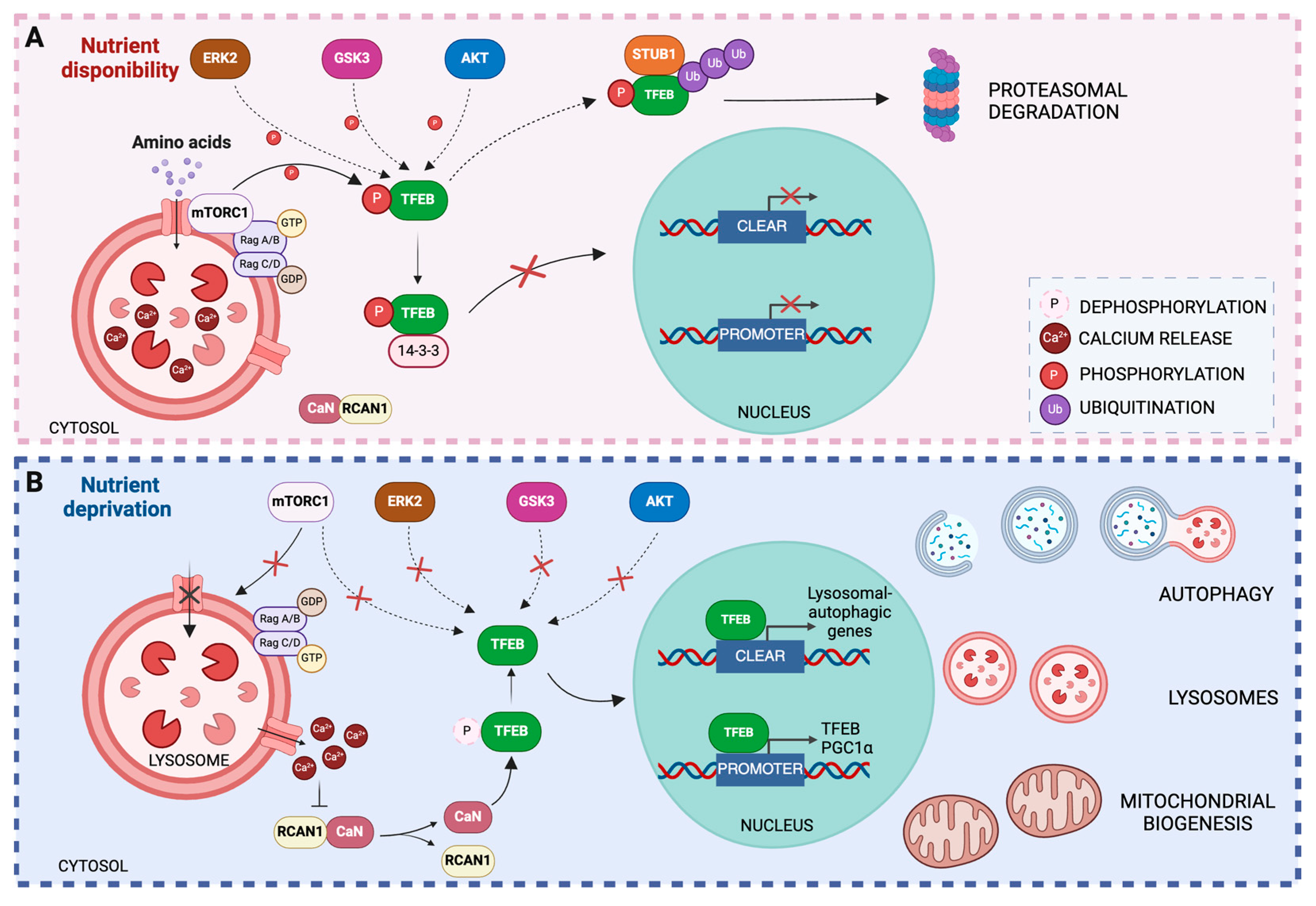
3.1. Regulation of TFEB Activity
3.2. TFEB in Huntington’s Disease
4. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- MacDonald, M.E.; Ambrose, C.M.; Duyao, M.P.; Myers, R.H.; Lin, C.; Srinidhi, L.; Barnes, G.; Taylor, S.A.; James, M.; Groot, N.; et al. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell 1993, 72, 971–983. [Google Scholar] [CrossRef] [PubMed]
- Saudou, F.; Humbert, S. The Biology of Huntingtin. Neuron 2016, 89, 910–926. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Feigin, A. Huntington’s Disease: New Frontiers in Therapeutics. Curr. Neurol. Neurosci. Rep. 2021, 21, 10. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Clabough, E.B.D.; Sarkar, S.; Futter, M.; Rubinsztein, D.C.; Zeitlin, S.O. Deletion of the huntingtin polyglutamine stretch enhances neuronal autophagy and longevity in mice. PLoS Genet. 2010, 6, e1000838. [Google Scholar] [CrossRef] [PubMed]
- Sanchez Mejia, R.O.; Friedlander, R.M. Caspases in Huntington’s disease. Neuroscientist 2001, 7, 480–489. [Google Scholar] [CrossRef]
- Yushchenko, T.; Deuerling, E.; Hauser, K. Insights into the Aggregation Mechanism of PolyQ Proteins with Different Glutamine Repeat Lengths. Biophys. J. 2018, 114, 1847–1857. [Google Scholar] [CrossRef] [PubMed]
- Christodoulou, C.C.; Papanicolaou, E.Z. Integrated Bioinformatics Analysis of Shared Genes, miRNA, Biological Pathways and Their Potential Role as Therapeutic Targets in Huntington’s Disease Stages. Int. J. Mol. Sci. 2023, 24, 4873. [Google Scholar] [CrossRef] [PubMed]
- Greco, T.M.; Secker, C.; Ramos, E.S.; Federspiel, J.D.; Liu, J.P.; Perez, A.M.; Al-Ramahi, I.; Cantle, J.P.; Carroll, J.B.; Botas, J.; et al. Dynamics of huntingtin protein interactions in the striatum identifies candidate modifiers of Huntington disease. Cell Syst. 2022, 13, 304–320.e5. [Google Scholar] [CrossRef] [PubMed]
- Cicchetti, F.; Lacroix, S.; Cisbani, G.; Vallières, N.; Saint-Pierre, M.; St-Amour, I.; Tolouei, R.; Skepper, J.N.; Hauser, R.A.; Mantovani, D.; et al. Mutant huntingtin is present in neuronal grafts in Huntington disease patients. Ann. Neurol. 2014, 76, 31–42. [Google Scholar] [CrossRef]
- Luo, F.; Sandhu, A.F.; Rungratanawanich, W.; Williams, G.E.; Akbar, M.; Zhou, S.; Song, B.J.; Wang, X. Melatonin and autophagy in aging-related neurodegenerative diseases. Int. J. Mol. Sci. 2020, 21, 7174. [Google Scholar] [CrossRef]
- Kim, A.; Lalonde, K.; Truesdell, A.; Welter, P.G.; Brocardo, P.S.; Rosenstock, T.R.; Gil-mohapel, J. New Avenues for the Treatment of Huntington’s Disease. Int. J. Mol. Sci. 2021, 22, 8363. [Google Scholar] [CrossRef] [PubMed]
- Croce, K.R.; Yamamoto, A. A role for autophagy in Huntington’s disease. Neurobiol. Dis. 2019, 122, 16–22. [Google Scholar] [CrossRef]
- Ha, A.D.; Fung, V.S.C. Huntington’s disease. Curr. Opin. Neurol. 2012, 25, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Koyuncu, S.; Fatima, A.; Gutierrez-Garcia, R.; Vilchez, D. Proteostasis of huntingtin in health and disease. Int. J. Mol. Sci. 2017, 18, 1568. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Taguchi, K.; Tanaka, M. Ubiquitin, Autophagy and Neurodegenerative Diseases. Cells 2020, 9, 2022. [Google Scholar] [CrossRef]
- Martin, D.D.O.; Heit, R.J.; Yap, M.C.; Davidson, M.W.; Hayden, M.R.; Berthiaume, L.G. Identification of a post-translationally myristoylated autophagy-inducing domain released by caspase cleavage of huntingtin. Hum. Mol. Genet. 2014, 23, 3166–3179. [Google Scholar] [CrossRef]
- Martinez-Vicente, M.; Talloczy, Z.; Wong, E.; Tang, G.; Koga, H.; Kaushik, S.; De Vries, R.; Arias, E.; Harris, S.; Sulzer, D.; et al. Cargo recognition failure is responsible for inefficient autophagy in huntington’s disease. Nat. Neurosci. 2010, 13, 567. [Google Scholar] [CrossRef] [PubMed]
- Hipp, M.S.; Patel, C.N.; Bersuker, K.; Riley, B.E.; Kaiser, S.E.; Shaler, T.A.; Brandeis, M.; Kopito, R.R. Indirect inhibition of 26S proteasome activity in a cellular model of Huntington’s disease. J. Cell Biol. 2012, 196, 573–587. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Tsujimura, A.; Taguchi, K.; Tanaka, M. HSF1 stress response pathway regulates autophagy receptor SQSTM1/p62-associated proteostasis. Autophagy 2017, 13, 133–148. [Google Scholar] [CrossRef]
- Jing, L.; Cheng, S.; Pan, Y.; Liu, Q.; Yang, W.; Li, S.; Li, X.J. Accumulation of Endogenous Mutant Huntingtin in Astrocytes Exacerbates Neuropathology of Huntington Disease in Mice. Mol. Neurobiol. 2021, 58, 5112–5126. [Google Scholar] [CrossRef]
- Yang, H.; Yang, S.; Jing, L.; Huang, L.; Chen, L.; Zhao, X.; Yang, W.; Pan, Y.; Yin, P.; Qin, Z.S.; et al. Truncation of mutant huntingtin in knock-in mice demonstrates exon1 huntingtin is a key pathogenic form. Nat. Commun. 2020, 11, 2582. [Google Scholar] [CrossRef] [PubMed]
- Miguez, A.; Gomis, C.; Vila, C.; Monguió-Tortajada, M.; Fernández-García, S.; Bombau, G.; Galofré, M.; García-Bravo, M.; Sanders, P.; Fernández-Medina, H.; et al. Soluble mutant huntingtin drives early human pathogenesis in Huntington’s disease. Cell. Mol. Life Sci. 2023, 80, 238. [Google Scholar] [CrossRef] [PubMed]
- Kamitsuka, P.J.; Ghanem, M.M.; Ziar, R.; McDonald, S.E.; Thomas, M.G.; Kwakye, G.F. Defective Mitochondrial Dynamics and Protein Degradation Pathways Underlie Cadmium-Induced Neurotoxicity and Cell Death in Huntington’s Disease Striatal Cells. Int. J. Mol. Sci. 2023, 24, 7178. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; Luo, X.; Fang, D.; Song, H.; Fang, W.; Shan, H.; Liu, P.; Lu, B.; Yin, X.-M.; Hong, L.; et al. Discovery of an autophagy inducer J3 to lower mutant huntingtin and alleviate Huntington’s disease-related phenotype. Cell Biosci. 2022, 12, 167. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Kang, J.H.; Lee, S. Autophagy in Neurodegenerative Diseases: A Hunter for Aggregates. Int. J. Mol. Sci. 2020, 21, 3369. [Google Scholar] [CrossRef]
- Chaudhary, S.; Dhiman, A.; Dilawari, R.; Chaubey, G.K.; Talukdar, S.; Modanwal, R.; Patidar, A.; Malhotra, H.; Raje, C.I.; Raje, M. Glyceraldehyde-3-Phosphate Dehydrogenase Facilitates Macroautophagic Degradation of Mutant Huntingtin Protein Aggregates. Mol. Neurobiol. 2021, 58, 5790–5798. [Google Scholar] [CrossRef] [PubMed]
- Fox, L.M.; Kim, K.; Johnson, C.W.; Chen, S.; Croce, K.R.; Victor, M.B.; Eenjes, E.; Bosco, J.R.; Randolph, L.K.; Dragatsis, I.; et al. Huntington’s Disease Pathogenesis Is Modified In Vivo by Alfy/Wdfy3 and Selective Macroautophagy. Neuron 2020, 105, 813–821.e6. [Google Scholar] [CrossRef] [PubMed]
- Rui, Y.N.; Xu, Z.; Patel, B.; Chen, Z.; Chen, D.; Tito, A.; David, G.; Sun, Y.; Stimming, E.F.; Bellen, H.J.; et al. Huntingtin Functions as a Scaffold for Selective Macroautophagy. Nat. Cell Biol. 2015, 17, 262. [Google Scholar] [CrossRef] [PubMed]
- Riguet, N.; Mahul-Mellier, A.-L.; Maharjan, N.; Burtscher, J.; Croisier, M.; Knott, G.; Hastings, J.; Patin, A.; Reiterer, V.; Farhan, H.; et al. Nuclear and cytoplasmic huntingtin inclusions exhibit distinct biochemical composition, interactome and ultrastructural properties. Nat. Commun. 2021, 12, 6579. [Google Scholar] [CrossRef]
- Stonebraker, A.R.; Hankin, R.; Kapp, K.L.; Li, P.; Valentine, S.J.; Legleiter, J. Charge within Nt17 peptides modulates huntingtin aggregation and initial lipid binding events. Biophys. Chem. 2023, 303, 107123. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Petroni, G.; Amaravadi, R.K.; Baehrecke, E.H.; Ballabio, A.; Boya, P.; Bravo-San Pedro, J.M.; Cadwell, K.; Cecconi, F.; Choi, A.M.K.; et al. Autophagy in major human diseases. EMBO J. 2021, 40, e108863. [Google Scholar] [CrossRef]
- Rippin, I.; Bonder, K.; Joseph, S.; Sarsor, A.; Vaks, L.; Eldar-Finkelman, H. Inhibition of GSK-3 ameliorates the pathogenesis of Huntington’s disease. Neurobiol. Dis. 2021, 154, 105336. [Google Scholar] [CrossRef]
- Del Toro, D.; Alberch, J.; Lazaro-Dieguez, F.; Martin-Ibaflez, R.; Xifro, X.; Egea, G.; Canals, J.M. Mutant Huntingtin Impairs Post-Golgi Trafficking to Lysosomes by Delocalizing Optineurin/Rab8 Complex from the Golgi Apparatus. Mol. Biol. Cell 2009, 20, 1478. [Google Scholar] [CrossRef] [PubMed]
- Del Toro, D.; Canals, J.M.; Ginés, S.; Kojima, M.; Egea, G.; Alberch, J. Mutant huntingtin Impairs the Post-Golgi Trafficking of Brain-Derived Neurotrophic Factor But Not Its Val66Met Polymorphism. J. Neurosci. 2006, 26, 12748. [Google Scholar] [CrossRef]
- Erie, C.; Sacino, M.; Houle, L.; Lu, M.L.; Wei, J. Altered lysosomal positioning affects lysosomal functions in a cellular model of Huntington’s disease. Eur. J. Neurosci. 2015, 42, 1941–1951. [Google Scholar] [CrossRef] [PubMed]
- Wong, Y.C.; Holzbaur, E.L.F. The regulation of autophagosome dynamics by huntingtin and HAP1 is disrupted by expression of mutant huntingtin, leading to defective cargo degradation. J. Neurosci. 2014, 34, 1293–1305. [Google Scholar] [CrossRef]
- Gauthier, L.R.; Charrin, B.C.; Borrell-Pagès, M.; Dompierre, J.P.; Rangone, H.; Cordelières, F.P.; De Mey, J.; MacDonald, M.E.; Lessmann, V.; Humbert, S.; et al. Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell 2004, 118, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Orr, A.L.; Li, S.; Wang, C.-E.; Li, H.; Wang, J.; Rong, J.; Xu, X.; Mastroberardino, P.G.; Greenamyre, J.T.; Li, X.-J. N-terminal mutant huntingtin associates with mitochondria and impairs mitochondrial trafficking. J. Neurosci. 2008, 28, 2783–2792. [Google Scholar] [CrossRef]
- Sardiello, M.; Palmieri, M.; Di Ronza, A.; Medina, D.L.; Valenza, M.; Gennarino, V.A.; Di Malta, C.; Donaudy, F.; Embrione, V.; Polishchuk, R.S.; et al. A gene network regulating lysosomal biogenesis and function. Science 2009, 325, 473–477. [Google Scholar] [CrossRef]
- Napolitano, G.; Ballabio, A. TFEB at a glance. J. Cell Sci. 2016, 129, 2475–2481. [Google Scholar] [CrossRef]
- Steingrímsson, E.; Copeland, N.G.; Jenkins, N.A. Melanocytes and the microphthalmia transcription factor network. Annu. Rev. Genet. 2004, 38, 365–411. [Google Scholar] [CrossRef] [PubMed]
- Beckmann, H.; Su, L.K.; Kadesch, T. TFE3: A helix-loop-helix protein that activates transcription through the immunoglobulin enhancer muE3 motif. Genes Dev. 1990, 4, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Roberts, K.; Gambino, G.; Cook, A.; Kouzarides, T.; Goding, C.R. CBP/p300 as a co-factor for the Microphthalmia transcription factor. Oncogene 1997, 14, 3083–3092. [Google Scholar] [CrossRef]
- Palmieri, M.; Impey, S.; Kang, H.; di Ronza, A.; Pelz, C.; Sardiello, M.; Ballabio, A. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum. Mol. Genet. 2011, 20, 3852–3866. [Google Scholar] [CrossRef] [PubMed]
- Martina, J.A.; Diab, H.I.; Lishu, L.; Jeong-A, L.; Patange, S.; Raben, N.; Puertollano, R. The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci. Signal. 2014, 7, ra9. [Google Scholar] [CrossRef] [PubMed]
- Puertollano, R.; Ferguson, S.M.; Brugarolas, J.; Ballabio, A. The complex relationship between TFEB transcription factor phosphorylation and subcellular localization. EMBO J. 2018, 37, e98804. [Google Scholar] [CrossRef] [PubMed]
- Martina, J.A.; Chen, Y.; Gucek, M.; Puertollano, R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 2012, 8, 903–914. [Google Scholar] [CrossRef] [PubMed]
- Roczniak-Ferguson, A.; Petit, C.S.; Froehlich, F.; Qian, S.; Ky, J.; Angarola, B.; Walther, T.C.; Ferguson, S.M. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci. Signal. 2012, 5, ra42. [Google Scholar] [CrossRef]
- Settembre, C.; Zoncu, R.; Medina, D.L.; Vetrini, F.; Erdin, S.; Erdin, S.; Huynh, T.; Ferron, M.; Karsenty, G.; Vellard, M.C.; et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012, 31, 1095–1108. [Google Scholar] [CrossRef]
- Martina, J.A.; Puertollano, R. Rag GTPases mediate amino acid-dependent recruitment of TFEB and MITF to lysosomes. J. Cell Biol. 2013, 200, 475–491. [Google Scholar] [CrossRef]
- Cui, Z.; Joiner, A.M.N.; Jansen, R.M.; Hurley, J.H. Amino acid sensing and lysosomal signaling complexes. Curr. Opin. Struct. Biol. 2023, 79, 102544. [Google Scholar] [CrossRef]
- Settembre, C.; Di Malta, C.; Polito, V.A.; Arencibia, M.G.; Vetrini, F.; Erdin, S.; Erdin, S.U.; Huynh, T.; Medina, D.; Colella, P.; et al. TFEB links autophagy to lysosomal biogenesis. Science 2011, 332, 1429–1433. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, M.; Pal, R.; Nelvagal, H.R.; Lotfi, P.; Stinnett, G.R.; Seymour, M.L.; Chaudhury, A.; Bajaj, L.; Bondar, V.V.; Bremner, L.; et al. mTORC1-independent TFEB activation via Akt inhibition promotes cellular clearance in neurodegenerative storage diseases. Nat. Commun. 2017, 8, 14338. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xu, M.; Ding, X.; Yan, C.; Song, Z.; Chen, L.; Huang, X.; Wang, X.; Jian, Y.; Tang, G.; et al. Protein kinase C controls lysosome biogenesis independently of mTORC1. Nat. Cell Biol. 2016, 18, 1065–1077. [Google Scholar] [CrossRef]
- Sha, Y.; Rao, L.; Settembre, C.; Ballabio, A.; Eissa, N.T. STUB1 regulates TFEB-induced autophagy-lysosome pathway. EMBO J. 2017, 36, 2544–2552. [Google Scholar] [CrossRef] [PubMed]
- Tong, Y.; Song, F. Intracellular calcium signaling regulates autophagy via calcineurin-mediated TFEB dephosphorylation. Autophagy 2015, 11, 1192–1195. [Google Scholar] [CrossRef] [PubMed]
- Settembre, C.; Ballabio, A. Lysosome: Regulator of lipid degradation pathways. Trends Cell Biol. 2014, 24, 743–750. [Google Scholar] [CrossRef] [PubMed]
- Erlich, A.T.; Brownlee, D.M.; Beyfuss, K.; Hood, D.A. Exercise induces TFEB expression and activity in skeletal muscle in a PGC-1α-dependent manner. Am. J. Physiol. Cell Physiol. 2018, 314, C62–C72. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Gong, S.; Zhang, H.; Chen, Y.; Liu, Y.; Hao, J.; Liu, H.; Li, X. From the regulatory mechanism of TFEB to its therapeutic implications. Cell Death Discov. 2024, 10, 84. [Google Scholar] [CrossRef]
- Polito, V.A.; Li, H.; Martini-Stoica, H.; Wang, B.; Yang, L.; Xu, Y.; Swartzlander, D.B.; Palmieri, M.; di Ronza, A.; Lee, V.M.; et al. Selective clearance of aberrant tau proteins and rescue of neurotoxicity by transcription factor EB. EMBO Mol. Med. 2014, 6, 1142–1160. [Google Scholar] [CrossRef]
- Yang, C.; Su, C.; Iyaswamy, A.; Krishnamoorthi, S.K.; Zhu, Z.; Yang, S.; Tong, B.C.; Liu, J.; Sreenivasmurthy, S.G.; Guan, X.; et al. Celastrol enhances transcription factor EB (TFEB)-mediated autophagy and mitigates Tau pathology: Implications for Alzheimer’s disease therapy. Acta Pharm. Sin. B 2022, 12, 1707–1722. [Google Scholar] [CrossRef] [PubMed]
- Bao, J.; Zheng, L.; Zhang, Q.; Li, X.; Zhang, X.; Li, Z.; Bai, X.; Zhang, Z.; Huo, W.; Zhao, X.; et al. Deacetylation of TFEB promotes fibrillar Aβ degradation by upregulating lysosomal biogenesis in microglia. Protein Cell 2016, 7, 417–433. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Q.; Yan, P.; Ma, X.; Liu, H.; Perez, R.; Zhu, A.; Gonzales, E.; Burchett, J.M.; Schuler, D.R.; Cirrito, J.R.; et al. Enhancing astrocytic lysosome biogenesis facilitates Aβ clearance and attenuates amyloid plaque pathogenesis. J. Neurosci. 2014, 34, 9607–9620. [Google Scholar] [CrossRef] [PubMed]
- Decressac, M.; Mattsson, B.; Weikop, P.; Lundblad, M.; Jakobsson, J.; Björklund, A. TFEB-mediated autophagy rescues midbrain dopamine neurons from α-synuclein toxicity. Proc. Natl. Acad. Sci. USA 2013, 110, E1817–E1826. [Google Scholar] [CrossRef] [PubMed]
- Kilpatrick, K.; Zeng, Y.; Hancock, T.; Segatori, L. Genetic and chemical activation of TFEB mediates clearance of aggregated α-synuclein. PLoS ONE 2015, 10, e0120819. [Google Scholar] [CrossRef] [PubMed]
- Vodicka, P.; Chase, K.; Iuliano, M.; Tousley, A.; Valentine, D.T.; Sapp, E.; Kegel-Gleason, K.B.; Sena-Esteves, M.; Aronin, N.; Difiglia, M. Autophagy Activation by Transcription Factor EB (TFEB) in Striatum of HDQ175/Q7 Mice. J. Huntingtons. Dis. 2016, 5, 249–260. [Google Scholar] [CrossRef] [PubMed]
- Tsunemi, T.; Ashe, T.D.; Morrison, B.E.; Soriano, K.R.; Au, J.; Roque, R.A.V.; Lazarowski, E.R.; Damian, V.A.; Masliah, E.; La Spada, A.R. PGC-1α rescues Huntington’s disease proteotoxicity by preventing oxidative stress and promoting TFEB function. Sci. Transl. Med. 2012, 4, 142ra97. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.W.; Oh, Y.M.; Victor, M.B.; Yang, Y.; Chen, S.; Strunilin, I.; Dahiya, S.; Dolle, R.E.; Pak, S.C.; Silverman, G.A.; et al. Longitudinal modeling of human neuronal aging reveals the contribution of the RCAN1-TFEB pathway to Huntington’s disease neurodegeneration. Nat. Aging 2024, 4, 95–109. [Google Scholar] [CrossRef] [PubMed]
- Brattås, P.L.; Hersbach, B.A.; Madsen, S.; Petri, R.; Jakobsson, J.; Pircs, K. Impact of differential and time-dependent autophagy activation on therapeutic efficacy in a model of Huntington disease. Autophagy 2021, 17, 1316–1329. [Google Scholar] [CrossRef]
- Yang, J.; Xu, H.; Zhang, C.; Yang, X.; Cai, W.; Chen, X. A prion-like domain of TFEB mediates the co-aggregation of TFEB and mHTT. Autophagy 2023, 19, 544–550. [Google Scholar] [CrossRef]
- Li, Z.; Zhu, C.; Ding, Y.; Fei, Y.; Lu, B. ATTEC: A potential new approach to target proteinopathies. Autophagy 2020, 16, 185–187. [Google Scholar] [CrossRef] [PubMed]
| Methods of Targeting TFEB | Model | Phenotype | Study |
|---|---|---|---|
| TFEB overexpression | Striatum of Q175/Q7 HD mice | Clearance of polyQ-HTT | [66] |
| TFEB expression is promoted by inducing PGC-1α expression | N171-82Q transgenic mice | Reduction of protein HTT aggregation | [67] |
| Enhanced calcineurin activity promotes nuclear localization of TFEB | Patient-derived striatal medium spiny neurons | Clearance of HTT inclusion bodies and neuronal survival | [68] |
| Overexpression of human TFEB | AAV-based model of HD | No change in the amount of mHTT aggregates | [69] |
| TFEB overexpression | HdhQ140 mice | Co-aggregation of TFEB and mHTT | [70] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ojalvo-Pacheco, J.; Yakhine-Diop, S.M.S.; Fuentes, J.M.; Paredes-Barquero, M.; Niso-Santano, M. Role of TFEB in Huntington’s Disease. Biology 2024, 13, 238. https://doi.org/10.3390/biology13040238
Ojalvo-Pacheco J, Yakhine-Diop SMS, Fuentes JM, Paredes-Barquero M, Niso-Santano M. Role of TFEB in Huntington’s Disease. Biology. 2024; 13(4):238. https://doi.org/10.3390/biology13040238
Chicago/Turabian StyleOjalvo-Pacheco, Javier, Sokhna M. S. Yakhine-Diop, José M. Fuentes, Marta Paredes-Barquero, and Mireia Niso-Santano. 2024. "Role of TFEB in Huntington’s Disease" Biology 13, no. 4: 238. https://doi.org/10.3390/biology13040238
APA StyleOjalvo-Pacheco, J., Yakhine-Diop, S. M. S., Fuentes, J. M., Paredes-Barquero, M., & Niso-Santano, M. (2024). Role of TFEB in Huntington’s Disease. Biology, 13(4), 238. https://doi.org/10.3390/biology13040238







