An Efficient Homologous Recombination-Based In Situ Protein-Labeling Method in Verticillium dahliae
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal and Bacterial Strains
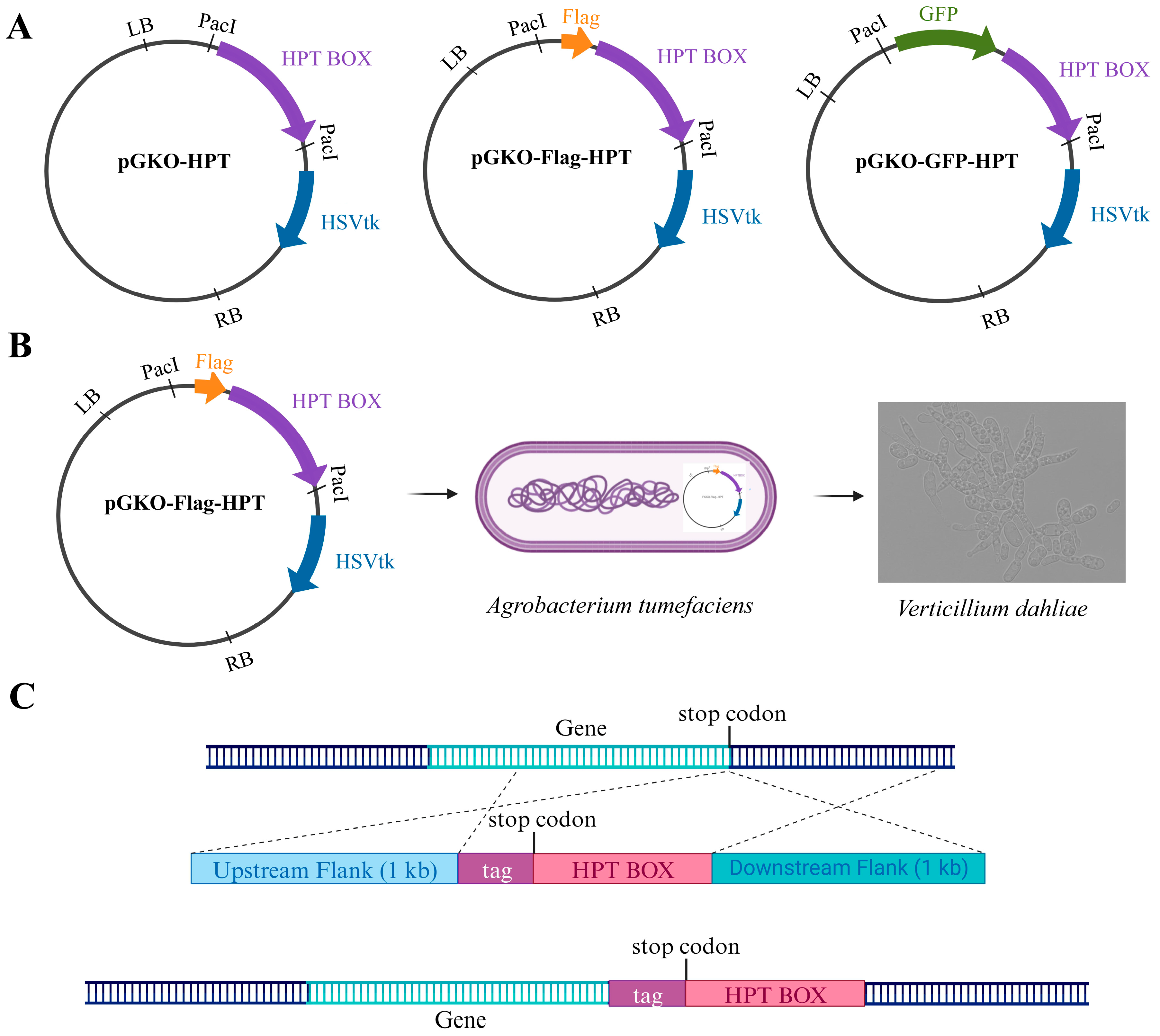
2.2. Vectors
2.3. Construction and Transformation
2.4. Confocal Laser Scanning Microscopy (CLSM)
2.5. Protein Detection via Western Blotting
3. Results
3.1. The General Strategy for In Situ C-Terminus Tagging in V. dahliae
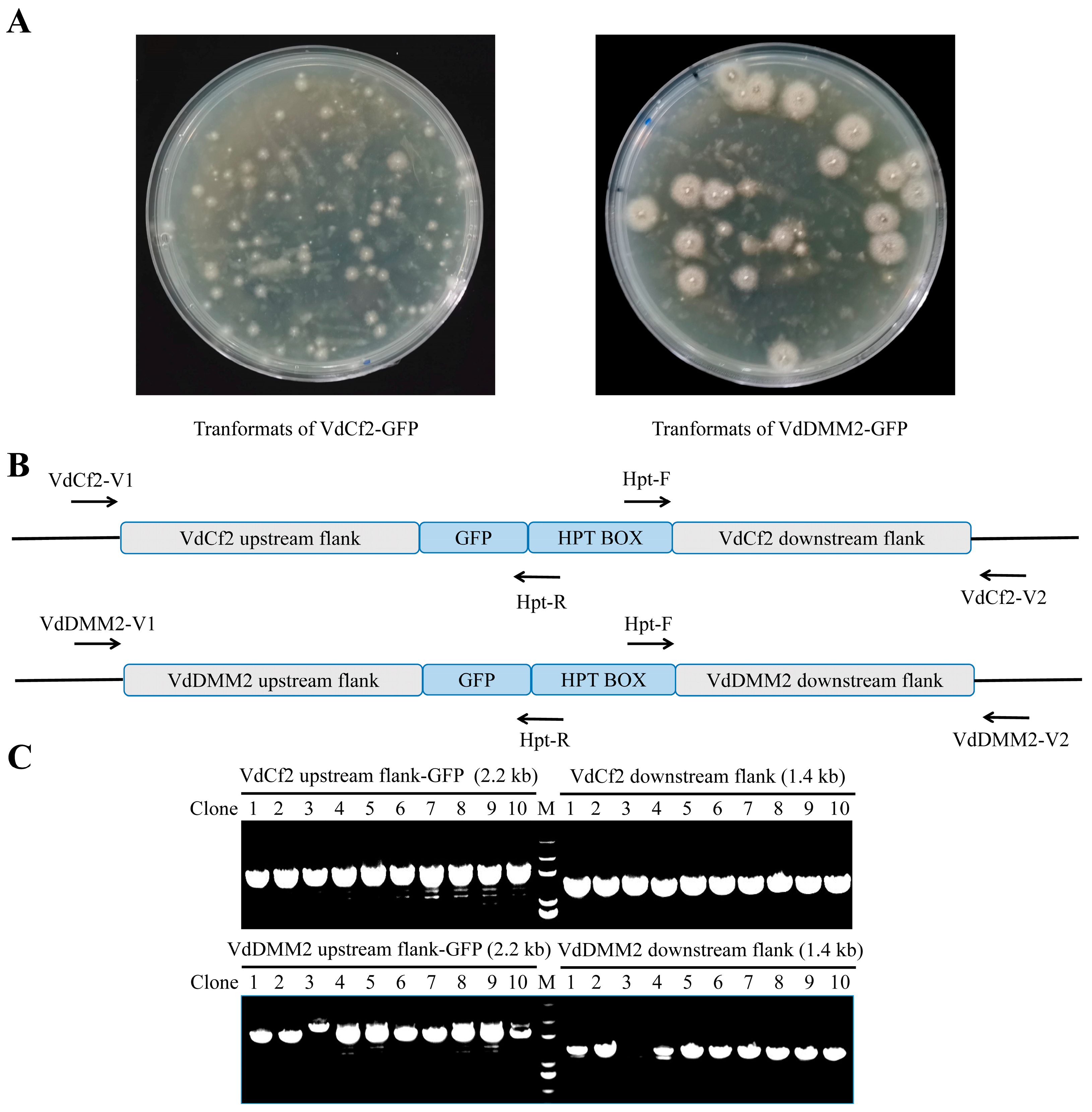
3.2. In Situ GFP Tagging of VdCf2 and VdDMM2 Using the pGKO-GFP-HPT Vector
3.3. Molecular Confirmation of C-Terminal GFP Tagging for VdCf2 and VdDMM2 in V. dahliae
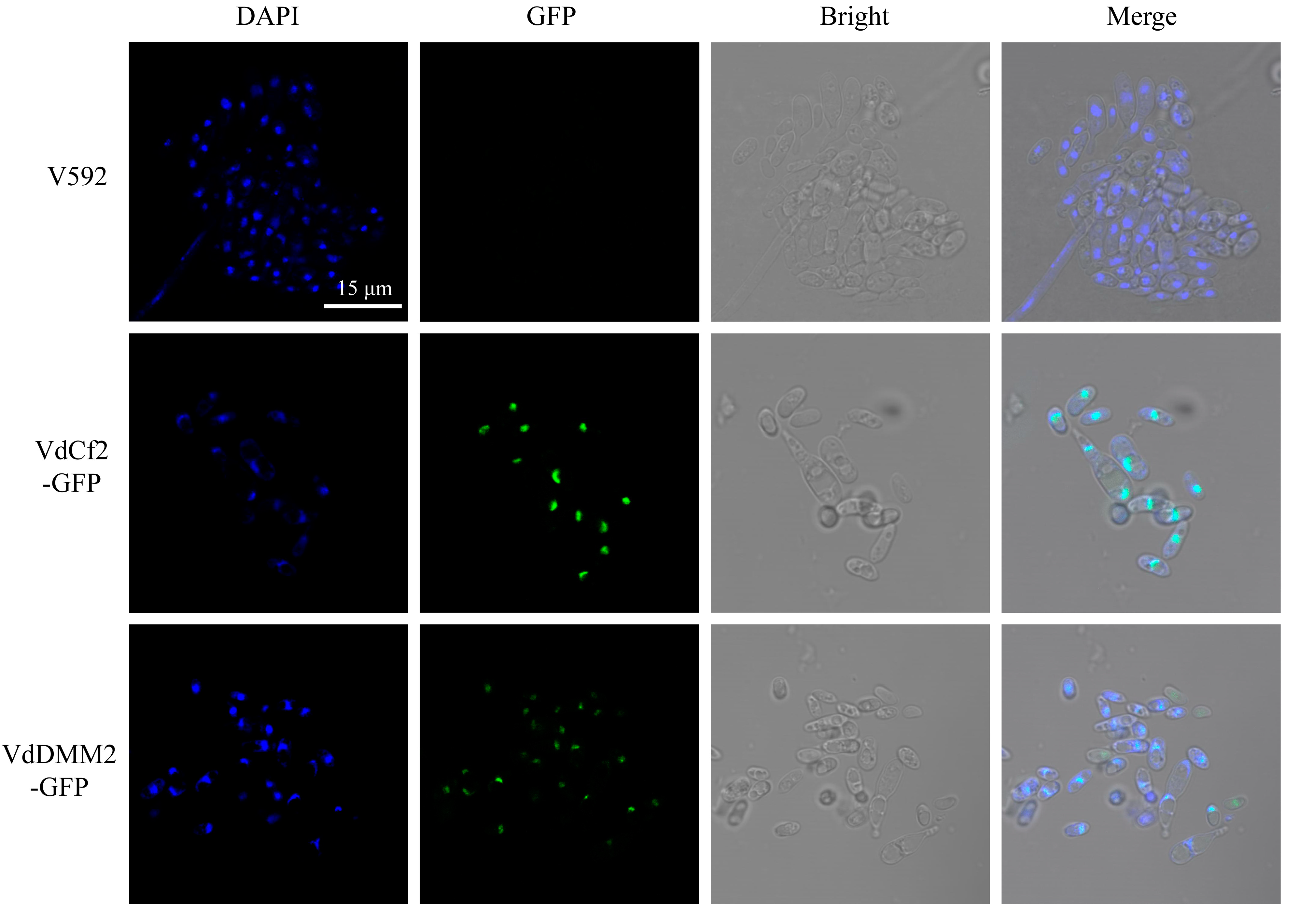
3.4. Subcellular Localization for In Situ Tagged Endogenous VdCf2-GFP and VdDMM2-GFP in V. dahliae
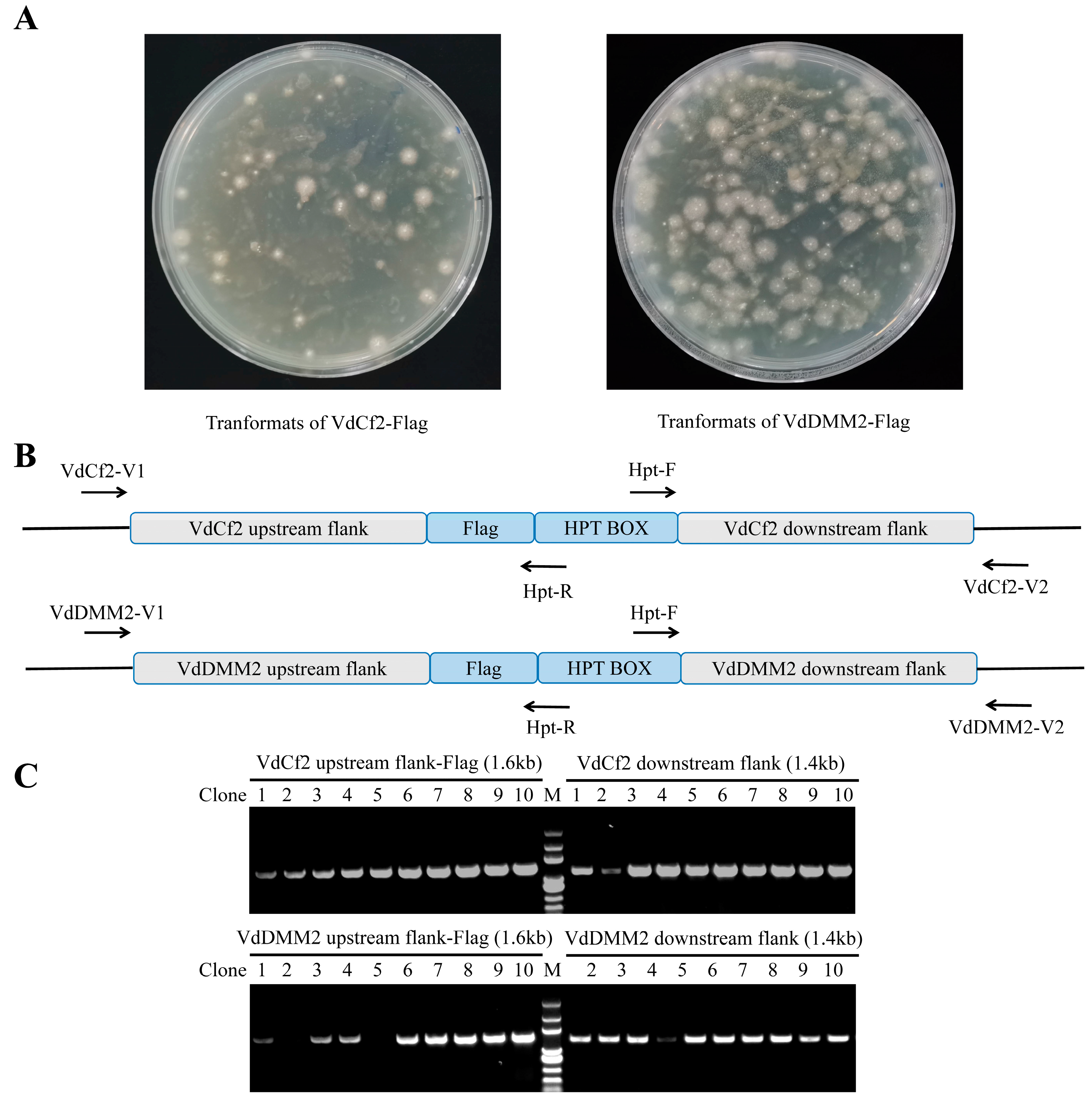
3.5. Assessment of Versatility for in In Situ Tagging System within V. dahliae
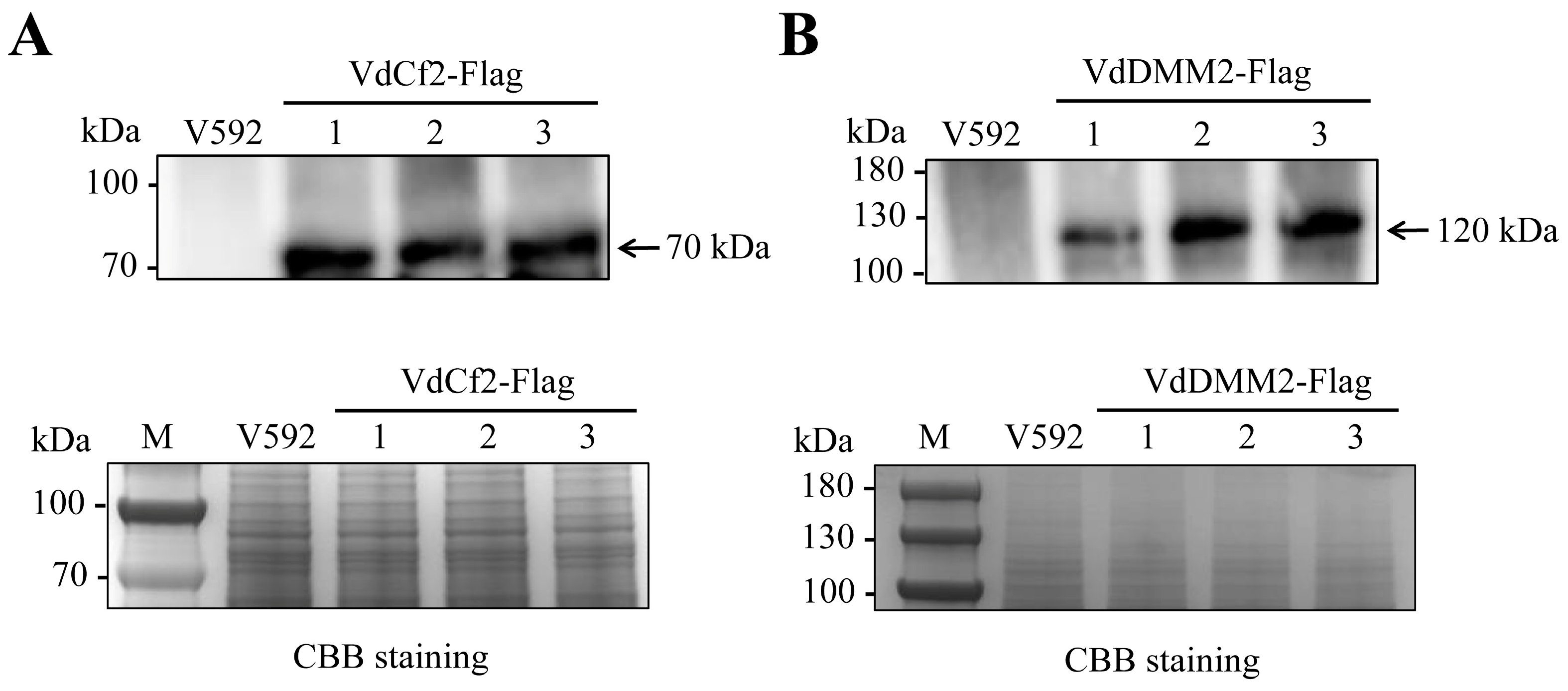
3.6. Validation of Endogenous VdCf2-Flag and VdDMM2-Flag Fusion Proteins in V. dahliae by Western Blot Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hu, X.P.; Gurung, S.; Short, D.P.G.; Sandoya, G.V.; Shang, W.J.; Hayes, R.J.; Davis, R.M.; Subbarao, K.V. Nondefoliating and Defoliating Strains from Cotton Correlate with Races 1 and 2 of Verticillium dahliae. Plant Dis. 2015, 99, 1713–1720. [Google Scholar] [CrossRef]
- Xie, C.; Li, Q.; Yang, X. Characterization of VdASP F2 Secretory Factor from Verticillium dahliae by a Fast and Easy Gene Knockout System. Mol. Plant-Microbe Interact. 2017, 30, 444–454. [Google Scholar] [CrossRef]
- Klosterman, S.J.; Atallah, Z.K.; Vallad, G.E.; Subbarao, K.V. Diversity, pathogenicity, and management of verticillium species. Annu. Rev. Phytopathol. 2009, 47, 39–62. [Google Scholar] [CrossRef]
- Snelders, N.C.; Rovenich, H.; Petti, G.C.; Rocafort, M.; van den Berg, G.C.M.; Vorholt, J.A.; Mesters, J.R.; Seidl, M.F.; Nijland, R.; Thomma, B.P.H.J. Microbiome manipulation by a soil-borne fungal plant pathogen using effector proteins. Nat. Plants 2020, 6, 1365–1374. [Google Scholar] [CrossRef]
- Chen, J.Y.; Zhang, D.D.; Huang, J.Q.; Li, R.; Wang, D.; Song, J.; Puri, K.D.; Yang, L.; Kong, Z.-Q.; Tong, B.-Z.; et al. Dynamics of Verticillium dahliae race 1 population under managed agricultural ecosystems. BMC Biol. 2021, 19, 131. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Chen, J.Y.; Song, J.; Li, J.J.; Klosterman, S.J.; Li, R.; Kong, Z.Q.; Subbarao, K.V.; Dai, X.F.; Zhang, D.D. Cytotoxic function of xylanase VdXyn4 in the plant vascular wilt pathogen Verticillium dahliae. Plant Physiol. 2021, 187, 409–429. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Zhao, M.; Li, T.; Wang, L.; Liao, C.; Liu, D.; Zhang, H.; Zhao, Y.; Liu, L.; Ge, X.; et al. Interactions between Verticillium dahliae and cotton: Pathogenic mechanism and cotton resistance mechanism to Verticillium wilt. Front. Plant Sci. 2023, 14, 1174281. [Google Scholar] [CrossRef]
- Wen, H.G.; Zhao, J.H.; Zhang, B.S.; Gao, F.; Wu, X.M.; Yan, Y.S.; Zhang, J.; Guo, H.S. Microbe-induced gene silencing boosts crop protection against soil-borne fungal pathogens. Nat. Plants 2023, 9, 1409–1418. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.M.; Zhang, B.S.; Zhao, Y.L.; Wu, H.W.; Gao, F.; Zhang, J.; Zhao, J.H.; Guo, H.S. DeSUMOylation of a Verticillium dahliae enolase facilitates virulence by derepressing the expression of the effector VdSCP8. Nat. Commun. 2023, 14, 4844. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Qin, J.; Rong, W.; Ni, H.; Guo, H.S.; Zhang, J. Cellophane surface-induced gene, VdCSIN1, regulates hyphopodium formation and pathogenesis via cAMP-mediated signalling in Verticillium dahliae. Mol. Plant Pathol. 2019, 20, 323–333. [Google Scholar] [CrossRef]
- Song, R.; Li, J.; Xie, C.; Jian, W.; Yang, X. An Overview of the Molecular Genetics of Plant Resistance to the Verticillium Wilt Pathogen Verticillium dahliae. Int. J. Mol. Sci. 2020, 21, 1120. [Google Scholar] [CrossRef]
- Miao, Z.; Wang, H.; Tu, X.; Huang, Z.; Huang, S.; Zhang, X.; Wang, F.; Huang, Z.; Li, H.; Jiao, Y.; et al. GetPrimers: A generalized PCR-based genetic targeting primer designer enabling easy and standardized targeted gene modification across multiple systems. Yeast 2023. [Google Scholar] [CrossRef]
- Teubl, F.; Schwank, K.; Ohmayer, U.; Griesenbeck, J.; Tschochner, H.; Milkereit, P. Tethered MNase Structure Probing as Versatile Technique for Analyzing RNPs Using Tagging Cassettes for Homologous Recombination in Saccharomyces cerevisiae. In Ribosome Biogenesis: Methods and Protocols; Entian, K.D., Ed.; Humana: New York, NY, USA, 2022; Volume 2533, pp. 127–145. [Google Scholar] [CrossRef]
- Kira, S.; Noda, T. A CRISPR/Cas9-based method for seamless N-terminal protein tagging in Saccharomyces cerevisiae. Yeast 2021, 38, 592–600. [Google Scholar] [CrossRef] [PubMed]
- Khmelinskii, A.; Meurer, M.; Duishoev, N.; Delhomme, N.; Knop, M. Seamless gene tagging by endonuclease-driven homologous recombination. PLoS ONE 2011, 6, e23794. [Google Scholar] [CrossRef] [PubMed]
- Sung, M.K.; Ha, C.W.; Huh, W.K. A vector system for efficient and economical switching of C-terminal epitope tags in Saccharomyces cerevisiae. Yeast 2008, 25, 301–311. [Google Scholar] [CrossRef] [PubMed]
- Kanca, O.; Zirin, J.; Hu, Y.; Tepe, B.; Dutta, D.; Lin, W.W.; Ma, L.; Ge, M.; Zuo, Z.; Liu, L.P.; et al. An expanded toolkit for Drosophila gene tagging using synthesized homology donor constructs for CRISPR-mediated homologous recombination. eLife 2022, 11, e76077. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Liu, J.L. Genomic Tagging of AGO1 Using CRISPR/Cas9-Mediated Homologous Recombination. Methods Mol. Biol. 2018, 1680, 217–235. [Google Scholar] [CrossRef] [PubMed]
- Baena-Lopez, L.A.; Alexandre, C.; Mitchell, A.; Pasakarnis, L.; Vincent, J.P. Accelerated homologous recombination and subsequent genome modification in Drosophila. Development 2013, 140, 4818–4825. [Google Scholar] [CrossRef]
- Xiang, X.; Li, C.; Chen, X.; Dou, H.; Li, Y.; Zhang, X.; Luo, Y. CRISPR/Cas9-Mediated Gene Tagging: A Step-by-Step Protocol. In CRISPR Gene Editing: Methods and Protocols; Luo, Y., Ed.; Humana Press: New York, NY, USA, 2019; Volume 1961, pp. 255–269. [Google Scholar] [CrossRef]
- Shan, C.M.; Wang, J.; Xu, K.; Chen, H.; Yue, J.X.; Andrews, S.; Moresco, J.J.; Yates, J.R.; Nagy, P.L.; Tong, L.; et al. A histone H3K9M mutation traps histone methyltransferase Clr4 to prevent heterochromatin spreading. eLife 2016, 5, e17903. [Google Scholar] [CrossRef]
- Shan, C.M.; Kim, J.K.; Wang, J.; Bao, K.; Sun, Y.; Chen, H.; Yue, J.X.; Stirpe, A.; Zhang, Z.; Lu, C.; et al. The histone H3K9M mutation synergizes with H3K14 ubiquitylation to selectively sequester histone H3K9 methyltransferase Clr4 at heterochromatin. Cell Rep. 2021, 35, 109137. [Google Scholar] [CrossRef]
- Wang, S.; Xing, H.; Hua, C.; Guo, H.S.; Zhang, J. An Improved Single-Step Cloning Strategy Simplifies the Agrobacterium tumefaciens-Mediated Transformation (ATMT)-Based Gene-Disruption Method for Verticillium dahliae. Phytopathology 2016, 106, 645–652. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.H.; Jun, S.; Hur, H.S.; Ryu, J.J.; Kim, J. Candida albicans protein analysis during hyphal differentiation using an integrative HA-tagging method. Biochem. Biophys. Res. Commun. 2005, 337, 784–790. [Google Scholar] [CrossRef]
- Gola, S.; Martin, R.; Walther, A.; Dünkler, A.; Wendland, J. New modules for PCR-based gene targeting in Candida albicans: Rapid and efficient gene targeting using 100 bp of flanking homology region. Yeast 2003, 20, 1339–1347. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.; Wang, W.; Igarashi, Y.; Luo, F.; Chen, J. Efficient vector systems for economical and rapid epitope-tagging and overexpression in Candida albicans. J. Microbiol. Methods 2018, 149, 14–19. [Google Scholar] [CrossRef]
- Hammond, T.M.; Xiao, H.; Rehard, D.G.; Boone, E.C.; Perdue, T.D.; Pukkila, P.J.; Shiu, P.K. Fluorescent and bimolecular-fluorescent protein tagging of genes at their native loci in Neurospora crassa using specialized double-joint PCR plasmids. Fungal Genet. Biol. 2011, 48, 866–873. [Google Scholar] [CrossRef]
- Wang, Q.; Coleman, J.J. CRISPR/Cas9-mediated endogenous gene tagging in Fusarium oxysporum. Fungal Genet. Biol. 2019, 126, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Azizi-Dargahlou, S.; Pouresmaeil, M. Agrobacterium tumefaciens-Mediated Plant Transformation: A Review. Mol. Biotechnol. 2023. [Google Scholar] [CrossRef]
- Schrammeijer, B.; den Dulk-Ras, A.; Vergunst, A.C.; Jurado Jácome, E.; Hooykaas, P.J. Analysis of Vir protein translocation from Agrobacterium tumefaciens using Saccharomyces cerevisiae as a model: Evidence for transport of a novel effector protein VirE3. Nucleic Acids Res. 2003, 31, 860–868. [Google Scholar] [CrossRef]
- de Groot, M.J.; Bundock, P.; Hooykaas, P.J.; Beijersbergen, A.G. Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat. Biotechnol. 1998, 16, 839–842. [Google Scholar] [CrossRef]
- Michielse, C.B.; Hooykaas, P.J.; van den Hondel, C.A.; Ram, A.F. Agrobacterium-mediated transformation as a tool for functional genomics in fungi. Curr. Genet. 2005, 48, 1–17. [Google Scholar] [CrossRef]
- Kunik, T.; Tzfira, T.; Kapulnik, Y.; Gafni, Y.; Dingwall, C.; Citovsky, V. Genetic transformation of HeLa cells by Agrobacterium. Proc. Natl. Acad. Sci. USA 2001, 98, 1871–1876. [Google Scholar] [CrossRef] [PubMed]
- Santhanam, P. Random Insertional Mutagenesis in Fungal Genomes to Identify Virulence Factors. In Plant Fungal Pathogens: Methods and Protocols; Bolton, M., Thomma, B., Eds.; Humana Press: Totowa, NJ, USA, 2012; Volume 835, pp. 509–517. [Google Scholar] [CrossRef]
- Kramer, H.M.; Cook, D.E.; van den Berg, G.C.M.; Seidl, M.F.; Thomma, B.P.H.J. Three putative DNA methyltransferases of Verticillium dahliae differentially contribute to DNA methylation that is dispensable for growth, development and virulence. Epigenetics Chromatin 2021, 14, 21. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Qin, J.; Cao, Y.; Subbarao, K.V.; Chen, J.; Mandal, M.K.; Xu, X.; Shang, W.; Hu, X. Transcription Factor VdCf2 Regulates Growth, Pathogenicity, and the Expression of a Putative Secondary Metabolism Gene Cluster in Verticillium dahliae. Appl. Environ. Microbiol. 2022, 88, e0138522. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Zhou, B.J.; Li, G.Y.; Jia, P.S.; Li, H.; Zhao, Y.L.; Zhao, P.; Xia, G.X.; Guo, H.S. A glutamic acid-rich protein identified in Verticillium dahliae from an insertional mutagenesis affects microsclerotial formation and pathogenicity. PLoS ONE 2010, 5, e15319. [Google Scholar] [CrossRef] [PubMed]
- Klosterman, S.J.; Subbarao, K.V.; Kang, S.; Veronese, P.; Gold, S.E.; Thomma, B.P.; Chen, Z.; Henrissat, B.; Lee, Y.H.; Park, J.; et al. Comparative genomics yields insights into niche adaptation of plant vascular wilt pathogens. PLoS Pathog. 2011, 7, e1002137. [Google Scholar] [CrossRef]
- Hua, C.; Zhao, J.H.; Guo, H.S. Trans-Kingdom RNA Silencing in Plant-Fungal Pathogen Interactions. Mol. Plant 2018, 11, 235–244. [Google Scholar] [CrossRef]
- Uetz, P.; Giot, L.; Cagney, G.; Mansfield, T.A.; Judson, R.S.; Knight, J.R.; Lockshon, D.; Narayan, V.; Srinivasan, M.; Pochart, P.; et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 2000, 403, 623–627. [Google Scholar] [CrossRef]
- Davidson, R.C.; Blankenship, J.R.; Kraus, P.R.; de Jesus Berrios, M.; Hull, C.M.; D’Souza, C.; Wang, P.; Heitman, J. A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology 2002, 148, 2607–2615. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, J.; Liu, M.; Jiao, Y.; Guo, H.-S.; Shan, C.-M.; Wang, H. An Efficient Homologous Recombination-Based In Situ Protein-Labeling Method in Verticillium dahliae. Biology 2024, 13, 81. https://doi.org/10.3390/biology13020081
Yang J, Liu M, Jiao Y, Guo H-S, Shan C-M, Wang H. An Efficient Homologous Recombination-Based In Situ Protein-Labeling Method in Verticillium dahliae. Biology. 2024; 13(2):81. https://doi.org/10.3390/biology13020081
Chicago/Turabian StyleYang, Jie, Mengran Liu, Yue Jiao, Hui-Shan Guo, Chun-Min Shan, and Haiting Wang. 2024. "An Efficient Homologous Recombination-Based In Situ Protein-Labeling Method in Verticillium dahliae" Biology 13, no. 2: 81. https://doi.org/10.3390/biology13020081
APA StyleYang, J., Liu, M., Jiao, Y., Guo, H.-S., Shan, C.-M., & Wang, H. (2024). An Efficient Homologous Recombination-Based In Situ Protein-Labeling Method in Verticillium dahliae. Biology, 13(2), 81. https://doi.org/10.3390/biology13020081






