Myogenic Differentiation and Immunomodulatory Properties of Rat Adipose-Derived Mesenchymal Stem/Stromal Cells
Abstract
Simple Summary
Abstract
1. Introduction
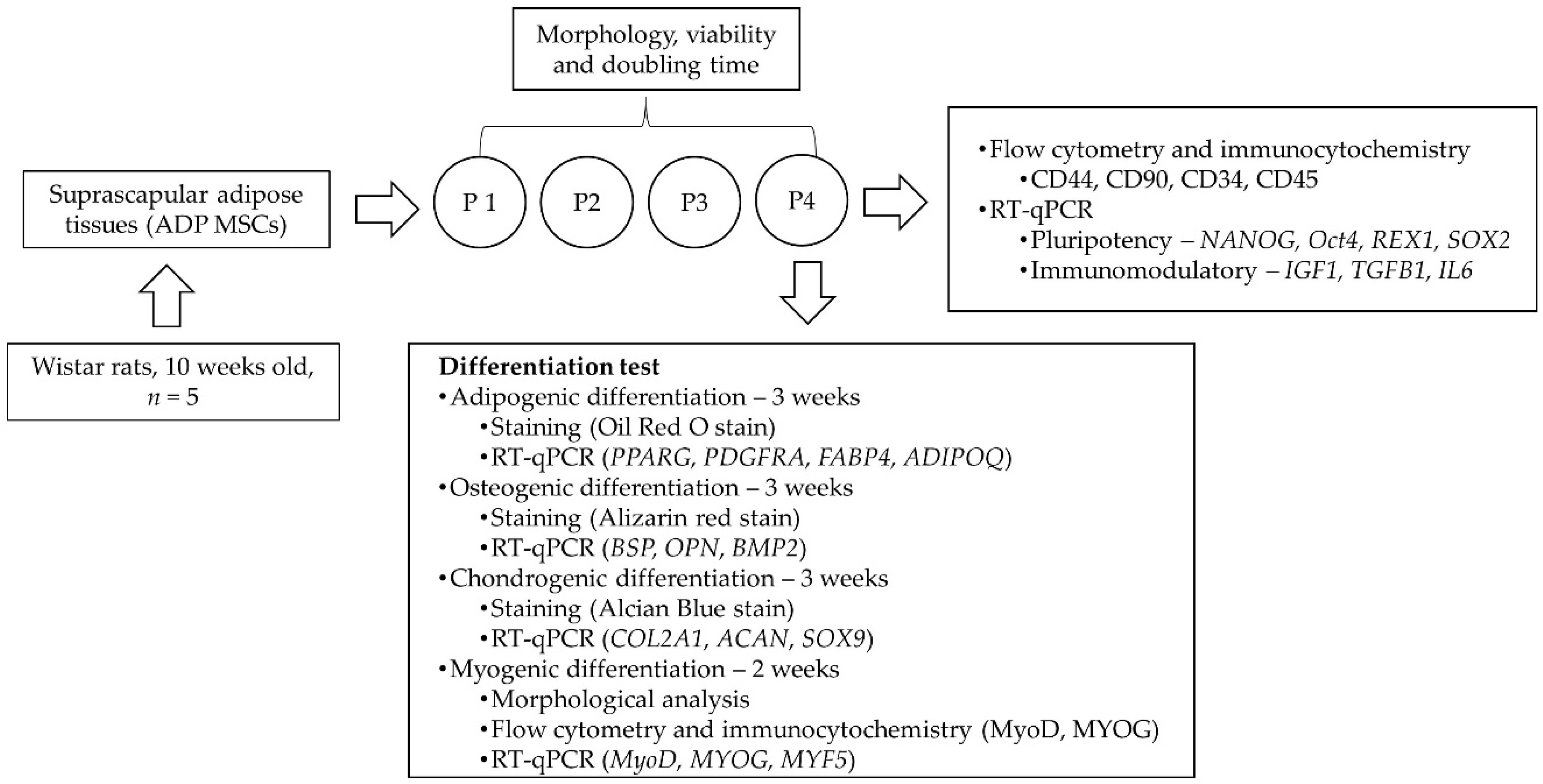
2. Materials and Methods
2.1. Animals and Study Design
2.2. Isolation and Culture of ADP MSCs
2.3. Assessment of ADP MSCs’ Morphology, Viability, and Proliferation
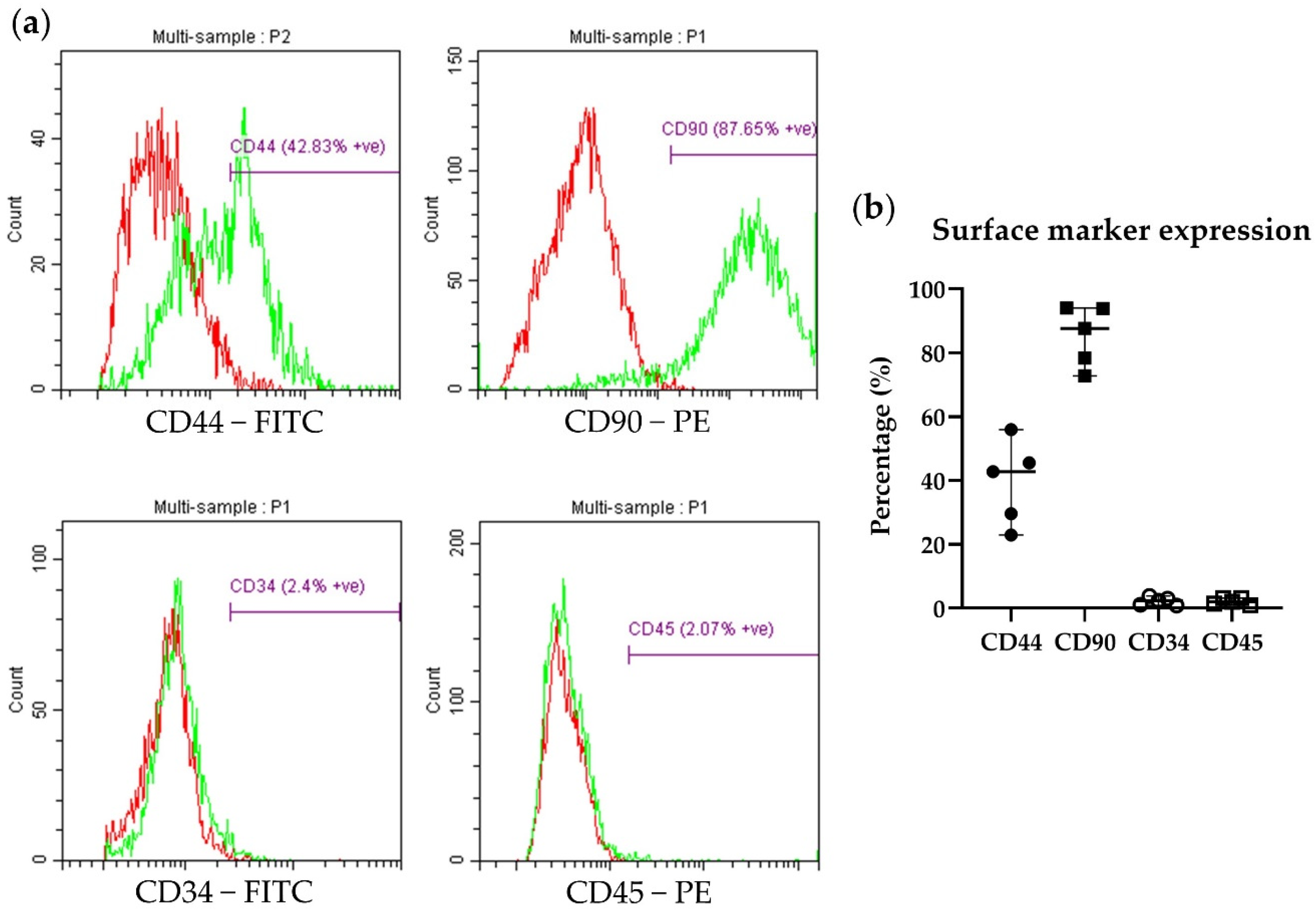
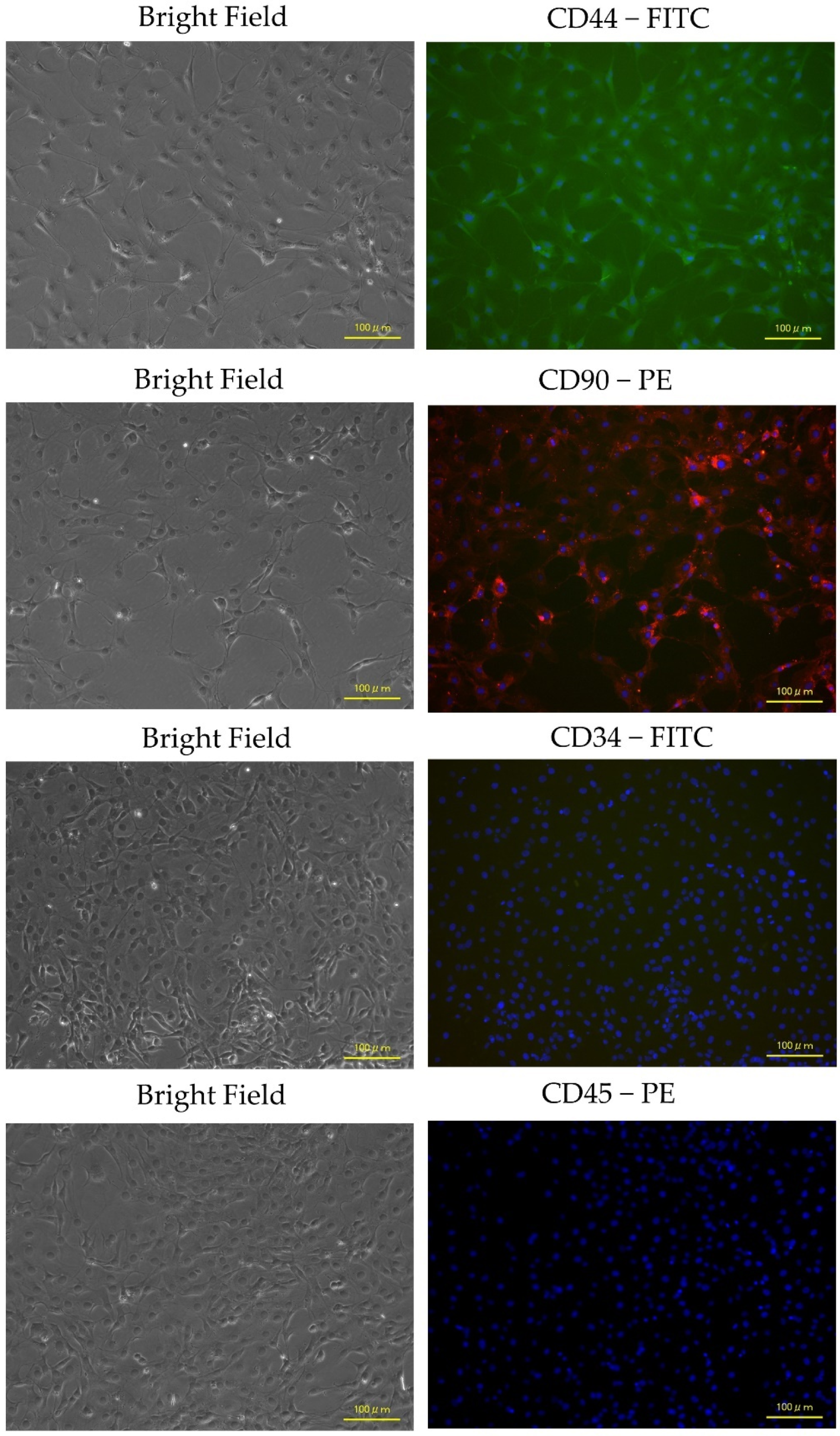
2.4. Flow Cytometry and Immunocytochemistry Analysis of ADP MSCs
2.5. Gene Expression of ADP MSCs by Reverse Transcription-Quantitative Polymerase Chain Reaction (RT-qPCR)
2.6. Trilineage and Myogenic Differentiation Potential of ADP MSCs
2.6.1. Induction of ADP MSCs into Adipocyte
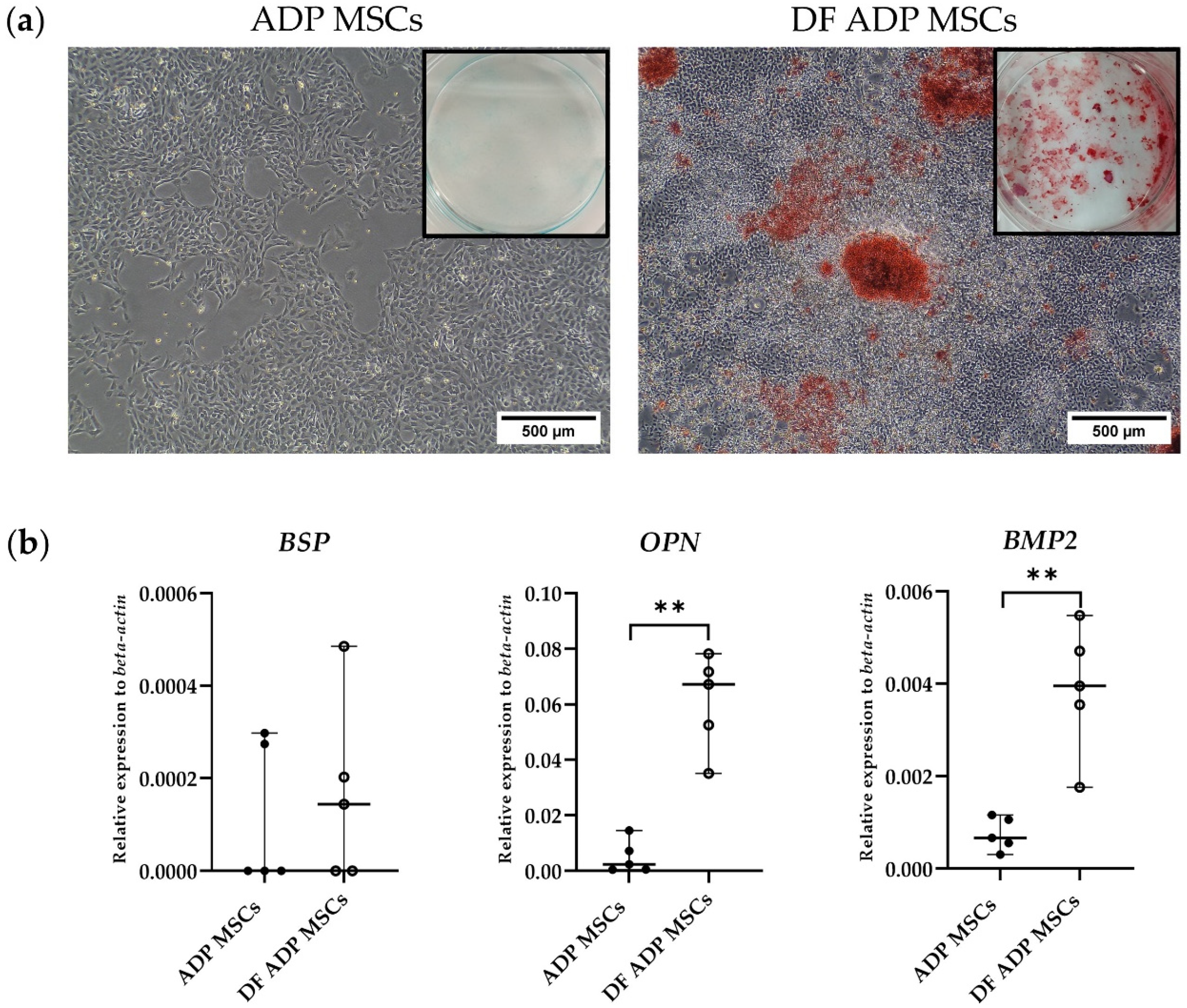
2.6.2. Induction of ADP MSCs into Osteocyte
2.6.3. Induction of ADP MSCs into Chondrocyte
2.6.4. Induction of ADP MSCs into Myocyte
2.6.5. Analysis of the Myogenic Differentiated ADP MSCs
2.7. Evaluation and Interpretation of Statistical Data
3. Results
3.1. Cell Morphology, Viability, Proliferation, and Gene Expression in Rat ADP MSCs
3.2. Immunophenotypic Characterization and Gene Expression of ADP MSCs
3.3. Differentiation Potential of Rat ADP MSCs
3.3.1. Adipogenic Potential
3.3.2. Osteogenic Potential
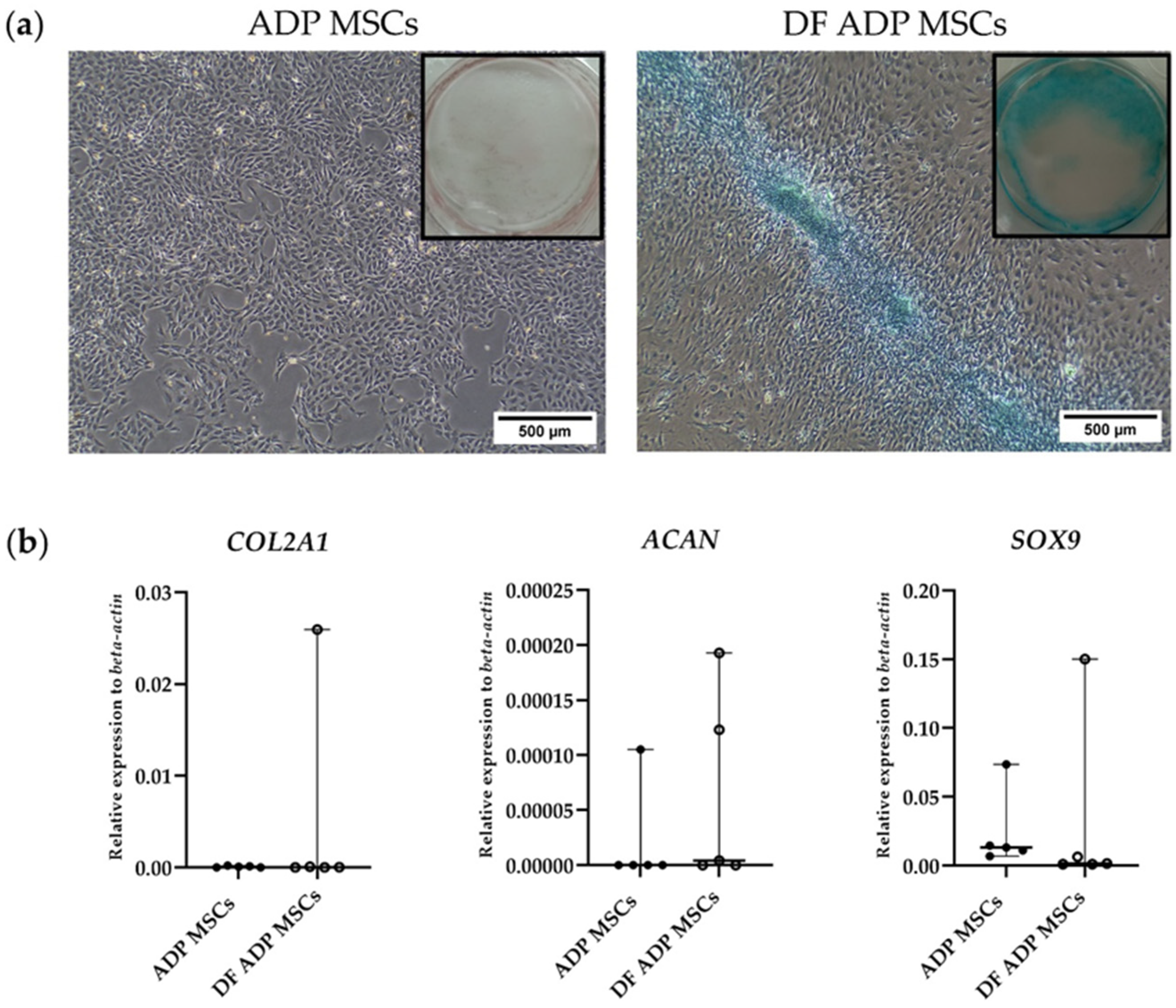
3.3.3. Chondrogenic Potential
3.3.4. Myogenic Potential
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Almalki, S.G.; Agrawal, D.K. Key transcription factors in the differentiation of mesenchymal stem cells. Differentiation 2016, 92, 41–51. [Google Scholar] [CrossRef]
- Bitto, F.F.; Klumpp, D.; Lange, C.; Boos, A.M.; Arkudas, A.; Bleiziffer, O.; Horch, R.E.; Kneser, U.; Beier, J.P. Myogenic differentiation of mesenchymal stem cells in a newly developed neurotised AV-loop model. Biomed Res. Int. 2013, 2013, 935046. [Google Scholar] [CrossRef]
- Kassis, I.; Vaknin-Dembinsky, A.; Karussis, D. Bone Marrow Mesenchymal Stem Cells: Agents of Immunomodulation and Neuroprotection. Curr. Stem Cell Res. Ther. 2011, 6, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Yuan, Q.; Xie, L. Mesenchymal Stem Cell-Based Immunomodulation: Properties and Clinical Application. Stem Cells Int. 2018, 2018, 3057624. [Google Scholar] [CrossRef] [PubMed]
- Kasper, G.; Dankert, N.; Tuischer, J.; Hoeft, M.; Gaber, T.; Glaeser, J.D.; Zander, D.; Tschirschmann, M.; Thompson, M.; Matziolis, G.; et al. Mesenchymal stem cells regulate angiogenesis according to their mechanical environment. Stem Cells 2007, 25, 903–910. [Google Scholar] [CrossRef]
- Watt, S.M.; Gullo, F.; van der Garde, M.; Markeson, D.; Camicia, R.; Khoo, C.P.; Zwaginga, J.J. The angiogenic properties of mesenchymal stem/stromal cells and their therapeutic potential. Br. Med. Bull. 2013, 108, 25–53. [Google Scholar] [CrossRef]
- Takegaki, J.; Sase, K.; Kono, Y.; Nakano, D.; Fujita, T.; Konishi, S.; Fujita, S. Intramuscular injection of mesenchymal stem cells activates anabolic and catabolic systems in mouse skeletal muscle. Sci. Rep. 2021, 11, 21224. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yan, X.; Sun, Z.; Chen, B.; Han, Q.; Li, J.; Zhao, R.C. Flk-1+ Adipose-Derived Mesenchymal Stem Cells Differentiate into Skeletal Muscle Satellite Cells and Ameliorate Muscular Dystrophy in MDX Mice. Stem Cells Dev. 2007, 16, 695–706. [Google Scholar] [CrossRef]
- Nitahara-Kasahara, Y.; Nakayama, S.; Kimura, K.; Yamaguchi, S.; Kakiuchi, Y.; Nito, C.; Hayashi, M.; Nakaishi, T.; Ueda, Y.; Okada, T. Immunomodulatory amnion-derived mesenchymal stromal cells preserve muscle function in a mouse model of Duchenne muscular dystrophy. Stem Cell Res. Ther. 2023, 14, 108. [Google Scholar] [CrossRef]
- Wu, Y.; Peng, Y.; Gao, D.; Feng, C.; Yuan, X.; Li, H.; Wang, Y.; Yang, L.; Huang, S.; Fu, X. Mesenchymal stem cells suppress fibroblast proliferation and reduce skin fibrosis through a TGF-β3-dependent activation. Int. J. Low. Extrem. Wounds 2015, 14, 50–62. [Google Scholar] [CrossRef]
- Flück, M.; Kasper, S.; Benn, M.C.; Clement Frey, F.; von Rechenberg, B.; Giraud, M.N.; Meyer, D.C.; Wieser, K.; Gerber, C. Transplant of Autologous Mesenchymal Stem Cells Halts Fatty Atrophy of Detached Rotator Cuff Muscle After Tendon Repair: Molecular, Microscopic, and Macroscopic Results from an Ovine Model. Am. J. Sports Med. 2021, 49, 3970–3980. [Google Scholar] [CrossRef] [PubMed]
- Sevivas, N.; Teixeira, F.G.; Portugal, R.; Araújo, L.; Carriço, L.F.; Ferreira, N.; Vieira da Silva, M.; Espregueira-Mendes, J.; Anjo, S.; Manadas, B.; et al. Mesenchymal Stem Cell Secretome: A Potential Tool for the Prevention of Muscle Degenerative Changes Associated with Chronic Rotator Cuff Tears. Am. J. Sports Med. 2017, 45, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Xu, J. Immune modulation by mesenchymal stem cells. Cell Prolif. 2020, 53, e12712. [Google Scholar] [CrossRef] [PubMed]
- Park, C.W.; Kim, K.S.; Bae, S.; Son, H.K.; Myung, P.K.; Hong, H.J.; Kim, H. Cytokine secretion profiling of human mesenchymal stem cells by antibody array. Int. J. Stem Cells 2009, 2, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhang, W.; Jiang, X.; Mao, N. Human-placenta-derived mesenchymal stem cells inhibit proliferation and function of allogeneic immune cells. Cell Tissue Res. 2007, 330, 437–446. [Google Scholar] [CrossRef]
- Heszele, M.F.C.; Price, S.R. Insulin-Like Growth Factor I: The Yin and Yang of Muscle Atrophy. Endocrinology 2004, 145, 4803–4805. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Husmann, I.; Soulet, L.; Gautron, J.; Martelly, I.; Barritault, D. Growth factors in skeletal muscle regeneration. Cytokine Growth Factor Rev. 1996, 7, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Saeedi, P.; Halabian, R.; Imani Fooladi, A.A. A revealing review of mesenchymal stem cells therapy, clinical perspectives and Modification strategies. Stem Cell Investig. 2019, 6, 34. [Google Scholar] [CrossRef]
- Hendawy, H.; Kaneda, M.; Metwally, E.; Shimada, K.; Tanaka, T.; Tanaka, R. A Comparative Study of the Effect of Anatomical Site on Multiple Differentiation of Adipose-Derived Stem Cells in Rats. Cells 2021, 10, 2469. [Google Scholar] [CrossRef]
- Koung Ngeun, S.; Shimizu, M.; Kaneda, M. Characterization of Rabbit Mesenchymal Stem/Stromal Cells after Cryopreservation. Biology 2023, 12, 1312. [Google Scholar] [CrossRef]
- Beier, J.P.; Bitto, F.F.; Lange, C.; Klumpp, D.; Arkudas, A.; Bleiziffer, O.; Boos, A.M.; Horch, R.E.; Kneser, U. Myogenic differentiation of mesenchymal stem cells co-cultured with primary myoblasts. Cell Biol. Int. 2011, 35, 397–406. [Google Scholar] [CrossRef]
- Witt, R.; Weigand, A.; Boos, A.M.; Cai, A.; Dippold, D.; Boccaccini, A.R.; Schubert, D.W.; Hardt, M.; Lange, C.; Arkudas, A.; et al. Mesenchymal stem cells and myoblast differentiation under HGF and IGF-1 stimulation for 3D skeletal muscle tissue engineering. BMC Cell Biol. 2017, 18, 15. [Google Scholar] [CrossRef] [PubMed]
- Bayati, V.; Hashemitabar, M.; Gazor, R.; Nejatbakhsh, R.; Bijannejad, D. Expression of surface markers and myogenic potential of rat bone marrow- and adipose-derived stem cells: A comparative study. Anat. Cell Biol. 2013, 46, 113–121. [Google Scholar] [CrossRef] [PubMed]
- de la Garza-Rodea, A.S.; van der Velde-van Dijke, I.; Boersma, H.; Gonçalves, M.A.; van Bekkum, D.W.; de Vries, A.A.; Knaän-Shanzer, S. Myogenic properties of human mesenchymal stem cells derived from three different sources. Cell Transplant. 2012, 21, 153–173. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Sharma, A.K.; Wolfrum, C. Novel insights into adipose tissue heterogeneity. Rev. Endocr. Metab. Disord. 2022, 23, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Mo, Q.; Salley, J.; Roshan, T.; Baer, L.A.; May, F.J.; Jaehnig, E.J.; Lehnig, A.C.; Guo, X.; Tong, Q.; Nuotio-Antar, A.M.; et al. Identification and characterization of a supraclavicular brown adipose tissue in mice. JCI Insight 2017, 2, e93166. [Google Scholar] [CrossRef] [PubMed]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.C.; Krause, D.S.; Deans, R.J.; Keating, A.; Prockop, D.J.; Horwitz, E.M. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef]
- Lotfy, A.; Salama, M.; Zahran, F.; Jones, E.; Badawy, A.; Sobh, M. Characterization of mesenchymal stem cells derived from rat bone marrow and adipose tissue: A comparative study. Int. J. Stem Cells 2014, 7, 135–142. [Google Scholar] [CrossRef]
- Bagno, L.L.; Carvalho, D.; Mesquita, F.; Louzada, R.A.; Andrade, B.; Kasai-Brunswick, T.H.; Lago, V.M.; Suhet, G.; Cipitelli, D.; Werneck-De-Castro, J.P.; et al. Sustained IGF-1 Secretion by Adipose-Derived Stem Cells Improves Infarcted Heart Function. Cell Transplant. 2016, 25, 1609–1622. [Google Scholar] [CrossRef]
- Sobh, M.A. Adipogenesis of Sprague Dawely rats mesenchymal stem cells: A morphological, immunophenotyping and gene expression follow-up study. Anat. Cell Biol. 2014, 47, 83–90. [Google Scholar] [CrossRef]
- Hendawy, H.; Uemura, A.; Ma, D.; Namiki, R.; Samir, H.; Ahmed, M.F.; Elfadadny, A.; El-Husseiny, H.M.; Chieh-Jen, C.; Tanaka, R. Tissue Harvesting Site Effect on the Canine Adipose Stromal Vascular Fraction Quantity and Quality. Animals 2021, 11, 460. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wu, X.; Dietrich, M.A.; Polk, P.; Scott, L.K.; Ptitsyn, A.A.; Gimble, J.M. Yield and characterization of subcutaneous human adipose-derived stem cells by flow cytometric and adipogenic mRNA analyzes. Cytotherapy 2010, 12, 538–546. [Google Scholar] [CrossRef] [PubMed]
- Chambers, I.; Tomlinson, S.R. The transcriptional foundation of pluripotency. Development 2009, 136, 2311–2322. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.; Smith, A. Capturing pluripotency. Cell 2008, 132, 532–536. [Google Scholar] [CrossRef] [PubMed]
- Pierantozzi, E.; Gava, B.; Manini, I.; Roviello, F.; Marotta, G.; Chiavarelli, M.; Sorrentino, V. Pluripotency regulators in human mesenchymal stem cells: Expression of NANOG but not of OCT-4 and SOX-2. Stem Cells Dev. 2011, 20, 915–923. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.-M.; Guo, Y.; Wang, Q.; Xu, Y.; Wang, M.; Chen, H.-N.; Shen, W.-M. Over-expression of PPAR-γ2 gene enhances the adipogenic differentiation of hemangioma-derived mesenchymal stem cells in vitro and in vivo. Oncotarget 2017, 8, 115817–115828. [Google Scholar] [CrossRef] [PubMed]
- Martin, G.; Poirier, H.; Crombie, D.; Fruchart, J.C.; Heyman, R.A.; Besnard, P.; Auwerx, J.; Hennuyer, N. Induction of the Fatty Acid Transport Protein 1 and Acyl-CoA Synthase Genes by Dimer-selective Rexinoids Suggests That the Peroxisome Proliferator-activated Receptor-Retinoid X Receptor Heterodimer Is Their Molecular Target. J. Biol. Chem. 2000, 275, 12612–12618. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Abuna, R.P.F.; De Oliveira, F.S.; Santos, T.D.S.; Guerra, T.R.; Rosa, A.L.; Beloti, M.M. Participation of TNF-α in Inhibitory Effects of Adipocytes on Osteoblast Differentiation. J. Cell. Physiol. 2016, 231, 204–214. [Google Scholar] [CrossRef]
- Uezumi, A.; Fukada, S.; Yamamoto, N.; Ikemoto-Uezumi, M.; Nakatani, M.; Morita, M.; Yamaguchi, A.; Yamada, H.; Nishino, I.; Hamada, Y.; et al. Identification and characterization of PDGFRα+ mesenchymal progenitors in human skeletal muscle. Cell Death Dis. 2014, 5, e1186. [Google Scholar] [CrossRef]
- Lian, J.B.; Stein, G.S. Development of the osteoblast phenotype: Molecular mechanisms mediating osteoblast growth and differentiation. Iowa Orthop. J. 1995, 15, 118–140. [Google Scholar]
- Valenti, M.T.; Dalle Carbonare, L.; Donatelli, L.; Bertoldo, F.; Zanatta, M.; Lo Cascio, V. Gene expression analysis in osteoblastic differentiation from peripheral blood mesenchymal stem cells. Bone 2008, 43, 1084–1092. [Google Scholar] [CrossRef]
- Li, Y.; Fu, G.; Gong, Y.; Li, B.; Li, W.; Liu, D.; Yang, X. BMP-2 promotes osteogenic differentiation of mesenchymal stem cells by enhancing mitochondrial activity. J. Musculoskelet. Neuronal Interact. 2022, 22, 123–131. [Google Scholar] [PubMed]
- Rustamov, V.; Keller, F.; Klicks, J.; Hafner, M.; Rudolf, R. Bone Sialoprotein Shows Enhanced Expression in Early, High-Proliferation Stages of Three-Dimensional Spheroid Cell Cultures of Breast Cancer Cell Line MDA-MB-231. Front. Oncol. 2019, 9, 36. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, Y.; Wang, S.; Ogata, Y. Transcriptional regulation of bone sialoprotein gene by CO2 laser irradiation. J. Oral Sci. 2011, 53, 51–59. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nenna, R.; Turchetti, A.; Mastrogiorgio, G.; Midulla, F. COL2A1 Gene Mutations: Mechanisms of Spondyloepiphyseal Dysplasia Congenita. Appl. Clin. Genet. 2019, 12, 235–238. [Google Scholar] [CrossRef] [PubMed]
- Mwale, F.; Stachura, D.; Roughley, P.; Antoniou, J. Limitations of using aggrecan and type X collagen as markers of chondrogenesis in mesenchymal stem cell differentiation. J. Orthop. Res. 2006, 24, 1791–1798. [Google Scholar] [CrossRef] [PubMed]
- Gibson, B.G.; Briggs, M.D. The aggrecanopathies; an evolving phenotypic spectrum of human genetic skeletal diseases. Orphanet J. Rare Dis. 2016, 11, 86. [Google Scholar] [CrossRef] [PubMed]
- Jo, A.; Denduluri, S.; Zhang, B.; Wang, Z.; Yin, L.; Yan, Z.; Kang, R.; Shi, L.L.; Mok, J.; Lee, M.J.; et al. The versatile functions of Sox9 in development, stem cells, and human diseases. Genes Dis. 2014, 1, 149–161. [Google Scholar] [CrossRef]
- Stöckl, S.; Bauer, R.J.; Bosserhoff, A.K.; Göttl, C.; Grifka, J.; Grässel, S. Sox9 modulates cell survival and adipogenic differentiation of multipotent adult rat mesenchymal stem cells. J. Cell Sci. 2013, 126, 2890–2902. [Google Scholar] [CrossRef]
- Bi, W.; Deng, J.M.; Zhang, Z.; Behringer, R.R.; de Crombrugghe, B. Sox9 is required for cartilage formation. Nat. Genet. 1999, 22, 85–89. [Google Scholar] [CrossRef]
- Bailes, J.; Soloviev, M. Insulin-Like Growth Factor-1 (IGF-1) and Its Monitoring in Medical Diagnostic and in Sports. Biomolecules 2021, 11, 217. [Google Scholar] [CrossRef]
- Sadat, S.; Gehmert, S.; Song, Y.H.; Yen, Y.; Bai, X.; Gaiser, S.; Klein, H.; Alt, E. The cardioprotective effect of mesenchymal stem cells is mediated by IGF-I and VEGF. Biochem. Biophys. Res. Commun. 2007, 363, 674–679. [Google Scholar] [CrossRef]
- Haque, S.; Morris, J.C. Transforming growth factor-β: A therapeutic target for cancer. Hum. Vaccines Immunother. 2017, 13, 1741–1750. [Google Scholar] [CrossRef] [PubMed]
- Scheller, J.; Chalaris, A.; Schmidt-Arras, D.; Rose-John, S. The pro- and anti-inflammatory properties of the cytokine interleukin-6. Biochim. Biophys. Acta 2011, 1813, 878–888. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Narazaki, M.; Kishimoto, T. IL-6 in inflammation, immunity, and disease. Cold Spring Harb. Perspect. Biol. 2014, 6, a016295. [Google Scholar] [CrossRef] [PubMed]
- Yi, T.; Song, S.U. Immunomodulatory properties of mesenchymal stem cells and their therapeutic applications. Arch. Pharm. Res. 2012, 35, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Florini, J.R.; Ewton, D.Z.; Coolican, S.A. Growth hormone and the insulin-like growth factor system in myogenesis. Endocr. Rev. 1996, 17, 481–517. [Google Scholar] [CrossRef] [PubMed]
- Gaipa, G.; Cazzaniga, G.; Valsecchi, M.G.; Panzer-Grümayer, R.; Buldini, B.; Silvestri, D.; Karawajew, L.; Maglia, O.; Ratei, R.; Benetello, A.; et al. Time point-dependent concordance of flow cytometry and real-time quantitative polymerase chain reaction for minimal residual disease detection in childhood acute lymphoblastic leukemia. Haematologica 2012, 97, 1582–1593. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Hernández, J.M.; García-González, E.G.; Brun, C.E.; Rudnicki, M.A. The myogenic regulatory factors, determinants of muscle development, cell identity and regeneration. Semin. Cell Dev. Biol. 2017, 72, 10–18. [Google Scholar] [CrossRef]
- Komori, T. Regulation of osteoblast differentiation by transcription factors. J. Cell. Biochem. 2006, 99, 1233–1239. [Google Scholar] [CrossRef]
- Gang, E.J.; Jeong, J.A.; Hong, S.H.; Hwang, S.H.; Kim, S.W.; Yang, I.H.; Ahn, C.; Han, H.; Kim, H. Skeletal myogenic differentiation of mesenchymal stem cells isolated from human umbilical cord blood. Stem Cells 2004, 22, 617–624. [Google Scholar] [CrossRef] [PubMed]
- Stern-Straeter, J.; Bonaterra, G.A.; Juritz, S.; Birk, R.; Goessler, U.R.; Bieback, K.; Bugert, P.; Schultz, J.; Hörmann, K.; Kinscherf, R.; et al. Evaluation of the effects of different culture media on the myogenic differentiation potential of adipose tissue- or bone marrow-derived human mesenchymal stem cells. Int. J. Mol. Med. 2014, 33, 160–170. [Google Scholar] [CrossRef] [PubMed]
- Kretlow, J.D.; Jin, Y.Q.; Liu, W.; Zhang, W.J.; Hong, T.H.; Zhou, G.; Baggett, L.S.; Mikos, A.G.; Cao, Y. Donor age and cell passage affects differentiation potential of murine bone marrow-derived stem cells. BMC Cell Biol. 2008, 9, 60. [Google Scholar] [CrossRef] [PubMed]
- Péault, B.; Rudnicki, M.; Torrente, Y.; Cossu, G.; Tremblay, J.P.; Partridge, T.; Gussoni, E.; Kunkel, L.M.; Huard, J. Stem and Progenitor Cells in Skeletal Muscle Development, Maintenance, and Therapy. Mol. Ther. 2007, 15, 867–877. [Google Scholar] [CrossRef]





| Gene Name | Direction | Primer Sequences (5′–3′) | |
|---|---|---|---|
| Housekeeping gene | beta-actin | Forward | GCAGGAGTACGATGAGTCCG |
| Reverse | ACGCAGCTCAGTAACAGTCC | ||
| Pluripotent marker genes | NANOG | Forward | TACCTCAGCCTCCAGCAGAT |
| Reverse | CATTGGTTTTTCTGCCACCT | ||
| Oct4 | Forward | CGAACCTGGCTAAGCTTCCA | |
| Reverse | GCCATCCCTCCACAGAACTC | ||
| REX1 | Forward | GCTCCGGCGGAATCGAGTGG | |
| Reverse | GCACGTGTTGCTTGGCGACC | ||
| SOX2 | Forward | CTCGCAGACCTACATGAAC | |
| Reverse | TCGGACTTGACCACAGAG | ||
| Immunomodulatory marker genes | IGF1 | Forward | TGGTGGACGCTCTTCAGTTC |
| Reverse | TCCGGAAGCAACACTCATCC | ||
| TGFB1 | Forward | ATGCCAACTTCTGTCTGGGG | |
| Reverse | GGTTGTAGAGGGCAAGGACC | ||
| IL6 | Forward | CCACCCACAACAGACCAGTA | |
| Reverse | TCTGACAGTGCATCATCGCT | ||
| Adipogenic genes | PPARG | Forward | AGCTCTGTGGACCTCTCTGT |
| Reverse | GTCAGCTCTTGTGAACGGGA | ||
| PDGFRA | Forward | AGTGCTTGGTCGGATCTTGG | |
| Reverse | GAGCATCTTCACAGCCACCT | ||
| FABP4 | Forward | AACTGGGCGTGGAATTCGAT | |
| Reverse | CACATGTACCAGGACCCCAC | ||
| ADIPOQ | Forward | TAATTCAGAGCAGCCCGTAG | |
| Reverse | TGGGGATAACACTCAGAACC | ||
| Osteogenic genes | BSP | Forward | AGGCTACGAGGGTCAGGATT |
| Reverse | GCACCTTCCTGAGTTGAGCT | ||
| OPN | Forward | GAAGAGCCAGGAGTCCGATG | |
| Reverse | CTTCCCGTTGCTGTCCTGAT | ||
| BMP2 | Forward | CAGGTCTTTGCACCAAGATG | |
| Reverse | GCTGGACTTAAGACGCTTCC | ||
| Chondrogenic genes | COL2A1 | Forward | TCCTAAGGGTGCCAATGGTGA |
| Reverse | AGGACCAACTTTGCCTTGAGGAC | ||
| ACAN | Forward | CTCTGCCTCCCGTGAAAC | |
| Reverse | TGAAGTGCCTGCATCTATGT | ||
| SOX9 | Forward | GAAAGACCACCCCGATTACAAG | |
| Reverse | AAGATGGCGTTAGGAGAGATGTG | ||
| Myogenic genes | MyoD | Forward | CGACTCTTCAGGCTTGGGTT |
| Reverse | TGTCGCAAAGGAGCAGAGAG | ||
| MYOG | Forward | GGCAATGCACTGGAGTTTGG | |
| Reverse | CGTAAGGGAGTGCAGGTTGT | ||
| MYF5 | Forward | ATGGACATGACGGACAGCTG | |
| Reverse | TGCGACTCTTGGCTCAAACT | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koung Ngeun, S.; Shimizu, M.; Kaneda, M. Myogenic Differentiation and Immunomodulatory Properties of Rat Adipose-Derived Mesenchymal Stem/Stromal Cells. Biology 2024, 13, 72. https://doi.org/10.3390/biology13020072
Koung Ngeun S, Shimizu M, Kaneda M. Myogenic Differentiation and Immunomodulatory Properties of Rat Adipose-Derived Mesenchymal Stem/Stromal Cells. Biology. 2024; 13(2):72. https://doi.org/10.3390/biology13020072
Chicago/Turabian StyleKoung Ngeun, Sai, Miki Shimizu, and Masahiro Kaneda. 2024. "Myogenic Differentiation and Immunomodulatory Properties of Rat Adipose-Derived Mesenchymal Stem/Stromal Cells" Biology 13, no. 2: 72. https://doi.org/10.3390/biology13020072
APA StyleKoung Ngeun, S., Shimizu, M., & Kaneda, M. (2024). Myogenic Differentiation and Immunomodulatory Properties of Rat Adipose-Derived Mesenchymal Stem/Stromal Cells. Biology, 13(2), 72. https://doi.org/10.3390/biology13020072






